Abstract
Fungal pathogens can rapidly evolve virulence towards resistant crops in agricultural ecosystems. Gains in virulence are often mediated by the mutation or deletion of a gene encoding a protein recognized by the plant immune system. However, the loci and the mechanisms of genome evolution enabling rapid virulence evolution are poorly understood. We performed genome-wide association mapping on a global collection of 106 strains of Zymoseptoria tritici, the most damaging pathogen of wheat in Europe, to identify polymorphisms linked to virulence on two wheat varieties. We found 25 distinct genomic loci associated with reproductive success of the pathogen. However, no locus was shared between the host genotypes, suggesting host specialization. The main locus associated with virulence encoded a highly expressed, small secreted protein. Population genomic analyses showed that the gain in virulence was explained by a segregating gene deletion polymorphism. The deletion was likely adaptive by preventing detection of the encoded protein. Comparative genomics of closely related species showed that the locus emerged de novo since speciation. A large cluster of transposable elements in direct proximity to the locus generated extensive rearrangements leading to multiple independent gene losses. Our study demonstrates that rapid turnover in the chromosomal structure of a pathogen can drive host specialization.
Similar content being viewed by others
Introduction
Fungal plant pathogens cause major damage in agricultural ecosystems and threaten food security worldwide (Fisher et al., 2012). Many epidemics on crop plants are triggered by an evolutionary innovation in the pathogen that prevents the host immune system from recognizing the pathogen. However, the evolutionary mechanisms by which pathogen populations gain the ability to circumvent the host immune system are poorly understood. A key component of the plant immune system is based on resistance genes (R-genes) encoding cytosolic or membrane proteins that directly or indirectly detect the presence of pathogen avirulence effector proteins and trigger immune defenses (Flor, 1971; Jones and Dangl, 2006). Effectors are often small secreted proteins (SSPs) that are highly expressed during infection and contain a high proportion of cysteines (Stergiopoulos and de Wit, 2009). Recognition of effectors can severely limit a pathogen's ability to cause disease (Oliveira-Garcia and Valent, 2015). Hence, selection pressure on pathogens favors mutations that enable the encoded effectors to escape recognition.
As host genotypes often differ in their composition in R-genes, selection pressure on corresponding pathogen populations can be highly heterogeneous, generating a long-term co-evolutionary dynamic that simultaneously favors multiple pathogen genotypes that each has evolved virulence on a specific subset of hosts (Stukenbrock and McDonald, 2009; Presti et al., 2015). Sequence diversification or deletion of effector genes were linked to an increase in virulence on host genotypes carrying corresponding R-genes in various plant pathogenic fungi and oomycetes (Jiang and Tyler, 2012; Ghanbarnia et al., 2015; Schmidt et al., 2015). Knowledge of effector gene evolution in pathogen genomes and natural fungal populations is essential for developing effective breeding strategies that lead to durable resistance. For instance, the presence of dispensable or rapidly evolving fungal effectors could be monitored in pathogen populations and included in R-gene deployment strategies (Boyd et al., 2012; Vleeshouwers and Oliver, 2014). Plant pathogen effectors can also have a role in the degradation of the plant cell wall and the lysis of cell components during infection (Esquerré-Tugayé et al., 2000; De Jonge et al., 2011). In addition to effectors, proteins involved in cellular transport and nutrition were shown to have an important role in the ability of a pathogen to cause disease (Stergiopoulos et al., 2003; Oh et al., 2008; Ipcho et al., 2012). However, the contribution of non-effector proteins to overall virulence is largely quantitative and their role in virulence evolution remains poorly understood.
Wheat is a major crop supplying 20% of the calorie needs of the human population and is threatened by multiple fungal pathogens. The major pathogen of wheat in Europe is Zymoseptoria tritici (syn. Mycosphaerella graminicola; Fones and Gurr, 2015). The genetic basis of virulence in Z. tritici is poorly understood (Rudd et al., 2015; Kettles and Kanyuka, 2016). This sexually reproducing fungus is found in temperate wheat production areas around the world (Eyal et al., 1987) and populations are highly polymorphic (Zhan et al., 2003; McDonald et al., 2016). A major reason why the Z. tritici–wheat interaction is poorly understood is that virulence of Z. tritici is mainly quantitative. Similarly, resistance of wheat to Z. tritici has both gene-for-gene and quantitative components. Eighteen Stb R-genes and 167 resistance quantitative trait loci were identified (Brown et al., 2015). Host resistance can be broken down rapidly, most likely as a result of effector evolution (Cowger et al., 2000; Krenz et al., 2008). Known components of Z. tritici virulence include only two secreted effectors: Mg3LysM encodes a protein that prevents chitin binding and protects against defense response triggered in the host (Marshall et al., 2011); the rapidly evolving gene Zt80707 contributes to virulence based on unknown mechanisms (Poppe et al., 2015). Expression profiling and in silico sequence analyses identified hundreds of candidate effectors with unknown contributions to virulence (Morais do Amaral et al., 2012; Mirzadi Gohari et al., 2015; Palma-Guerrero et al., 2016). However, in silico approaches to identify genes contributing to virulence face challenges because many effectors may have redundant functions or may be specific to particular host genotypes.
We aimed to circumvent these challenges by performing an unbiased genomic screen using association mapping to directly identify loci in the pathogen genome contributing to virulence. Genome-wide association studies (GWAS) link polymorphism in the genome with phenotypic variation and are particularly powerful in species with high standing genetic variation and frequent sexual reproduction (Bergelson and Roux, 2010; Korte and Farlow, 2013). The population genetic structure of Z. tritici makes this pathogen particularly suitable for GWAS. First, we determined the genetic architecture of quantitative and qualitative virulence in a set of pathogen populations spanning the global distribution range. We assessed the production of asexual fruiting bodies called pycnidia on wheat leaves as a measure of virulence and pathogen fitness (Zhan et al., 2005; Stewart and McDonald, 2014; Brown et al., 2015). Second, we dissected the mechanisms of chromosomal evolution generating variation in virulence among isolates of the pathogen. For this, we performed population and comparative genomics analyses of completely assembled genomes of the same species and closely related sister species. We found that the strongest contribution to virulence was a segregating effector gene deletion that arose multiple times through a complex series of chromosomal rearrangements.
Materials and Methods
Fungal isolate collection
We analyzed 106 isolates of Z. tritici collected from single wheat fields in four different locations: Australia, Israel, Switzerland and Oregon, USA. In Oregon, isolates were sampled from cultivars Madsen (Oregon R) and Stephens (Oregon S). All sampling was performed between 1990 and 2001. Spores were stored in anhydrous silica gel at −80 °C after sampling. Details regarding sampling years and locations of these five populations were described in Zhan et al. (2005). Previous analysis showed that there were no clonal genotypes among the isolates (Linde et al., 2002; Zhan et al., 2005).
Virulence assay to measure pycnidia production on two wheat cultivars
We used the virulence phenotypes obtained in the pathogenicity tests carried out by Zhan et al. (2005). Briefly, a greenhouse seedling assay was performed to assess the virulence of the Z. tritici isolates on two Swiss spring wheat cultivars: Greina (susceptible to Z. tritici) and Toronit (moderately resistant to Z. tritici; Brabant et al., 2006). Twenty-one days-old wheat seedlings were artificially inoculated with each isolate. Infected leaves were collected 22 days after inoculation and digitized. The percentage of leaf area covered by pycnidia (PLACP) was measured using the software Assess (Lamari, 2002). We repeated the inspection of digital images for accuracy. The PLACP values of two isolates assayed on Toronit were modified as a result. All other data points were retained as shown in Zhan et al. (2005). In total, phenotypic data was available for 96 isolates assayed on Toronit and 95 isolates assayed on Greina.
Whole-genome sequencing and variant calling procedure
Sequencing data was previously available for five Swiss isolates on the NCBI Short Read Archive under the BioProject PRJNA178194 (Torriani et al., 2011). We performed Illumina whole-genome sequencing of 101 additional isolates (Supplementary Table S1). We extracted high-quality genomic DNA from liquid cultures for the remaining isolates. Paired-end sequencing of 100 bp was performed on an Illumina HiSeq 2000. We obtained approximately 1 billion bp of raw sequence for each isolate. The raw sequencing data were deposited on the NCBI Short Read Archive under the BioProject PRJNA327615.
The sequencing reads were trimmed for remaining adapters and sequencing quality using the software Trimmomatic v0.32 (Bolger et al., 2014) with the following settings: illuminaclip=TruSeq3-PE.fa:2:30:10, leading=10, trailing=10, slidingwindow=5:10, minlen=50. Sequence data from all isolates was aligned to the reference genome IPO323 (Goodwin et al., 2011), which is fully assembled into chromosomes, contains no gaps and includes complete telomere sequences except for one small chromosome. We used the short read aligner Bowtie 2 version 2.2.3 (Langmead et al., 2009) with the —very-sensitive-local option. PCR duplicates were identified using Picard tools version 1.118 (http://broadinstitute.github.io/picard).
Single nucleotide polymorphism (SNP) calling was performed using the Genome Analysis Toolkit (GATK) version 3.3-0 (McKenna et al., 2010). HaplotypeCaller was run on each isolate individually with the following settings: -emitRefConfidence GVCF; —variant_index_type LINEAR; —variant_index_parameter 128000; —sample_ploidy 1. Joint variant calls were performed using GenotypeGVCFs on a merged gvcf variant file using the option -maxAltAlleles 2. For details on SNP call filtering, validation and genetic structure analyses see Supplementary Methods. The variant call format (VCF) file was deposited in the European Nucleotide Archive (ENA) under the accession numbers PRJEB15502/ERP017268 and the analysis number ERZ330467.
Model selection for genome-wide association study
Population structure and varying degrees of relatedness between individuals may be a source for p-value inflations and false positives due to non-random phenotype-genotype associations (Bergelson and Roux, 2010; Korte and Farlow, 2013). To account for this, we analyzed the population structure and genetic relatedness of all 106 isolates by performing a principal component analysis (PCA) and computing a kinship matrix, respectively (for full details see Supplementary Note S1). Then, we performed GWAS with three different statistical models commonly used for population structure and kinship corrections. We used a general linear model with the first three principal components (GLM Q), a mixed linear model with kinship effect only (MLM K), and a mixed linear model with kinship effect and principal components (MLM Q+K). Significance thresholds used for GWAS analyses were the following: Bonferroni threshold P<6.42e−08 (α=0.05), false discovery rate (FDR) thresholds of 5% and 10% calculated using the R package q-value (Storey and Tibshirani, 2003). For additional details on GWAS model selection see Supplementary Methods.
Analyses of linkage disequilibrium decay
We assessed the mapping power and resolution of GWAS by estimating linkage disequilibrium (LD) decay for SNPs on chromosome 1. We calculated the LD r2 between all marker pairs up to a distance of 20 kb using the option —hap-r2 in vcftools. We performed the analysis for each population separately to avoid r2 inflation due to demography. We fitted the decay of r2 over physical distance using a nonlinear model following the equation described by Ingvarsson (2005). We used the nls function (nonlinear least squares) in R software (R Core Team, 2014) to estimate the parameters of the model. We used the same procedure to estimate LD decay for specific loci associated with phenotypic traits. For this, we calculated LD within the region of the most significantly associated SNP up to a distance of ~200 kb.
Functional prediction of genes associated with virulence
Loci identified by GWAS were analyzed for their functional role. The average distance between genes in the Z. tritici genome is approximately 1 kb (Goodwin et al., 2011). Hence, we considered a SNP to be intergenic if the SNP was located at a distance greater than 1.2 kb from the 5′ or 3′ end of a gene. SNPs within 1.2 kb of a gene were considered up- or downstream variants.
For SNPs located in the coding sequence of a gene, we screened all gene annotations available for the reference genome IPO323 (Goodwin et al., 2011; Grandaubert et al., 2015; Rudd et al., 2015) and manually assessed the support for individual gene models using RNAseq data (Rudd et al., 2015). RNAseq data obtained in planta were used to assess differential gene regulation during infections (Rudd et al., 2015; Palma-Guerrero et al., 2016). Predicted amino acid sequences were analyzed for signatures of signal peptides and functional domains using SignalP 4.1 server (Petersen et al., 2011), the InterProScan server (Jones et al., 2014), the Uniprot server (Magrane and Consortium, 2011) and the NCBI Conserved Domain Blast server (Marchler-Bauer et al., 2015). We identified homologs of Z. tritici genes in three closely related species using blastn. The genomes of five isolates of Z. pseudotritici, 4 isolates of Z. ardabiliae and one isolate of Z. passerinii were downloaded from NCBI under the accession numbers PRJNA63035, PRJNA277173, PRJNA63037, PRJNA63039, PRJNA46489, PRJNA63043, PRJNA277174, PRJNA63045, PRJNA63047, PRJNA63049 (Robbertse and Tatusova, 2011; Stukenbrock et al., 2012a).
Identification of segmental deletions
We used CNVnator (Abyzov et al., 2011) to perform a statistical analysis of short read coverage along chromosomes to identify segmental deletions in the 106 resequenced isolates. We assessed deletions in bins of 100 bp as recommended, because nearly all resequenced isolates exceeded 20–30 × read coverage. We additionally validated evidence for gene deletion using blastn performed on the de novo genome assemblies of individual isolates (see below). For the isolate STAus01_1D5, we manually corrected a CNVnator deletion call for one gene as a blastn search yielded the correct gene sequence in the genome assembly. Segmental deletions were visualized using the R package {ggplot2} (Wickham, 2009). The detected gene deletion polymorphisms were validated by PCR (Supplementary Methods).
Analysis of the sequence diversity of the gene Zt_8_609
We obtained sequences of Zt_8_609 using blastn performed on de novo genome assemblies of each isolate. The Zt_8_609 sequence could not be retrieved for four isolates, most likely due to poor assembly in the region containing the locus. De novo assemblies of the Illumina reads were performed for each isolate using SPAdes 3.6.0 (Bankevich et al., 2012). We used the BayesHammer read correction prior to assembly and ran the assembly over a k-mer range of ‘21,29,37,45,53,61,79,87’. The assembly was polished using MismatchCorrector. De novo assemblies were used to build BLAST databases for each isolate. BLAST searches were performed using the blastn command of the ncbi-blast-2.2.30+ software (Camacho et al., 2009). Nucleotide diversity in Zt_8_609 was calculated using the R package {Ape} (Paradis et al., 2004) according to (Nei, 1987). We also analyzed large-scale synteny of the locus within the species based on completely assembled chromosomal sequences of the reference genome IPO323 (Goodwin et al., 2011) and the Swiss isolate ST99CH3D7 (Plissonneau et al., 2016). Synteny was analyzed using blastn and visualized using the R package genoPlotR v. 0.8.4 (Guy et al., 2010).
Results
Genome sequencing of highly polymorphic pathogen populations
To map the genetic architecture of virulence in natural populations of Z. tritici, we used the genomes of 106 isolates across the pathogen’s geographic range sequenced at a coverage of 8–29 × (haploid genome; Supplementary Table S1). We obtained a total of 779 178 high-confidence SNPs. Populations were highly polymorphic with an average SNP density of 20 SNPs per kb. Australia was the least polymorphic population with 294 249 segregating SNPs. In total, 96.4% of all SNPs were segregating in at least two populations (Figure 1a; Supplementary Figure S1). Genotypes clustered according to their population of origin (Figure 1b; Supplementary Note S1). We assessed the mapping power of GWAS by calculating LD decay for each population. LD decayed to r2<0.2 within 0.6–2.2 kb in the Swiss, Israel and Oregon populations, whereas LD decayed to r2<0.2 within 10.2 kb in the Australian population (Supplementary Figure S2). Given that the average distance between genes in Z. tritici is 1080 bp, we expected GWAS to resolve associations to individual genes.
Genetic polymorphism, population structure, and virulence phenotypes of 106 Zymoseptoria tritici isolates. (a) Sampling locations and number of SNPs segregating within populations. In Oregon (USA), two populations were collected from two different wheat cultivars growing in the same field. (b) Neighbor-joining tree of all isolates based on a supermatrix constructed from 779 178 SNPs. Branch colors refer to the geographic origin of the isolates. The scale bar indicates the proportion of the total number of polymorphic SNPs. (c) Symptoms caused by two Swiss Z. tritici isolates on Toronit 16 days after inoculation. The isolates are ST99CH3D7 (upper panel) and ST99CH3F3 (lower panel). (d) Distribution of the percentage leaf area covered by pycnidia (PLACP) for each population on the wheat cultivars Toronit and Greina. Center lines show medians, boxes indicate the 25th and 75th percentiles. (e) Correlation of PLACP produced on the cultivars Toronit and Greina for each isolate. The distributions of PLACP are shown at the panel edges including the median PLACP as a dotted line.
Complex genetic architecture and host specificity of pycnidia production
We performed a GWAS of 106 Z. tritici isolates assayed for pycnidia production on the Swiss spring wheat cultivars Greina and Toronit (Figures 1c–e). Greina and Toronit have been deployed since 1994 and 1996, respectively. Long-term monitoring of wheat fields showed that resistance to Z. tritici was intermediate in comparison to a reference panel of other commonly planted Swiss wheat cultivars (Brabant et al., 2006). In greenhouse assays performed with these two cultivars, virulence of Z. tritici was a quantitative trait. Consistent with field monitoring, Toronit was on average more resistant than Greina (Zhan et al., 2005). The quantitative nature of virulence suggests that fungal populations evolved a complex genetic architecture to overcome resistance. The difference in field resistance suggests that the pathogen evolved different mechanisms to overcome resistance to each of the two cultivars. Hence, identifying loci in the pathogen responsible for virulence on these two cultivars should provide a broad perspective of virulence evolution on wheat.
Population genetic analyses of the 106 isolates identified genetic subdivisions according to the population of origin (see Supplementary Note S1 for details). Hence, we used GWAS models that accounted for population structure and corrected for P-value inflations. All SNPs significantly associated with a phenotypic trait were validated using two independent SNP genotyping algorithms. We identified five significantly associated SNPs for the trait percent leaf area covered by pycnidia (PLACP) on the cultivar Toronit (Bonferroni α=0.05). We found 23 additional SNPs at a 5% FDR (Supplementary Note S2). All SNPs at the 5% FDR threshold clustered into 13 chromosomal regions. An additional 69 SNPs were found at the 10% FDR threshold (Figure 2a; Supplementary Tables S2 and S3). The strongest association was at 1.844 Mb on chromosome 8 with snp_chr8_1844714 being the most significant SNP. The association was robust as a GWAS performed only on a single population recovered the association at a 10% FDR (Oregon n=45; Supplementary Figure S3). GWAS performed for PLACP on the cultivar Greina identified overall weaker associations than for Toronit (Figure 2b; Supplementary Tables S4 and S5). No association was significant at Bonferroni α=0.05 or at 5% FDR. We identified 50 significantly associated SNPs at the 10% FDR. All these SNPs clustered into 12 regions on chromosomes 1–6, 10 and 11.
GWAS for virulence of Z. tritici on two wheat cultivars. GWAS based on a mixed linear model for percentage leaf area covered by pycnidia (PLACP) on the cultivar Toronit (a) and cultivar Greina (b). The horizontal dotted line represents the Bonferroni significance threshold (α=0.05). The horizontal dashed line and solid gray line represent the FDR significance thresholds of 5% and 10%, respectively. For each GWAS analysis, the associated loci were characterized as intragenic or linked to a gene.
PLACP was weakly but significantly correlated between the two cultivars (Pearson's correlation coefficient r=0.34, P<0.002; Figure 1e). However, no significantly associated SNP for PLACP was shared among the two tested cultivars. In particular, the strongest association identified for PLACP on Toronit (snp_chr8_1844714) was not significant for PLACP on Greina. The allelic effect of snp_chr8_1844714 was high on Toronit (16.3 difference in PLACP) but low on Greina (3.31 difference in PLACP). Overall, allelic effects between cultivars were not correlated (Pearson's correlation coefficient r=0.17; Supplementary Figure S4).
Functional analyses of loci linked to virulence
Nine associations for PLACP on Toronit were either in coding regions of a gene or within 1.2 kb of a gene (the average distance between genes in the genome is ~1 kb). Associations found in close proximity to genes may be linked to variation in transcriptional regulation of the locus. Identified candidate genes included genes encoding a peptidase C19, an uncharacterized secreted protein and a beta-galactosidase (glycoside hydrolase family 35). Furthermore, we found associations in proximity to genes encoding a major facilitator superfamily transporter up-regulated during advanced stages of the infection (that is, during necrotrophy) and a pectinase. The pectinase is a member of the cell wall degrading enzymes shown to be under purifying selection in the species (Brunner et al., 2013). Four chromosomal regions associated with PLACP on Toronit were located in intergenic regions (⩾1.2 kb from the closest gene; Supplementary Table S3,Supplementary Note S2). These intergenic associations included the strongest association (snp_chr8_1844714). For PLACP on the cultivar Greina, nine out of the 12 identified associations (10% FDR) were found in coding sequences or within 1.2 kb of the nearest gene. Candidate genes included two genes encoding glycosyltransferases belonging to families 22 and 71 (Supplementary Table S5; Supplementary Note S2).
Genome-wide association linked to a segregating gene deletion polymorphism
We investigated the chromosomal region in which we identified the strongest association for PLACP on Toronit. The associated chromosomal region on chromosome 8 was intergenic and comprised four SNPs above the Bonferroni threshold (Supplementary Table S2). The allele frequency of the allele at snp_chr8_1844714 associated with higher virulence was 33% (0% in Australia, 9% in Oregon, 65% in Switzerland and 94% in Israel). Population differentiation for snp_chr8_1844714 was higher (FST=0.59) than the genome-wide population differentiation (FST=0.24; Supplementary Note S1).
To investigate polymorphism linked to the strongest associated SNP, we screened the sequenced isolates for evidence of segmental deletions using normalized read depth. We found that all 34 isolates carrying the alternative allele (higher PLACP) had a segmental deletion affecting the neighboring gene Zt_8_609 located at a distance of 1486 bp. Conversely, all 51 isolates carrying the reference allele (lower PLACP) did not show any evidence for segmental deletions of the gene. The deletion polymorphism of Zt_8_609 was in complete linkage disequilibrium with the snp_chr8_1844714 (Figure 3a). We found no other gene deletion polymorphism in linkage disequilibrium with snp_chr8_1844714. The length of the segmental deletions affecting Zt_8_609 was at least 3 kb. We confirmed the deletion polymorphism of Zt_8_609 by PCR (Figure 3b; Supplementary Table S6).
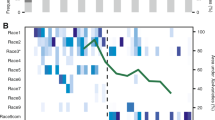
Segmental gene deletion polymorphism of a gene encoding a SSP correlated with the strongest association for virulence of Z. tritici. (a) The left panel shows segmental deletions detected within 3 kb of the most significant association (snp_chr8_1844714) for virulence. The length of segmental deletions detected per isolate is indicated by red rectangles. Each row corresponds to one isolate. The Zt_8_609 gene affected by deletion polymorphism is indicated with black rectangles. The right panel shows the percentage leaf area covered by pycnidia for each isolate. Barplots are colored by genotypes at snp_chr8_1844714. (b) Polymerase chain reaction (PCR) screening for the presence of Zt_8_609 in six Z. tritici isolates originating from Switzerland, Israel and Oregon. (c) Expression profile of Zt_8_609 based on RNAseq transcription data collected throughout the entire time course of leaf infection. Transcription levels are shown as mapped reads per kilobase of exon per million mapped reads (RPKM). (d) Amino acid sequence of the protein encoded by Zt_8_609. Cysteine residues are highlighted in pink.
We used RNAseq data generated for IPO323 (Rudd et al., 2015) to validate the gene model of Zt_8_609 and could identify the missing stop codon (Supplementary Figure S5; Supplementary Note S3). Zt_8_609 was 204 bp long and composed of four exons encoding a protein of 67 amino acids. The immature protein contained a signal peptide and six cysteines (9%; Figure 3d) and had no homology to any other known protein. The gene was strongly up-regulated in IPO323 infecting wheat with a peak expression at 9 days post inoculation (Figure 3c). This corresponds to the onset of visible disease symptoms (that is, lesions) and indicates the switch to a necrotrophic lifestyle (Rudd et al., 2015; Sánchez-Vallet et al., 2015). All characteristics of Zt_8_609 strongly suggest that the gene encodes an SSP that acts as an effector during wheat infection.
An effector gene located in proximity to a highly polymorphic genome compartment
The gene Zt_8_609 was located at the border region between a gene-poor (1.774-1.844 Mb) and a gene-dense region (1.848-1.944 Mb) of chromosome 8 (Figures 4a and b). The gene-poor region was rich in transposable elements (TEs) including long terminal repeat retrotransposons (Gypsy, Copia, AFUT1-I) and non-long terminal repeat retrotransposons (LINE-like Tad1; Figure 4c). We identified a large number of segmental deletions among isolates (Figure 4d). Individual segmental deletions ranged from 0.2-66 kb and were affecting chromosomal regions with a minimum distance of 1 kb to snp_chr8_1844714. The total length of segmental deletions per isolate was on average 41 kb. The maximum total deletion length was 68 kb and only two Swiss isolates lacked any deletions in the region. Deletion profiles differed substantially among isolates and nearly all isolates were of a unique deletion genotype. Furthermore, segmental deletions showed evidence for population structure. Isolates from the Oregon populations had the highest total length of segmental deletions with an average of 55 kb per isolate. Deletions spanning 1.784–1.794 Mb and 1.825–1.830 Mb were fixed among all isolates from Oregon and Israel. The Swiss and Israel populations showed a similar range of total segmental deletion with an average of 35 and 37 kb per isolate, respectively. The total deletion length was lowest in Australia with an average of 19 kb per isolate.
Population genomic analyses for the most significant GWAS locus associated with virulence of Z. tritici. (a) Manhattan plot for the percent leaf area covered by pycnidia on Toronit based on a mixed linear model with kinship matrix (MLM K). (b) Location of genes in the reference genome. (c) Location of transposable elements in the reference genome. (d) Segmental deletions (shown in red) identified among 106 sequenced isolates compared with the reference genome. Each row represents the segmental deletions identified in one isolate. (e) LD r2 decay for snp_chr8_1844714 with all neighboring SNPs shown for each population separately.
We investigated linkage disequilibrium decay in both the gene-poor and the gene-dense regions adjacent to Zt_8_609 (Figure 4e). For this, we calculated LD decay of all pairs of SNPs that included snp_chr8_1844714 and found that LD r2 decayed rapidly in the gene-dense 1.844–1.944 Mb region (r2<0.2 within 2 kb). However, the highly polymorphic 1.744–1.844 Mb region showed a much slower LD decay (r2<0.2 within 13, 12 and 19 kb in the Oregon, Swiss and Israel populations, respectively). The slower LD decay in the 1.744–1.844 Mb region is most likely explained by the fact that physical marker distances were shorter than expected from the reference genome because the region was rich in segmental deletions. However, the high degree of structural variation could also have led to reduced recombination rates (Otto and Lenormand, 2002). Alternatively, selective sweeps associated with Zt_8_609 could also have led to higher LD.
Evidence for partial gene deletions and loss-of-function mutations
We analyzed sequence diversity of Zt_8_609 in the 56 isolates carrying the gene using de novo genome assemblies (Supplementary Table S7). The gene sequence was completely conserved in 37 isolates from the Swiss, Israel, and Oregon populations. We found 6 SNPs in 33 out of the 37 isolates within 700 bp of the 5′ upstream region. We identified substantial sequence polymorphism in the remaining 19 isolates. Segmental deletions affecting one or more exons were found in five Swiss and Oregon isolates. In all five isolates the flanking sequences were conserved. The deletions affected either the first two exons including the sequence encoding the signal peptide or the last exon including the stop codon. The gene was highly polymorphic in Australian isolates (π=0.05). We found that all except one Australian isolate had a start codon mutation from AUG to AUA. Two isolates had a segmental deletion affecting the last exon. Isolate STAus01_1D8 had a 1 bp deletion in the first exon leading to a truncated reading frame. The high frequency of start codon mutations and high nucleotide diversity among Australian isolates suggests that the gene is largely non-functional in this population. We found that virulence on Toronit was higher in isolates found to have non-functional alleles of Zt_8_609 compared with the isolates that have the conserved allele. Isolates carrying alleles with partial gene deletions (mean PLACP=5.5; n=7) and alleles with start or stop codon mutations (mean PLACP=5.5; n=12) were of intermediate virulence compared with the isolates with the conserved Zt_8_609 allele (mean PLACP=2.5; n=37) and isolates missing Zt_8_609 (mean PLACP=14.8; n=36; Figure 5). We found no impact of Zt_8_609 deletions on PLACP on Greina (Figure 5b).
Correlation of virulence of Z. tritici on Toronit and Greina with major mutations in the Zt_8_609 gene. (a) Gene structure and loss-of-function mutations in the Zt_8_609 gene are shown schematically. In total, six major variants of the locus were identified. Missing exons are shown in gray. Conserved exons are shown in black. Mutations in the start codon are shown with a red cross. (b) Bars show the mean values of percentage leaf area covered by pycnidia on the cultivars Toronit (blue) and Greina (red) according to the six major mutations in the gene structure. Bars show the standard error of the mean.
Comparative genomic analyses of the non-homologous chromosomal region
We characterized the segmental deletion affecting Zt_8_609 using comparative genomics analyses. For this, we analyzed chromosomal synteny between the reference genome IPO323 (Goodwin et al., 2011) and the completely assembled Swiss isolate ST99CH3D7 which lacks Zt_8_609 (Figure 6a; Plissonneau et al., 2016). We found that the 1.78–1.84 Mb region in IPO323 which includes Zt_8_609 had no homology in the ST99CH3D7 genome. The loss of Zt_8_609 was accompanied by the deletion of a 65 kb cluster of TEs. At the homologous position, ST99CH3D7 did not harbor any significant non-syntenic sequences compared with IPO323. The chromosomal sequences adjacent to the TE cluster (1.76–1.78 Mb and 1.84–1.88 Mb) showed high degrees of synteny between the two genomes with the exception of four non-syntenic sequences of ST99CH3D7. The length of the non-syntenic sequences varied from 0.1 to 1 kb and contained small clusters of TEs. The large segmental deletion in ST99CH3D7 confirmed that physical distances between SNPs in the 1.776–1.844 Mb region of the reference genome were indeed shorter in homologous chromosomes of the species.
The evolutionary history and structural variation at the major virulence locus detected by GWAS. (a) Chromosomal synteny analysis between the two completely assembled genomes of Z. tritici. The genome of ST99CH3D7 lacking the gene Zt_8_609 is shown in the upper panel and the reference genome IPO323 is shown in the lower panel. The synteny analyses comprised the region of 1.76–1.9 Mb on chromosome 8. Homologous regions between the two genomes are shown by red areas. The location of genes and transposable elements are shown by black and orange rectangles, respectively. (b) Comparative genomic analyses of the three closely related sister species Z. tritici, Z. pseudotritici, Z. ardabiliae and Z. passerinii. The presence of Zt _8_609 and eight neighboring genes is indicated by gray rectangles. Genes missing in any of the genomes are indicated by red rectangles. Haplotypes of specific combinations of presence and absence of individual genes are shown with their frequency relative to the population (in Z. tritici) or species (all other species).
The evolutionary history of the chromosomal region among sister species
We investigated the origin of Zt_8_609 by performing homology searches in genomes of the closest known sister species. We found no ortholog of Zt_8_609 in any genomes of the three closest known sister species Z. pseudotritici, Z. ardabiliae and Z. passerinii from which Z. tritici diverged 10 500, 18 500 and 68 500 years ago, respectively (Stukenbrock et al., 2006, 2012b). Given the absence of Zt_8_609 in any of the three closest known relatives of Z. tritici, the gene was likely gained in Z. tritici after speciation.
We extended the homology search to the four neighboring genes of Zt_8_609 in each direction (Figure 6b). In total, we found that five out of eight genes were present in all analyzed genomes of the sister species. Three genes (Zt_8_606, Zt_8_608 and Zt_8_611) were missing in at least one of the species. For Zt_8_611, we found no ortholog in any sister species. Zt_8_611 is located in a gene-dense region and is predicted to encode an F-box domain protein. Furthermore, we found no orthologs for Zt_8_608 in the most closely related species Z. pseudotritici, but the gene was present in the two more distant species Z. ardabiliae and Z. passerinii. Zt_8_608 was located similarly to Zt_8_609 at the border of the large TE cluster between 1.774 and 1.844 Mb and encodes a protein of unknown function. Zt_8_606 was found in one out of five Z. pseudotritici isolates and in all Z. ardabiliae isolates. Zt_8_606 encodes a protein of unknown function. The lack of Zt_8_608 and Zt_8_611 in several Z. tritici sister species shows that the chromosomal region containing Zt_8_609 underwent significant sequence rearrangements in its recent evolutionary history. In Z. tritici, both Zt_8_608 and Zt_8_611 were not present in all isolates. Zt_8_611 was affected by deletions in all populations except Australia, ranging from a frequency of 10% in Israel to 30% in the Swiss and Oregon populations. Zt_8_608 was deleted in one isolate from Israel. These comparative genomic analyses showed that the virulence locus was embedded in a highly dynamic chromosomal compartment that may have favored the emergence of evolutionary novelty.
Discussion
Virulence of field isolates of Z. tritici had a complex genetic basis, which included at least 25 distinct genomic regions. Significantly associated SNPs were located in genes associated with plant cell wall degradation, cell transport, and fungal metabolism. Virulence in Z. tritici was suggested to be a largely quantitative trait despite evidence for gene-for-gene interactions (Brading et al., 2002; Brown et al., 2015). Polymorphism linked to pycnidia production segregated in most populations with the exception of Australia. The Australian population was monomorphic for most of the associated SNPs and the low level of genetic diversity was proposed to be due to a bottleneck that occurred during the introduction of wheat to Australia (Zhan et al., 2005).
The diversity and distribution of genes associated with pycnidia production confirmed that virulence was largely quantitative. Some associated genes encoded protein functions that were previously associated with virulence in Z. tritici or other plant pathogens. For instance, we identified two genes encoding cell wall degrading enzymes. One cell wall degrading enzyme likely has a role in galactan hydrolysis. The second one was a pectinase previously found to be under purifying selection in Z. tritici populations (Brunner et al., 2013). Cell wall degrading enzymes have a crucial role in host cell wall degradation and pathogen nutrition during the infection (Esquerré-Tugayé et al., 2000; De Jonge et al., 2011). In addition, we identified associations in proximity to genes encoding major facilitator superfamily transporters. Major facilitator superfamily transporters are involved in nutrient uptake, the secretion of effectors, or protection against plant defense compounds (Pao et al., 1998). Some major facilitator superfamily transporters are known virulence factors in plant pathogens, including Z. tritici (Stergiopoulos et al., 2003; Oh et al., 2008). We also identified adaptive genetic variation in genes encoding proteins involved in nutrient acquisition and general metabolism which were not previously associated with fungal virulence.
Host specificity of virulence genes
We found strong evidence for host specificity in the genes affecting virulence, as there was no overlap in significant associations between the two cultivars. The strength of the associations was generally higher for SNPs linked to PLACP on Toronit than for PLACP on Greina. A lack of overlap in associated chromosomal regions for PLACP among cultivars may partially be due to a lack in power to identify variants of small effect (Korte and Farlow, 2013). However, there was no meaningful correlation among association p-values for PLACP on the two different cultivars. Host specificity in virulence may evolve through multiple evolutionary scenarios. In several well studied host-pathogen systems, host specificity is determined largely by the matching of resistance or host susceptibility genes and their corresponding effectors (Faris et al., 2010; Liu et al., 2013; Bourras et al., 2015). However, we found that host specificity in Z. tritici included an important quantitative component based on a large number of small effect loci. Virulence-associated loci likely included polymorphism associated with variation in gene regulation, adding further complexity to the host-specific expression of virulence.
Deletion of a gene encoding a SSP strongly increased virulence
The strongest GWAS association on Toronit was linked to the deletion polymorphism of Zt_8_609. The gene encodes a SSP which is highly expressed during the infection. The absence of a recognizable domain in Zt_8_609 is a characteristic shared with many plant pathogen effectors (Presti et al., 2015). The peak of expression of Zt_8_609 was during the necrotrophic phase, which suggests that the encoded protein is involved in plant defense activation or recognition, or the induction of necrosis in the plant cell. The absence of Zt_8_609 was correlated with significantly higher pycnidia production. This strongly suggests that Zt_8_609 is involved in a gene-for-gene interaction with a corresponding R-gene in the host. Zt_8_609 likely encodes an avirulence effector that largely prevents the fungus from successfully reproducing on its host. In addition to complete gene deletions, we found loss-of-function mutations and multiple exon deletions in Australian isolates. Interestingly, the isolates carrying loss-of-function alleles were on average more virulent than isolates carrying the conserved copy of the gene, suggesting that partial deletions are sufficient to evade detection.
Despite the strong association of the gene deletion polymorphism and virulence, multiple isolates carrying Zt_8_609 were able to produce intermediate levels of pycnidia on Toronit. Therefore, the presence of the protein encoded by Zt_8_609 does not completely prevent disease in all genetic backgrounds. The importance of the genetic background for the function of effectors was already shown in the oil seed rape pathogen Leptosphaeria maculans, where the presence of the allele of the avirulence gene AvrLm4-7 suppressed the Rlm3-mediated recognition of AvrLm3 (Plissonneau et al., 2015). Similar interactions among effectors could explain how strains carrying Zt_8_609 are virulent. Furthermore, differential regulation of Zt_8_609 among strains could explain the incomplete association of Zt_8_609 and suppressed virulence.
The evolution of an avirulence gene in a plant pathogen
The comparative genomics analyses among the closest known sister species indicated that Zt_8_609 was likely gained in Z. tritici after speciation. Horizontal gene transfer has been recently suggested as an important source of evolutionary innovation in fungal pathogens (Friesen et al., 2006; Gardiner et al., 2012; Soanes and Richards, 2014). However, the chromosomal region surrounding Zt_8_609 showed no evidence for horizontal transfer such as unusual variation in GC-content. Alternatively, the gene could have evolved from a paralogous gene copy. Gene duplications in effector gene families are a known driver of effector gene evolution in plant pathogenic fungi (Khang et al., 2008; Pendleton et al., 2014). However, we identified no paralogs in Z. tritici genomes. As the coding sequence was extremely short (204 bp), the gene may also have evolved de novo from a primitive open reading frame through a step-wise fixation of mutations. Extremely short genes are likely to evolve more readily than genes encoding structurally complex proteins. The de novo gene evolution from non-coding sequences was shown in the Baker's yeast Saccharomyces cerevisiae (Carvunis et al., 2012; McLysaght and Guerzoni, 2015). However, the presence of multiple short introns in Zt_8_609 suggests that the open reading frame required a complex series of adaptive mutations to become functional. Identifying the evolutionary trajectory of effector genes lacking any known homolog in sister species will be an important step toward understanding processes driving rapid adaptive evolution in plant pathogens.
Effector gene deletions as an adaptive response to host recognition
Zt_8_609 and multiple neighboring genes were segregating gene deletion polymorphisms in all populations except Australia. The trajectory of Zt_8_609 was different in the Australian population, as the gene accumulated loss-of-function mutations and is likely pseudogenized. The deletion frequency of Zt_8_609 varied extensively among populations. The high degree of differentiation of the gene (FST=0.59) compared with the genome-wide average (FST=0.24) suggests that selection may have reinforced population differentiation in deletion frequencies. Effector gene deletions are a key evolutionary step in plant pathogens to evade host immunity (Stukenbrock and McDonald, 2009; Presti et al., 2015). The fact that we identified alleles of Zt_8_609 in at least one isolate in all analyzed populations suggests that the gene may confer a fitness benefit in the absence of recognition by the host (that is, a host not expressing a cognate R-gene product). For example the protein may modulate host immune system responses as shown for a number of effector proteins (De Jonge et al., 2012). The observed variation in Zt_8_609 deletion frequencies may reflect geographic patterns in the deployment of cultivars sharing the same R-gene. However, R-gene deployment is poorly documented for Stb R-genes (Brown et al., 2015). The segregating effector gene polymorphisms found in field populations highlight the evolutionary potential of fungal pathogens to rapidly respond to host-driven selection.
Genomic rearrangements as a driver to modulate virulence in Z. tritici populations
Synteny analyses of the two completely assembled genomes of the same species showed that Zt_8_609 was located in proximity to a large chromosomal rearrangement. This confirmed the extensive segmental deletions observed within the species. Given the highly polymorphic region adjacent to Zt_8_609, it is likely that the gene was independently lost multiple times through transposable element-mediated rearrangements. As the gene deletion likely led to higher virulence on hosts carrying the corresponding R-gene, the chromosomal rearrangements were likely adaptive and favored by selection. Chromosomal rearrangements associated with virulence evolution were reported for closely related species or asexual lineages of the same pathogen (Raffaele and Kamoun, 2012; De Jonge et al., 2013). Our study identified structural variation linked to virulence segregating within a sexually reproducing plant pathogen. The high levels of recombination observed in the species (Croll et al., 2015) likely had a significant impact on the extensive and rapid diversification observed for Zt_8_609.
Fungal plant pathogen genomes are often compartmentalized into gene-sparse, repeat-rich and gene-dense compartments similar to more complex eukaryotic genomes (Lynch and Walsh, 2007). In multiple species, the gene-sparse and repeat-rich compartments were enriched in effector genes and referred to as the ‘two-speed genome’ (Dong et al., 2015). In Z. tritici, genome compartmentalization was suggested for core and accessory chromosomes (Stukenbrock et al., 2010; Croll and McDonald, 2012), but no enrichment of effector genes according to genomic compartments was identified to date (Rudd et al., 2015). The avirulence gene Zt_8_609 was located on a core chromosome and was not within an island of TEs. Our comparative genomics analyses of two complete genomes showed that the loss of Zt_8_609 was associated with the loss of a massive cluster of TEs. Unlike most genomes analyzed to date, the location of heterochromatic or TE-rich regions was not conserved within Z. tritici. The mechanisms leading to the complete excision or insertion of TE clusters is poorly understood. The localization of genes in proximity to TEs likely imposes higher mutation loads due to the increased likelihood of non-homologous recombination. Furthermore, fungal genomes evolved genomic defenses to prevent the activity of TEs. The best known defense mechanism is repeat-induced point mutation (RIP) that prevents the spread of TEs by mutating near identical sequences in the genome (Galagan and Selker, 2004). RIP was involved in the emergence and diversification of effector genes in L. maculans (Rouxel et al., 2011) and effector inactivation in Fusarium oxysporum (Inami et al., 2012). RIP is active in Z. tritici (Goodwin et al., 2011) and may have contributed to the high levels of Zt_8_609 diversity found among Australian isolates. TEs may also affect transcription of Zt_8_609 by mutating promoter sequences or through epigenetic silencing (Ali et al., 2014; Elbarbary et al., 2016).
Conclusions
We identified a major association between virulence on a wheat cultivar and a deletion polymorphism for a gene encoding an SSP. The gene deletion likely enabled the pathogen to escape recognition by hosts carrying a cognate R-gene. The gene encoding the SSP was located in a rapidly evolving, TE-rich region of the genome in which multiple neighboring genes were similarly affected by gene deletions. The gene was not found in any of the three known sister species and we found no evidence for acquisition by horizontal gene transfer. Hence, the gene may have evolved recently de novo from non-coding DNA. The rapid evolution of open reading frames encoding effectors may be an underappreciated mechanism of virulence evolution in plant pathogens. Each sequenced genome showed a unique haplotype of segmental deletions in the TE-rich region. Strong directional selection for gene loss and non-homologous recombination likely acted jointly to create the extant structural variation at the locus. Virulence evolution driven by the loss of a recognized effector gene is accelerated by segregating structural variation. Hence, chromosomal rearrangements can be a major driver of virulence evolution.
References
Abyzov A, Urban AE, Snyder M, Gerstein M . (2011). CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21: 974–984.
Ali S, Laurie JD, Linning R, Cervantes-Chávez JA, Gaudet D, Bakkeren G . (2014). An immunity-triggering effector from the barley smut fungus Ustilago hordei resides in an Ustilaginaceae-specific cluster bearing signs of transposable element-assisted evolution. PLoS Pathog 10: e1004223–19.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477.
Bergelson J, Roux F . (2010). Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev 11: 867–879.
Bolger AM, Lohse M, Usadel B . (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120.
Bourras S, McNally KE, Ben-David R, Parlange F, Roffler S, Praz CR et al. (2015). Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 27: 2991–3012.
Boyd LA, Ridout C, O'Sullivan DM, Leach JE, Leung H . (2012). Plant–pathogen interactions: disease resistance in modern agriculture. Trends Genet 29: 1–8.
Brabant C, Fossati D, Kleijer G . (2006). La sélection du blé de printemps en Suisse. Rev Suisse Agric 38: 73–80.
Brading PA, Verstappen ECP, Kema GHJ, Brown JKM . (2002). A gene-for-gene relationship between wheat and Mycosphaerella graminicola, the Septoria tritici blotch pathogen. Phytopathology 92: 439–445.
Brown JKM, Chartrain L, Lasserre-Zuber P, Saintenac C . (2015). Genetics of resistance to Zymoseptoria tritici and applications to wheat breeding. Fungal Genet Biol 79: 33–41.
Brunner PC, Torriani SFF, Croll D, Stukenbrock EH, McDonald BA . (2013). Coevolution and life cycle specialization of plant cell wall degrading enzymes in a hemibiotrophic pathogen. Mol Biol Evol 30: 1337–1347.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10: 421–429.
Carvunis A-R, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N et al. (2012). Proto-genes and de novo gene birth. Nature 487: 370–374.
Cowger C, Hoffer ME, Mundt CC . (2000). Specific adaptation by Mycosphaerella graminicola to a resistant wheat cultivar. Plant Pathol 49: 445–451.
Croll D, Lendenmann MH, Stewart E, McDonald BA . (2015). The impact of recombination hotspots on genome evolution of a fungal plant pathogen. Genetics 201: 1213–1228.
Croll D, McDonald BA . (2012). The accessory genome as a cradle for adaptive evolution in pathogens. PLoS Pathog 8: e1002608.
De Jonge R, van Esse HP, Maruthachalam K, Bolton MD, Santhanam P, Saber MK et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci USA 109: 5110–5115.
De Jonge R, Bolton MD, Kombrink A, van den Berg GCM, Yadeta KA, Thomma BPHJ . (2013). Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res 23: 1271–1282.
De Jonge R, Bolton MD, Thomma BP . (2011). How filamentous pathogens co-opt plants: the ins and outs of fungal effectors. Curr Opin Plant Biol 14: 400–406.
Dong S, Raffaele S, Kamoun S . (2015). The two-speed genomes of filamentous pathogens: waltz with plants. Curr Opin Genetics Dev 35: 57–65.
Elbarbary RA, Lucas BA, Maquat LE . (2016). Retrotransposons as regulators of gene expression. Science 351: aac7247.
Esquerré-Tugayé MT, Boudart G, Dumas B . (2000). Cell wall degrading enzymes, inhibitory proteins, and oligosaccharides participate in the molecular dialogue between plants and pathogens. Plant Physiol Biochem 38: 157–163.
Eyal Z, Scharen AL, Prescott JM, Van Ginkel M . (1987) The Septoria Diseases of Wheat: Concepts and Methods of Disease Management. CIMMYT: Mexico.
Faris JD, Zhang Z, Lu H, Lu S, Reddy L, Cloutier S et al. (2010). A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc Natl Acad Sci USA 107: 13544–13549.
Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nat Rev 484: 186–194.
Flor HH . (1971). Current status of gene-for-gene concept. Annu Rev Phytopathol 9: 275–296.
Fones H, Gurr S . (2015). The impact of Septoria tritici Blotch disease on wheat: an EU perspective. Fungal Genet Biol 79: 3–7.
Friesen TL, Stukenbrock EH, Liu Z, Meinhardt S, Ling H, Faris JD et al. (2006). Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet 38: 953–956.
Galagan JE, Selker EU . (2004). RIP: the evolutionary cost of genome defense. Trends Genet 20: 417–423.
Gardiner DM, McDonald MC, Covarelli L, Solomon PS, Rusu AG, Marshall M et al. (2012). Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog 8: e1002952.
Ghanbarnia K, Fudal I, Larkan NJ, Links MG, Balesdent M-H, Profotova B et al. (2015). Rapid identification of the Leptosphaeria maculans avirulence gene AvrLm2 using an intraspecific comparative genomics approach. Mol Plant Pathol 16: 699–709.
Goodwin SB, Ben M'Barek S, Dhillon B, Wittenberg AHJ, Crane CF, Hane JK et al. (2011). Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet 7: e1002070–17.
Grandaubert J, Bhattacharyya A, Stukenbrock EH . (2015). RNA-seq-based gene annotation and comparative genomics of four fungal grass pathogens in the genus Zymoseptoria identify novel orphan genes and species-specific invasions of transposable elements. G3 5: 1323–1333.
Guy L, Kultima JR, Andersson SGE . (2010). genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26: 2334–2335.
Inami K, Yoshioka-Akiyama C, Morita Y, Yamasaki M, Teraoka T, Arie T . (2012). A genetic mechanism for emergence of races in Fusarium oxysporum f. sp. lycopersici: inactivation of avirulence gene AVR1 by transposon insertion. PLoS One 7: e44101.
Ingvarsson PK . (2005). Nucleotide polymorphism and linkage disequilibrium within and among natural populations of European aspen (Populus tremula L., Salicaceae). Genetics 169: 945–953.
Ipcho SVS, Hane JK, Antoni EA, Ahren D, Henrissat B, Friesen TL et al. (2012). Transcriptome analysis of Stagonospora nodorum: gene models, effectors, metabolism and pantothenate dispensability. Mol Plant Pathol 13: 531–545.
Jiang RHY, Tyler BM . (2012). Mechanisms and evolution of virulence in oomycetes. Annu Rev Phytopathol 50: 295–318.
Jones JDG, Dangl JL . (2006). The plant immune system. Nat Rev 444: 323–329.
Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240.
Kettles GJ, Kanyuka K . (2016). Dissecting the molecular interactions between wheat and the fungal pathogen Zymoseptoria tritici. Front Plant Sci 7: 508.
Khang CH, Park S-Y, Lee Y-H, Valent B, Kang S . (2008). Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol Plant Microbe Interact 21: 658–670.
Korte A, Farlow A . (2013). The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9: 29.
Krenz JE, Sackett KE, Mundt CC . (2008). Specificity of incomplete resistance to Mycosphaerella graminicola in wheat. Phytopathology 98: 555–561.
Lamari L . (2002) Assess: Image Analysis Software for Plant Disease Quantification. St. Paul, MN, USA: Amer Phytopathological Society.
Langmead B, Trapnell C, Pop M, Salzberg SL . (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
Linde CC, Zhan J, McDonald BA . (2002). Population structure of Mycosphaerella graminicola: from lesions to continents. Phytopathology 92: 946–955.
Liu W, Liu J, Ning Y, Ding B, Wang X, Wang Z et al. (2013). Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae. Mol Plant 6: 605–620.
Lynch M, Walsh B . (2007) The Origins of Genome Architecture. Sunderland: MA, USA: Sinauer Associates.
Magrane M,, UniProt Consortium. (2011). UniProt Knowledgebase: a hub of integrated protein data. Database 2011: bar009.
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res 43: D222–D226.
Marshall R, Kombrink A, Motteram J, Loza-Reyes E, Lucas J, Hammond-Kosack KE et al. (2011). Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol 156: 756–769.
McDonald MC, McGinness L, Hane JK, Willams AH, Milgate A, Solomon PS . (2016). Utilizing gene tree variation to identify candidate effector genes in Zymoseptoria tritici. G3 (Bethesda) 6: 779–791.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303.
McLysaght A, Guerzoni D . (2015). New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos Trans R Soc B 370: 20140332–20140338.
Mirzadi Gohari A, Ware SB, Wittenberg AHJ, Mehrabi R, Ben M'Barek S, Verstappen ECP et al. (2015). Effector discovery in the fungal wheat pathogen Zymoseptoria tritici. Mol Plant Pathol 16: 931–945.
Morais do Amaral A, Antoniw J, Rudd JJ, Hammond-Kosack KE . (2012). Defining the predicted protein secretome of the fungal wheat leaf pathogen Mycosphaerella graminicola. PLoS One 7: e49904.
Nei M . (1987) Molecular Evolutionary Genetics. New York, NY, USA: Columbia University Press.
Oh Y, Donofrio N, Pan H, Coughlan S, Brown DE, Meng S et al. (2008). Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol 9: 1–24.
Oliveira-Garcia E, Valent B . (2015). How eukaryotic filamentous pathogens evade plant recognition. Curr Opin Microbiol 26: 92–101.
Otto SP, Lenormand T . (2002). Resolving the paradox of sex and recombination. Nat Rev Genet 3: 252–261.
Palma-Guerrero J, Torriani SFF, Zala M, Carter D, Courbot M, Rudd JJ et al. (2016). Comparative transcriptomic analyses of Zymoseptoria tritici strains show complex lifestyle transitions and intraspecific variability in transcription profiles. Mol Plant Pathol 17: 845–859.
Pao SS, Paulsen IT, Saier MH . (1998). Major facilitator superfamily. FEMS Microbiol Rev 62: 1–34.
Paradis E, Claude J, Strimmer K . (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290.
Pendleton AL, Smith KE, Feau N, Martin FM, Grigoriev IV, Hamelin R et al. (2014). Duplications and losses in gene families of rust pathogens highlight putative effectors. Front Plant Sci 5: 299.
Petersen TN, Brunak S, Heijne von G, Nielsen H . (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Meth 8: 785–786.
Plissonneau C, Daverdin G, Ollivier B, Blaise F, Degrave A, Fudal I et al. (2015). A game of hide and seek between avirulence genes AvrLm4-7and AvrLm3 in Leptosphaeria maculans. New Phytol 209: 1613–1624.
Plissonneau C, Stürchler A, Croll D . (2016). The Evolution of Orphan Regions in Genomes of a Fungal Pathogen of Wheat. mBio 7: e01231–16.
Poppe S, Dorsheimer L, Happel P, Stukenbrock EH . (2015). Rapidly evolving genes are key players in host specialization and virulence of the fungal wheat pathogen Zymoseptoria tritici Mycosphaerella graminicola. PLoS Pathog 11: e1005055–21.
Presti LoL, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M et al. (2015). Fungal effectors and plant susceptibility. Annu Rev Plant Biol 66: 513–545.
R Core Team. (2014) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, Available at http://www.R-project.org/.
Raffaele S, Kamoun S . (2012). Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol 10: 417–430.
Robbertse B, Tatusova T . (2011). Fungal genome resources at NCBI. Mycology 2: 142–160.
Rouxel T, Grandaubert J, Hane JK, Hoede C, van de Wouw AP, Couloux A et al. (2011). Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat Commun 2: 202.
Rudd JJ, Kanyuka K, Hassani-Pak K, Derbyshire M, Andongabo A, Devonshire J et al. (2015). Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol 167: 1158–1185.
Sánchez-Vallet A, McDonald MC, Solomon PS, McDonald BA . (2015). Is Zymoseptoria tritici a hemibiotroph? Fungal Genet Biol 79: 29–32.
Schmidt SM, Lukasiewicz J, Farrer R, van Dam P, Bertoldo C, Rep M . (2015). Comparative genomics of Fusarium oxysporum f. sp. melonis reveals the secreted protein recognized by the Fom-2 resistance gene in melon. New Phytol 209: 307–318.
Soanes D, Richards TA . (2014). Horizontal gene transfer in eukaryotic plant pathogens. Annu Rev Phytopathol 52: 583–614.
Stergiopoulos I, de Wit PJGM . (2009). Fungal effector proteins. Annu Rev Phytopathol 47: 233–263.
Stergiopoulos I, Zwiers L-H, De Waard MA . (2003). The ABC transporter MgAtr4 is a virulence factor of Mycosphaerella graminicola that affects colonization of substomatal cavities in wheat leaves. Mol Plant Microbe Interact 16: 689–698.
Stewart EL, McDonald BA . (2014). Measuring quantitative virulence in the wheat pathogen Zymoseptoria tritici using high-throughput automated image analysis. Phytopathology 104: 985–992.
Storey JD, Tibshirani R . (2003). Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445.
Stukenbrock EH, Banke S, Javan-Nikkhah M, McDonald BA . (2006). Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Mol Biol Evol 24: 398–411.
Stukenbrock EH, Christiansen FB, Hansen TT, Dutheil JY, Schierup MH . (2012a). Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proc Natl Acad Sci USA 109: 10954–10959.
Stukenbrock EH, Jørgensen FG, Zala M, Hansen TT, McDonald BA, Schierup MH . (2010). Whole-genome and chromosome evolution associated with host adaptation and speciation of the wheat pathogen Mycosphaerella graminicola. PLoS Genet 6: e1001189–13.
Stukenbrock EH, McDonald BA . (2009). Population genetics of fungal and oomycete effectors involved in gene-for-gene interactions. Mol Plant Microbe Interact 22: 371–380.
Stukenbrock EH, Quaedvlieg W, Javan-Nikhah M, Zala M, Crous PW, McDonald BA . (2012b). Zymoseptoria ardabiliae and Z. pseudotritici, two progenitor species of the septoria tritici leaf blotch fungus Z. tritici (synonym: Mycosphaerella graminicola. Mycologia 104: 1397–1407.
Torriani SFF, Stukenbrock EH, Brunner PC, McDonald BA, Croll D . (2011). Evidence for extensive recent intron transposition in closely related fungi. Curr Biol 21: 2017–2022.
Vleeshouwers VGAA, Oliver RP . (2014). Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol Plant Microbe Interact 27: 196–206.
Wickham H . (2009) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag: New York, NY, USA.
Zhan J, Linde CC, Jürgens T, Merz U, Steinebrunner F, McDonald BA . (2005). Variation for neutral markers is correlated with variation for quantitative traits in the plant pathogenic fungus Mycosphaerella graminicola. Mol Ecol 14: 2683–2693.
Zhan J, Pettway RE, McDonald BA . (2003). The global genetic structure of the wheat pathogen Mycosphaerella graminicola is characterized by high nuclear diversity, low mitochondrial diversity, regular recombination, and gene flow. Fungal Genet Biol 38: 286–297.
Acknowledgements
We acknowledge Ina Nissan and Christian Beisel for the sequencing at the Quantitative Genomics Facility at the Department of Biosystems Science and Engineering (D-BSSE), in ETH Zurich, Switzerland. Marcello Zala provided assistance in the laboratory. The research was supported by the ETH Zurich Research Grant 12-03.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Hartmann, F., Sánchez-Vallet, A., McDonald, B. et al. A fungal wheat pathogen evolved host specialization by extensive chromosomal rearrangements. ISME J 11, 1189–1204 (2017). https://doi.org/10.1038/ismej.2016.196
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.196
This article is cited by
-
A systematic screen for co-option of transposable elements across the fungal kingdom
Mobile DNA (2024)
-
Analysis of five near-complete genome assemblies of the tomato pathogen Cladosporium fulvum uncovers additional accessory chromosomes and structural variations induced by transposable elements effecting the loss of avirulence genes
BMC Biology (2024)
-
Quantitative pathogenicity and host adaptation in a fungal plant pathogen revealed by whole-genome sequencing
Nature Communications (2024)
-
Combined pangenomics and transcriptomics reveals core and redundant virulence processes in a rapidly evolving fungal plant pathogen
BMC Biology (2023)
-
Genome-wide expression QTL mapping reveals the highly dynamic regulatory landscape of a major wheat pathogen
BMC Biology (2023)