Abstract
The budding yeasts are prime models in genomics and cell biology, but the ecological factors that determine their success in non-human-associated habitats is poorly understood. In North America Saccharomyces yeasts are present on the bark of deciduous trees, where they feed on bark and sap exudates. In the North East, Saccharomyces paradoxus is found on maples, which makes maple sap a natural substrate for this species. We measured growth rates of S. paradoxus natural isolates on maple sap and found variation along a geographical gradient not explained by the inherent variation observed under optimal laboratory conditions. We used a functional genomic screen to reveal the ecologically relevant genes and conditions required for optimal growth in this substrate. We found that the allantoin degradation pathway is required for optimal growth in maple sap, in particular genes necessary for allantoate utilization, which we demonstrate is the major nitrogen source available to yeast in this environment. Growth with allantoin or allantoate as the sole nitrogen source recapitulated the variation in growth rates in maple sap among strains. We also show that two lineages of S. paradoxus display different life-history traits on allantoin and allantoate media, highlighting the ecological relevance of this pathway.
Similar content being viewed by others
Introduction
Unicellular eukaryotes such as yeast are well-established models for genetics and cell biology, but they are also powerful emerging models for ecology, evolution and ecotoxicology (Landry et al., 2006; Hittinger, 2013; Braconi et al., 2016). The budding yeasts of the genus Saccharomyces in particular are widely studied organisms. Its most studied species, S. cerevisiae, is the testbed of a large body of research in fundamental cell biology and genetics. However, there is little knowledge about Saccharomyces ecology, natural history and geographic distribution compared with our understanding of their behavior under optimal laboratory conditions (Goddard and Greig, 2015; Sylvester et al., 2015). A major limiting element is our lack of knowledge of fitness determinants under their natural environmental conditions. Overcoming this limitation would require either measuring fitness in the wild (Boynton et al., 2016) or elucidating the composition and availability of substrates in natural habitats (Samani et al., 2015), or at least growing yeast isolates on a natural substrate in controlled conditions (Bell, 2010; Giraldo-Perez and Goddard, 2013; Clowers et al., 2015; Kowallik et al., 2015). For example, nitrogen availability is an important ecological factor for microbial communities associated with trees (Kembel and Mueller, 2014; Kembel et al., 2014) and the ability to use specific nitrogen sources could also be critical for yeast growth in the wild. Microhabitats occupied by yeast such as decomposing fruits, flowering plant nectar and saps are rich in sugar but poor in nitrogen, with one or few dominating sources, mainly amino acids (Landry et al., 2006; Hittinger, 2013; Ibstedt et al., 2015).
In North America, Saccharomyces strains have been successfully isolated from deciduous tree bark such as oak and maple (Naumov et al., 1998; Charron et al., 2014a), where they likely feed on sugar-rich sap exudate and bark, as they can be grown in sap and bark tea (Bell, 2010; Kowallik et al., 2015). For instance, Saccharomyces paradoxus strains were successfully isolated from maple tree bark. This species is of particular interest because it is an undomesticated relative of S. cerevisiae and an emerging model system for speciation where distinct lineages have been well described (Leducq et al., 2016). The region where S. paradoxus has been isolated and characterized in North America vastly overlaps with the production area of maple syrup, a natural sweetener obtained from the concentration of maple sap, mostly from Acer saccharum (Filteau, 2011; Leducq et al., 2016). Maple sap composition varies over the spring flow period, along with maple syrup quality (Filteau et al., 2011, 2012). Numerous constituents have been reported, including sugars, organic acid, phenolic compounds, amino acids and minerals, but many compounds remain unidentified (Dumont, 1994a, b; Morselli and Whalen, 1996; Yoshikawa et al., 2013; Yuan et al., 2013). Therefore, aside from the high sugar content, mainly sucrose, other nutrients contributing to fitness determinants of yeast remain to be characterized.
The utilization of maple sap in the food industry makes it a readily available deciduous tree exudate, and thus a prime model substrate for yeast ecological studies. Methods have been optimized to extract and concentrate maple sap in large volumes so that it provides the opportunity to use it as a substrate in the laboratory. We took advantage of the availability of maple sap concentrates and syrups to study the growth of a diverse population of natural isolates of S. paradoxus that have been isolated directly on maple trees, their associated soils and on other substrates in regions where maple tree is known to occur. We identify key metabolic and regulatory pathways that are required for growth in this natural substrate, and at the same time, the major nitrogen source available, allantoate.
Materials and methods
Maple sap and syrup samples
This work was designed to identify yeast cellular functions specifically affected by the molecular composition of maple sap and maple syrup. Since maple syrup is a stable concentrate of maple sap, it was used as a biological replicate to monitor seasonal trends. Maple sap and syrups of every production day of the 2015 season were obtained from a sugaring plant in Edmundston, NB, Canada. Maple sap was concentrated by reverse osmosis before being processed into maple syrup. The sugar concentration of sap concentrate and syrups was measured at room temperature with a refractometer. Sap concentrates were rapidly frozen at −20 °C until processing time. Syrup samples were packaged as per the production norms and kept at room temperature. A total of 17 paired samples were collected (Supplementary Table S1). Six samples spanning the whole tapping period were selected for further experiments. Samples 16 and 17 were collected only 4 h apart, yet there were noticeable differences in the appearance of the syrup produced. In particular, syrup17 was turbid and had an increased viscosity, a defect known as ropy or stringy syrup caused by specific microbial activity in the sap. Before their use in experiments, the sap concentrates were thawed, adjusted to 10° Brix and then sterilized by 0.2 uM filtration. Syrups were handled aseptically and negative controls were included in each experiment, showing that no microbial contamination was present in the original samples. However, a bacterial contamination occurred in syrup1, preventing us to include this sample in subsequent experiments.
Saccharomyces strains
Strains used in this study are listed in Table 1.
Growth experiments
Rich undefined media (YPD) consisted of 1% yeast extract, 2% tryptone and 2% glucose while synthetic media was made with 0.175% yeast nitrogen base, 2% sucrose and 0.005% of nitrogen source, either ammonium sulfate, allantoin or allantoate. Solid medium also contained 2% agar. Sap concentrates and syrups were added to 2% sterilized agar at a final dilution factor of 1/5 and 1/33, respectively, which would correspond to the natural 2–3% sugar concentration of maple sap.
Strains were precultured for 2 days at room temperature on YPD at a density of 1 536 colonies per plate using a BM3-BC robot (S&P Robotics, Inc., North York, ON, Canada). Strains were then transferred on the desired media and grown at room temperature for 10 days. Time series images of the plates were analyzed using custom scripts written in ImageJ 1.45 s (National Institutes of Health, Bethesda, MD, USA; https://imagej.nih.gov/ij/) to measure colony sizes in pixel density as described in Diss et al. (2013). Colony sizes were log2 transformed and normalized by subtracting the value obtained after transfer to account for variation in the initial amount of cells transferred. A time series of 16 data points was used to construct growth curves. Growth curve analysis was performed in JMP12 (SAS Institute, Cary, NC, USA) individually for each replicate by fitting a three parameter Gompertz model. The growth rate and asymptote (efficiency) parameters were averaged for a minimum of six replicates for each strain, excluding colonies at border positions on agar plates. Pearson correlations are presented unless the data were non-normally distributed, then Spearman correlations are presented. A t-test was used to compare lineages since the data had similar variance. P-values correspond to the two-sided hypothesis.
Functional genomic screen
To characterize the ecologically relevant genes for growth in maple sap, we used a systems biology approach using the fitness of S. cerevisiae mutants as biological reporters. The approach consists of using a collection of strains in which the non-essential genes have been deleted individually and replaced with an antibiotic resistance cassette flanked with unique identifying DNA barcodes. These strains are pooled and grown together, and the change in relative frequency of each barcode between treatments is used to calculate fitness. The MATa yeast knockout collection (Giaever et al., 2002) was pooled as in Smith et al. (2011) with the following modification: The collection was replicated twice and selected colonies from the second set of plate were added to the pool to increase the amount of cells of slow growing strains. Following the same protocol, liquid assays were carried out in 96-deep-well plates. The control culture media was prepared with 1.75 g l−1 of yeast nitrogen base without amino acid or ammonium sulfate, 0.073 g l−1 of histidine, methionine and uracil, and 0.367 g l−1 leucine, for which the knockout strains are auxotroph, and 2% sucrose. For the maple media, sap concentrates and syrups substituted sucrose, with a final dilution factor of 5 and 33, respectively. For each sap or syrup sample, eight wells were inoculated with 0.08 OD units (~500 000 cells). To limit stochastic effects, aliquots from four wells were pooled to obtain two final replicates for each sample. Cultures were sampled after an average of 18 generations. DNA extractions were performed using the Blood and Tissue Kit (Qiagen, Hilden, Germany).
For the initial pool, two separate DNA extractions and two PCR were performed on each DNA sample to create four libraries. The same multiplexing tags were used for the two PCRs, which were sequenced on different chips to estimate technical variation. We used predefined tags (Faircloth and Glenn, 2012) to construct primers for multiplex sequencing with Ion Torrent technology. Oligos are listed in Supplementary Table S2. Each PCR reaction was performed in triplicate and PCR products were pooled to limit stochastic effects. Each PCR reaction had a final volume of 50 μl, which contained 1 × PCR buffer, 300 μM of each dNTP, 1 U of Kapa HiFi DNA polymerase (KAPA Biosystems Inc., Wilmington, MA, USA), 50 ng of genomic DNA and 200 nM of each primer. PCR amplification was conducted in a MasterCycler ProS (Eppendorf, Hamburg, Germany) with the following conditions: 3 min at 95 °C; 10 cycles of 20 s at 98 °C, 15 s at 52 °C and 15 s at 72 °C; followed by 15 cycles of 20 s at 98 °C, 15 s at 75 °C and 15 s at 72 °C, and a final extension time of 15 s at 72 °C.
Pooled PCR products were purified using Axyprep Mag PCR cleanup kit (Corning, Corning, NY, USA). The libraries were quantified using Picogreen (ThermoFisher, Waltham, MA, USA) and quality was assessed using a high-sensitivity chip (Agilent, Santa Clara, CA, USA) on a Bioanalyzer2100 (Agilent). Libraries were loaded on P1 sequencing chips using an IonChef (ThermoFisher) and sequenced (IBIS sequencing platform) on an Ion Proton (ThermoFisher) according to the manufacturer’s instructions. Sequencing data were analyzed with Geneious 6.1.8 software (Biomatters Inc., Newark, NJ, USA). All possible expected synthetic PCR products were concatenated with NNNNN spacers to create a reference on which the barcode reads were mapped using the default settings and excluding reads with multiple best matches. We used the two available versions of barcodes lists (Smith et al., 2009) to determine the best barcode for each strain and remapped the reads to this updated reference (Supplementary Table S3). We obtained an unambiguous assignment for 95% of reads with correlation coefficients of 0.92–0.97 among the initial pool replicates.
There were 4771 strains present in the prepared pool, among which 4606 were detected. The 4278 strains that had a total of more than 100 reads in the initial pool were considered for further analysis. Since the total number of reads obtained per library varied and correlated with the number of detected strains, we conservatively set the undetected strains in the end point at 0.5 reads to estimate their proportion in the library. Fitness was then calculated as in Qian et al. (2012) using the average of pseudogenes deletion strains (YCL074W, YCL075W, YDR134C, YFL056C, YIL170W, YIR044C, YLL017W) as the wild type. Duplicates were averaged resulting in a total of 4 250 ORFs.
Because the chemical composition of maple sap and maple syrup varies throughout the production season, we considered median values across samples to establish thresholds on P-values of Welch tests comparing fitness to the control minimal media. Strains having a median P-value lower than 0.1 and a difference in fitness greater than 0.04 were considered candidates and used for functional category enrichment analysis. Gene Ontology (GO) enrichments were performed with the ClueGO plug-in v2.2.3 (Bindea et al., 2009) in Cytoscape v3.2.1 (Shannon et al., 2003). The lists of sap and syrup candidate genes with significant effects on fitness were tested for enrichment in biological processes against the 4250 ORF recovered in the functional genomic experiment with a right-sided hypergeometric test. P-values were corrected with the Bonferonni stepdown method and other ClueGO parameters were kept to default settings.
Allantoin and allantoate quantification
Allantoin, allantoate and Allantoin-13C2, 15N4 were purchased from Cedarlane (Burlington, ON, Canada). Solvents and acids of LC-MS grade were purchased from EMD Millipore Chemicals (Billerica, MA, USA). Ultrapure water was obtained from a Milli-Q water purification system (Millipore).
Maple products, saps and syrups, previously diluted 3 × with water, were diluted 4 × with 90:10 (acetonitrile:water) containing allantoin-13C2,15N4 as an internal standard (1ppm), vortexed for 1 min and centrifuged at 16 000 g for 5 min at 4 °C. Supernatants were collected, filtered on 0.2 μm nylon filters and placed in a vial prior to injection. A UPLC-MSMS analysis was performed using a Waters Acquity H-Class Ultra-Performance LC system (Waters, Milford, CT, USA), equipped with a quaternary pump system. An Aquity BEH HILIC Column, 130 Å, 1.7 μm, 2.1 mm × 150 mm from Waters set at 40 °C was used. Allantoin and Allantoate were separated with a mobile phase that consisted of 0.5% formic acid (eluent A) and acetonitrile (eluent B). The flow rate was 0.4 ml min−1 and the gradient elution was 70% B; 0–1.5 min, 70–50% B; 1.5–3.0 min, 50–5% B; 3.0–3.1 min, 5–70% B with post time of 3.5 min. The MS analyses were carried out on a Xevo TQD mass spectrometer (Waters) equipped with a Z-spray electrospray interface. The analysis was performed in positive mode and the data were acquired through masses reactions monitoring, allantoin 158.71>61.04 (quantification) and 158.71>116.04 (identification confirmation), allantoate 176.78>61.03 (quantification) and 176.78>116.03 (identification confirmation). The ionization source parameters were capillary voltage 3.1 kV; source temperature 150°C; cone gas flow rate 10 l h−1 and desolvation gas flow rate 1000 l h−1; desolvation temperature 500°C. Nitrogen (99% purity) and argon (99% purity) were used as nebulizing and collision gases, respectively. Data acquisition was carried out with the MassLynx 4.1 software (Waters Corporation, Milford, MA, USA). Quantification was performed based on internal calibration.
DAL deletion strains construction
Four allantoin pathway genes were deleted in a previously isolated S. paradoxus strain with its HO locus replaced with the NATMX antibiotic resistance cassette (Table 1; Charron et al., 2014b). This strain is representative of the SpC wild populations found in the North-East of North America (Leducq et al., 2014, 2016). The deletions of DAL1, DAL2, DAL7 and DUR1, 2 were conducted using homologous recombination cassettes (Rothstein, 1991). The cassettes were amplified using lineage-specific oligonucleotides designed from available genomic data (Leducq et al., 2016) on the pFA-HphNT1 plasmid (Janke et al., 2004). Cassette amplification, transformation by homologous recombination and verification of cassette integration followed Leducq et al. (2012) with heat shock performed at 37 °C instead of 42 °C (primers listed in Supplementary Table S4). Verification of cassette integration was performed on the 5′ region of the gene. One verified transformant was kept for each gene deletion and stocked at −80 °C in 25% glycerol culture medium.
Data availability
Raw sequencing data are available at Bioproject number PRJNA342949 at http://www.ncbi.nlm.nih.gov/bioproject/.
Results
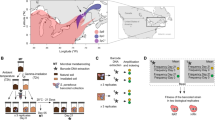
To investigate variation in fitness of natural yeast populations on maple sap, we performed a high-throughput screen to measure growth rate (normalized log2 colony size per hour) and efficiency (normalized log2 colony size) of 72 S. paradoxus strains and one wild S. cerevisiae strain on the selected maple samples (Supplementary Table S5). A principal component analysis of these growth parameters shows that variation in growth phenotypes on maple sap discriminates strains belonging to different S. paradoxus lineages (Figure 1a) with the first and second principal component (PC1 and PC2) positively reflecting efficiency and growth rate, respectively. Indeed, both PC1 and PC2 were significantly higher for the SpB than the SpC lineage (t-test P-value <0.001), indicating that in maple sap this set of SpB strains performed better than the SpC strains tested. A similar difference in efficiency was also observed on the standard laboratory rich undefined medium YPD (t-test P-value <0.001), but the growth rate difference was reversed, with SpC growing significantly faster (t-test P-value=0.01).
Wild strains of Saccharomyces show variation in growth parameters on maple sap. (a) Principal component analysis on correlations of growth rates and efficiency measured for 73 strains on six maple saps. PC1 and PC2 positively reflect growth efficiency and rate in maple sap samples, respectively. Colored symbols correspond to S. paradoxus lineages as defined in Leducq et al. (2016): SpA (green squares), SpB (red circles), SpC (blue triangles), SpC* (purple diamonds), SpBf (orange rectangle) and the empty square represents a wild S. cerevisiae strain. (b) Map showing the relationship between the median PC2 and the geographical origin of the strains. The area defined by the broken line represents the distribution of Acer saccharum Marsh (based on USGS.gov).
While there were no significant correlation between PC1 and geographical origin (P-value >0.14), PC2 correlated with latitude and longitude (Spearman ρ=−0.46 and −0.57, respectively, P-value <0.001; Figure 1b). Pairwise correlations between growth rates and coordinates are significant (Spearman ρ<−0.31, P-value <0.01) for all but the first maple sap sample, while there is no significant correlation between these parameters on YPD (P-value >0.23). Altogether these results indicate that maple sap is an ecologically relevant substrate for S. paradoxus in North America. The growth parameters measured on syrup correlated well with those measured on sap (Supplementary Figure S1), indicating that the transformation process might only subtly alter the chemical components influencing yeast growth.
To identify the ecologically relevant component of maple sap that may underlie this phenotypic variation, we performed a functional genomic screen via liquid competition growth assays of a pool of S. cerevisiae deletion strains. We used as control a minimal medium containing sucrose, and substituted sucrose with maple sap or maple syrup for the experimental media. The fitness of each deletion strain growing in one of the six sap or syrup samples was compared with their respective fitness in the control media. Differences in fitness result from the effect of one or more chemical compounds found only in the sap or syrup. Because maple syrup is a concentrate of maple sap, in most cases, seasonal changes should occur in parallel in maple saps and syrups.
We defined a list of candidate genes based on the median fitness difference with the control in each sample type and the median P-value of a Welch test (Figure 2a and Supplementary Table S6). Half of the 80 candidates were common to sap and syrup, a highly significant overlap (Fisher’s exact test, P-value=8.6e-67). The lists of genes with significant effect on growth were collectively enriched for connected biological processes required for nitrogen assimilation and its regulation (Figure 2b), including the allantoin catabolic pathway. This pathway is under the regulation of the nitrogen catabolic repression, and the 80 genes with significant effects were enriched in members of the nitrogen catabolic repression regulon (A-NCR) as defined by Godard et al. (2007; strongest enrichment for the candidates in sap or syrup Fisher’s exact test, right-sided P-value=4.1e-33). Fitness of strains lacking a gene involved in the allantoin catabolic pathway and its regulation (Figure 3) suggested that being able to use allantoate, and not allantoin, was most critical for growth on maple sap, suggesting that it is the major source of nitrogen in this substrate.
The most relevant genes for growth in maple samples are enriched in biological processes relevant for nitrogen utilization. (a) The lists of genes with significant effects were defined based on the median fitness difference with the control and the median P-value of a Welch test for maple saps and syrups. (b) Functionally grouped network with biological processes as nodes linked based on their κ-score level (⩾ 0.4). Node color represents the term enrichment specificity to sap, syrup or both gene lists and the node size reflects the significance.
The S. cerevisiae deletion strains for genes involved in the allantoin catabolic pathway and its regulation show consistent fitness differences in maple samples relative to the control media. (a) Members of the allantoin catabolic pathway and its known regulators, including genes regulating the nitrogen catabolic repression, found among the candidates are highlighted in bold. Genes are colored according to their overall median fitness difference from the control. (b) The detailed fitness effect of each gene deletion in the maple samples over the flow period connected by dotted lines (DAL pathway) or full lines (Regulators). The YKR033C ORF overlaps the DAL80 gene and was among the candidates.
Quantification of the ureides allantoin and allantoate in maple sap concentrates and syrups confirmed that allantoate is present in maple sap with concentrations that range between 0.6 and 24.1 mg l−1 in unconcentrated sap. Both compounds are present and increasing in concentration over the spring flow period in maple syrup (Figures 4a and b). These quantification results mirrored the general increase in efficiency observed for the wild Saccharomyces strains (Figure 4c), which support our overall findings that these ureides are the main nitrogen sources available in maple sap. To confirm this hypothesis, we constructed dal1 (allantoinase), dal2 (allantoicase), dal7 (malate synthase) and dur1, 2 (urea amidolyase) single knockout in a S. paradoxus wild isolate. We then measured their growth in maple saps and syrups along with the wild type. These genes are part of the allantoin catabolism pathway (Figure 3) and are reported as being required for the use of their upstream metabolites in S. cerevisiae, except for dal7, which has a functional paralog. All strains were able to grow on ammonium sulfate media as expected, but also on sap1 and on syrup17, indicating that nitrogen sources other than ureides were available in these media (Figure 5). There was also weak growth of the dal2 mutant in allantoate media, but not on most saps and syrups, reflecting a possible interaction with other properties of the media, a lower pH for example, since the allantoate media was not buffered.
Quantification of allantoin and allantoate in maple sap concentrates (a) and maple syrups (b) echoes growth efficiency on maple sap and syrup samples over the sampling flow period (c). Box plots represent the distribution of growth efficiency of 73 wild Saccharomyces strains in each sample. Whiskers indicate the 1.5 interquartile range.
Heat map showing the growth efficiency of S. paradoxus knockout strains of the allantoin catabolism pathway compared with the wild type (WT) on maple samples and synthetic media with allantoate (ALA), allantoin (ALN) or ammonium sulfate (AMS) as the nitrogen source. DAL2 and DUR1,2 are required for growth on most maple saps while DAL1 and DAL7 are not. These results indicate that deletion strains unable to utilize allantoate as a nitrogen source are unable to grow on most maple sap samples, underlying the absence of alternate nitrogen sources.
As allantoin and allantoate are the main nitrogen sources in most saps, the ability to use these molecules could be a major fitness determinant in the wild. Indeed, growth of the S. paradoxus (n=72) and S. cerevisiae (n=1) strains on allantoin and allantoate media correlated very well with growth in maple sap, even better than ammonium sulfate media (Figure 6 and Supplementary Table S7). Moreover, the correlation coefficients increased with the flow period, mirroring the increase of ureides, again suggesting that the ability to use these nitrogen sources is a major determinant of fitness in the wild.
Variation in growth rate among strains in maple saps does not correlate with growth rate in rich undefined media (YPD). Minimal media containing sucrose and ammonium sulfate (AMS) show intermediate correlations, which can be increased by using allantoate (ALA) or allantoin (ALN) as the nitrogen source. Dots are colored according to the correlation coefficient.
Given the apparent ecological relevance of allantoin and allantoate, we examined the life-history traits that are reflected in the correlation among the growth components of S. paradoxus SpB and SpC lineages. We observed no significant correlations between growth rate and efficiency for strains of the SpC lineage in maple sap, but all SpB correlations were significant (P-value<0.05; Figure 7). Similar correlations were recovered when the SpB strains where grown on allantoin and allantoate media, while they were only partially recovered on ammonium sulfate media, and weakest on YPD, implying different life-history relationships with allantoin and allantoate for these lineages.
Discussion
Budding yeasts of the genus Saccharomyces are the workhorses of the baking, brewing, wine making and biofuel industries. S. cerevisiae is also the best-characterized model eukaryotic species in genetics, genomics and cell biology. Understanding the evolution and phenotypic diversity of these species has been limited by our humble understanding of their ecology, and a lack of natural context in which to measure fitness components that would be relevant to natural conditions. The only major ecological parameters that have clearly been shown to play a role in shaping the ecological distribution of Saccharomyces sp. so far are temperature and survival to freeze and thaw cycles (Leducq et al., 2014; Sylvester et al., 2015). Here we exploited the resources of the maple syrup industry to measure genetic variation of growth parameters in populations of Saccharomyces isolated from maple trees and other substrates found in North American forests. We found that fitness in maple sap significantly differs among isolates and varies with geography to a larger extent than fitness on artificial laboratory conditions such as rich undefined medium. Our results show that maple sap from A. saccharum or growth medium reconstituted from concentrated and cooked maple sap (from syrup) provide a representative medium for measuring relevant fitness parameters in natural isolates of S. paradoxus.
We identified key genes involved in growth in maple sap and at the same time the most abundant source of nitrogen for yeast growth in this natural substrate, allantoate. In early experiments on A. saccharum, the main sap nitrogen content has been reported to be of organic origin, but could not be identified (Pollard and Sproston, 1954). In the following decade, allantoate has been reported as an important translocatory and metabolic nitrogen currency in other Acer species (Barnes, 1959, 1963). Our results indicate that A. saccharum shares this metabolic trait. Moreover, the ureide content of xylem sap of deciduous trees, such as Acer species, undergoes a seasonal cycle where ureides are stored in roots during winter and account for 70–100% of nitrogen in root bleeding sap in spring (Conn et al., 1990). Our results show that sugar maple saps collected during spring also follow this pattern, with values found for allantoate (0.6–24.1 mg l−1) matching the reported concentration of total nitrogen compounds (10–20 mg l−1; Dumont, 1994a, 1994b; Morselli and Whalen, 1996). Thus, allantoate is the major form of nitrogen compound in sugar maple sap during spring.
The ability to use allantoate may therefore be a major factor determining fitness for microbial populations growing on maple trees. Being such a major determinant of fitness, it may be surprising to find so much variation in growth rate and efficiency among natural strains using these key compounds. However, the yeast isolates used here come from various substrates, including different trees, which may constitute a different selective regime. In oak trees for instance the main nitrogen compound in xylem sap is asparagine and aspartate depending on the species (Barnes, 1963). The importance of using allantoate could also vary through the season. Considering both sample types, we observed a general increase in allantoin and allantoate as the spring progresses, which matches the general increase in efficiency observed for the wild Saccharomyces strains. This trend also matches the previously reported average bacterial and fungal contamination increase over the tapping period (Filteau et al., 2010). A change in the maple-associated communities could also influence nitrogen sources throughout the season. Here for instance we observed that the first sap sample could contain other nitrogen sources (Figure 5), which is most likely because the first flow washed away the dead biofilms present in the tubing systems (Lagacé et al., 2006), which contributed other nitrogen sources. Similarly, the syrup17 sample can be recognized as a ropy or stringy syrup defined as a maple syrup that forms strings and becomes gelatinous (Canadian Food Inspection Agency, 2007). This defect is attributable to the action of the bacterium Enterobacter (Aerobacter) aerogenes, a rare contaminant of maple sap (Filteau, 2011), which polymerizes the sugars in the syrup. The optimal growth condition of this bacterium occurs at temperatures between 24 and 32 °C, and is therefore more likely to cause problems during the last days of the collecting period. Thus microbial activity would convert part of the allantoate in other sources of nitrogen as illustrated by the growth of dal2 and dur1,2 mutants on syrup17 that has been sterilized by boiling, but not on sap17 that has been sterilized by filtration.
Although acknowledged to be a nitrogen source for Saccharomyces yeast, allantoate has seldom been used in studies investigating nitrogen metabolism and regulation on a broad scale (Godard et al., 2007; VanderSluis et al., 2014; Ibstedt et al., 2015). Here we show that the allantoin catabolic pathway genes and its main regulators are important factors for growth in maple sap. Nitrogen availability is a major determinant of the ecology of microbial species associated with trees (Kembel and Mueller, 2014; Kembel et al., 2014). Nitrogen use appears to have been a major determinant in the evolutionary history of budding yeasts, as suggested by the organization of the yeast genomes and from studies on the model S. cerevisiae. Most of the genes required for allantoin utilization form the largest metabolic gene cluster found in the budding yeast genome, which may be a co-adapted gene complex formed by epistatic selection (Wong and Wolfe, 2005). Variation in DAL1 and DAL4 has been reported as the genetic basis for nitrogen source use variation in S. cerevisiae (Ibstedt et al., 2015). Both genes are required for allantoin use, but are not essential for allantoate utilization. Also, the allantoate permease gene has been shown to be strain specific and DAL cluster gene expression to be variable among strains (Treu et al., 2014). Given these observations and the fact that allantoin and allantoate are found in several plants and excretion products of various organisms (de Bruijn, 2015; Atkins, 1982), it is likely that these ureides played a role in shaping the current ecology and evolution of budding yeast. How important this role is in the North American forests will require further investigations at the interface of ecology, genetics and evolution. However, using a recently proposed approach looking at correlation between fitness components over natural isolates (Ibstedt et al., 2015), we already observed differences among the incipient species SpB and SpC in their use of these nitrogen sources. Significant negative correlations between growth parameters in the SpB but not the SpC lineages in maple sap suggest that they may allocate energy differently in the presence of these molecules. While rapidly growing strains of SpB show a decline in achieved biomass compared to slow growing strains, no such strong trade-off exists in SpC, which may indicate different degree of adaptation to this environment. Moreover, we observed a correlation between the growth rate of Saccharomyces in maple sap and the geographic origin of the strains, but also a significant difference in growth rate between lineages, which are known to have different distributions (Leducq et al., 2016). How much genetic or ecological factors such as lineage and substrate adaptation contribute to this pattern is still unclear at this point and will require further targeted sampling efforts.
The main focus of this study was to investigate fitness variation of S. paradoxus populations on a natural substrate, maple sap. However, our results also bear important consequences for the maple syrup industry, which does not control tree physiology or their associated microbes. On the one hand, microbial growth in maple sap is intimately linked to the quality of the maple syrup produced (Filteau et al., 2012). The report that allantoate is the main nitrogen source in maple sap is therefore highly relevant for the understanding of the complex microbial community dynamics over the spring flow period (Filteau et al., 2011). On the other hand, part of the complex maple flavor comes from the Maillard reaction that takes place during the heating process in which compounds containing amino groups such as allantoin and allantoate can react with the reducing sugars present in the sap. An excess of these compounds in the presence of sufficient reducing sugars would lead to strong empyreumatic or burnt flavors. Knowledge of ureide concentration in maple saps could help predict maple syrup quality as producers currently monitor only glucose in their sap, and this variable alone cannot fully explain syrup quality (Lagacé and Charron, 2008). The current results highlight the importance of ureides both as a nitrogen source available for microbial growth in maple sap and as a Maillard reaction reactive. These findings stress the need for development of on-site monitoring tools of nitrogen compounds and bring forward new possibilities in maple sap quality control.
In conclusion, we exposed variation in the adaptation to a specific nitrogen source in closely related lineages of S. paradoxus, demonstrating its ecological relevance and illustrating the usefulness of our functional genomic approach in a natural substrate. Altogether our results bring us closer to a better understanding of yeast ecology and the maple microhabitat.
References
Atkins C . (1982). Ureide metabolism and the significance of ureides in legumes. In Subba Rao NS ed Advances in agricultural microbiology Elsevier: Amsterdam, Netherlands, pp 53–88.
Barnes RL . (1959). Formation of allantoin and allantoic acid from adenine in leaves of Acer saccharinum L. Nature 184: 1944–1944.
Barnes RL . (1963). Organic nitrogen compounds in tree xylem sap. Forest Sci 9: 98–102.
Bell G . (2010). Experimental genomics of fitness in yeast. Proc Biol Sci 277: 1459–1467.
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A et al. (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093.
Boynton PJ, Stelkens R, Kowallik V, Greig D . (2016). Measuring microbial fitness in a field reciprocal transplant experiment. Mol Ecol Resour e-pub ahead of print 22 June 2016 doi:10.1111/1755-0998.12562.
Braconi D, Bernardini G, Santucci A . (2016). Saccharomyces cerevisiae as a model in ecotoxicological studies: a post-genomics perspective. J Proteomics 137: 19–34.
Canadian Food Inspection Agency. (2007) Maple Products Establishment Inspection Manual. Government of Canada: Hamilton, ON, Canada.
Charron G, Leducq JB, Bertin C, Dubé AK, Landry CR . (2014a). Exploring the northern limit of the distribution of Saccharomyces cerevisiae and Saccharomyces paradoxus in North America. FEMS Yeast Res 14: 281–288.
Charron G, Leducq JB, Landry CR . (2014b). Chromosomal variation segregates within incipient species and correlates with reproductive isolation. Mol Ecol 17: 4362–4372.
Clowers KJ, Heilberger J, Piotrowski JS, Will JL, Gasch AP . (2015). Ecological and genetic barriers differentiate natural populations of Saccharomyces cerevisiae. Mol Biol Evol 32: 2317–2327.
Conn PM, Stumpf W, Miflin B, Lea PJ . (1990) Intermediary Nitrogen Metabolism vol. 16. Academic Press: San Diego, CA, USA.
de Bruijn FJ . (2015) Biological Nitrogen Fixation, 2. Volume set. Wiley: Hoboken, NJ, USA.
Diss G, Dubé AK, Boutin J, Gagnon-Arsenault I, Landry CR . (2013). A systematic approach for the genetic dissection of protein complexes in living cells. Cell Rep 3: 2155–2167.
Dumont J. (1994a). Caractéristiques chimiques et nutritives du sirop d’érable. Les acides aminés de la sève et du sirop d’érable Programme de développement des marchés des produits de l’érable Projet 3..
Dumont J. (1994b). L'eau d'érable. Centre de recherche de développement et de transfert technologique en acériculture (ACER) Publication: 1094.
Faircloth BC, Glenn TC . (2012). Not all sequence tags are created equal: designing and validating sequence identification tags robust to indels. PLoS One 7: e42543.
Filteau M, Lagacé L, LaPointe G, Roy D . (2010). Seasonal and regional diversity of maple sap microbiota revealed using community PCR fingerprinting and 16 S rRNA gene clone libraries. Syst Appl Microbiol 33: 165–173.
Filteau M. (2011). Étude du microbiote de la sève d'érable et de son impact sur la qualité du sirop. Ph.D. thesis, Université Laval, Québec.
Filteau M, Lagacé L, LaPointe G, Roy D . (2011). Correlation of maple sap composition with bacterial and fungal communities determined by multiplex automated ribosomal intergenic spacer analysis (MARISA). Food Microbiol 28: 980–989.
Filteau M, Lagacé L, LaPointe G, Roy D . (2012). Maple sap predominant microbial contaminants are correlated with the physicochemical and sensorial properties of maple syrup. Int J Food Microbiol 154: 30–36.
Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391.
Giraldo-Perez P, Goddard MR . (2013). A parasitic selfish gene that affects host promiscuity. Proc Biol Sci 280: 20131875.
Godard P, Urrestarazu A, Vissers S, Kontos K, Bontempi G, van Helden J et al. (2007). Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol 27: 3065–3086.
Goddard MR, Greig D . (2015). Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res 15: fov009.
Hittinger CT . (2013). Saccharomyces diversity and evolution: a budding model genus. Trends Genet 29: 309–317.
Ibstedt S, Stenberg S, Bages S, Gjuvsland AB, Salinas F, Kourtchenko O et al. (2015). Concerted evolution of life stage performances signals recent selection on yeast nitrogen use. Mol Biol Evol 32: 153–161.
Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H et al. (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962.
Kembel SW, Mueller RC . (2014). Plant traits and taxonomy drive host associations in tropical phyllosphere fungal communities. Botany 92: 303–311.
Kembel SW, O'Connor TK, Arnold HK, Hubbell SP, Wright SJ, Green JL . (2014). Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Nat Acad Sci USA 111: 13715–13720.
Kowallik V, Miller E, Greig D . (2015). The interaction of Saccharomyces paradoxus with its natural competitors on oak bark. Mol Ecol 24: 1596–1610.
Kuehne HA, Murphy HA, Francis CA, Sniegowski PD . (2007). Allopatric divergence, secondary contact, and genetic isolation in wild yeast populations. Curr Biol 17: 407–411.
Lagacé L, Jacques M, Mafu AA, Roy D . (2006). Compositions of maple sap microflora and collection system biofilms evaluated by scanning electron microscopy and denaturing gradient gel electrophoresis. Int J Food Microbiol 109: 9–18.
Lagacé L, Charron C . (2008) Evaluating maple sap quality—the glucometer. Maple factsheet no. 231A0508E. Centre Acer: St-Norbert d'Arthabaska, QC, Canada.
Landry CR, Townsend JP, Hartl DL, Cavalieri D . (2006). Ecological and evolutionary genomics of Saccharomyces cerevisiae. Mol Ecol 15: 575–591.
Leducq JB, Charron G, Diss G, Gagnon-Arsenault I, Dubé AK, Landry CR . (2012). Evidence for the robustness of protein complexes to inter-species hybridization. PLoS Genet 8: e1003161.
Leducq JB, Charron G, Samani P, Dubé AK, Sylvester K, James B et al. (2014). Local climatic adaptation in a widespread microorganism. Proc Biol Sci 281: 20132472.
Leducq JB, Nielly-Thibault L, Charron G, Eberlein C, Verta J-P, Samani P et al. (2016). Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nat Microbiol 1: 15003.
Morselli M, Whalen ML . (1996). Maple chemistry and quality. North American maple syrup producers manual, The Ohio State University, Columbus, OH, pp, 162–171..
Naumov GI . (1987). Genetic-basis for classification and identification of the ascomycetous yeasts. Stud Mycol 30: 469–475.
Naumov GI, Naumova ES, Sniegowski PD . (1998). Saccharomyces paradoxus and Saccharomyces cerevisiae are associated with exudates of North American oaks. Can J Microbiol 44: 1045–1050.
Pollard JK, Sproston T . (1954). Nitrogenous constituents of sap exuded from the sapwood of Acer saccharum. Plant Physiol 29: 360–364.
Qian W, Ma D, Xiao C, Wang Z, Zhang J . (2012). The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep 2: 1399–1410.
Rothstein R . (1991). Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol 194: 281–301.
Samani P, Low-Decarie E, McKelvey K, Bell T, Burt A, Koufopanou V et al. (2015). Metabolic variation in natural populations of wild yeast. Ecol Evol 5: 722–732.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504.
Smith AM, Heisler LE, Mellor J, Kaper F, Thompson MJ, Chee M et al. (2009). Quantitative phenotyping via deep barcode sequencing. Genome Res 19: 1836–1842.
Smith AM, Durbic T, Oh J, Urbanus M, Proctor M, Heisler LE et al. (2011). Competitive genomic screens of barcoded yeast libraries. J Vis Exp 54: 2864.
Sylvester K, Wang QM, James B, Mendez R, Hulfachor AB, Hittinger CT . (2015). Temperature and host preferences drive the diversification of Saccharomyces and other yeasts: a survey and the discovery of eight new yeast species. FEMS Yeast Res 15: fov002.
Treu L, Toniolo C, Nadai C, Sardu A, Giacomini A, Corich V et al. (2014). The impact of genomic variability on gene expression in environmental Saccharomyces cerevisiae strains. Environ Microbiol 16: 1378–1397.
VanderSluis B, Hess DC, Pesyna C, Krumholz EW, Syed T, Szappanos B et al. (2014). Broad metabolic sensitivity profiling of a prototrophic yeast deletion collection. Genome Biol 15: R64.
Wong S, Wolfe KH . (2005). Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat Genet 37: 777–782.
Yoshikawa K, Tani S, Baba C, Hashimoto T . (2013). Phenylpropanoid, sapnol A, lignan and neolignan sophorosides, saposides A and B, isolated from Canadian sugar maple sap. Molecules 18: 9641–9649.
Yuan T, Li L, Zhang Y, Seeram NP . (2013). Pasteurized and sterilized maple sap as functional beverages: Chemical composition and antioxidant activities. J Funct Foods 5: 1582–1590.
Acknowledgements
We are grateful to Patrick and Chantal Levesque from L’érablière de la Montagne Verte for providing the maple samples. This work was funded by a NSERC ENGAGE grant to MF and CRL (EGP 477513-14) and a Discovery grant to CRL. MF holds a MITACS accelerate scholarship. CRL holds the Canada Research Chair in Evolutionary Cell and Systems Biology. GC holds a NSERC A G Bell graduate scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Filteau, M., Charron, G. & Landry, C. Identification of the fitness determinants of budding yeast on a natural substrate. ISME J 11, 959–971 (2017). https://doi.org/10.1038/ismej.2016.170
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.170
This article is cited by
-
Mitochondrial-nuclear coadaptation revealed through mtDNA replacements in Saccharomyces cerevisiae
BMC Evolutionary Biology (2020)
-
Competition experiments in a soil microcosm reveal the impact of genetic and biotic factors on natural yeast populations
The ISME Journal (2020)
-
Hybridization is a recurrent evolutionary stimulus in wild yeast speciation
Nature Communications (2019)
-
A systems biology approach to explore the impact of maple tree dormancy release on sap variation and maple syrup quality
Scientific Reports (2018)
-
Evolutionary biology through the lens of budding yeast comparative genomics
Nature Reviews Genetics (2017)