Abstract
Cobalamin (vitamin B12) is a complex metabolite and essential cofactor required by many branches of life, including most eukaryotic phytoplankton. Algae and other cobalamin auxotrophs rely on environmental cobalamin supplied from a relatively small set of cobalamin-producing prokaryotic taxa. Although several Bacteria have been implicated in cobalamin biosynthesis and associated with algal symbiosis, the involvement of Archaea in cobalamin production is poorly understood, especially with respect to the Thaumarchaeota. Based on the detection of cobalamin synthesis genes in available thaumarchaeotal genomes, we hypothesized that Thaumarchaeota, which are ubiquitous and abundant in aquatic environments, have an important role in cobalamin biosynthesis within global aquatic ecosystems. To test this hypothesis, we examined cobalamin synthesis genes across sequenced thaumarchaeotal genomes and 430 metagenomes from a diverse range of marine, freshwater and hypersaline environments. Our analysis demonstrates that all available thaumarchaeotal genomes possess cobalamin synthesis genes, predominantly from the anaerobic pathway, suggesting widespread genetic capacity for cobalamin synthesis. Furthermore, although bacterial cobalamin genes dominated most surface marine metagenomes, thaumarchaeotal cobalamin genes dominated metagenomes from polar marine environments, increased with depth in marine water columns, and displayed seasonality, with increased winter abundance observed in time-series datasets (e.g., L4 surface water in the English Channel). Our results also suggest niche partitioning between thaumarchaeotal and cyanobacterial ribosomal and cobalamin synthesis genes across all metagenomic datasets analyzed. These results provide strong evidence for specific biogeographical distributions of thaumarchaeotal cobalamin genes, expanding our understanding of the global biogeochemical roles played by Thaumarchaeota in aquatic environments.
Similar content being viewed by others
Introduction
Cobalamin (vitamin B12) is an enzyme cofactor that many prokaryotic and eukaryotic species use to catalyze rearrangement-reduction or methyl transfer reactions involved primarily in amino acid synthesis (i.e., cobalamin-dependent methionine synthase), return of carbon to central metabolism via the tricarboxylic acid cycle (i.e., adenosylcobalamin-dependent methylmalonyl-CoA mutase) and synthesis of DNA (ribonucleotide reductase). Other cobalamin-dependent enzymes include ethanolamine ammonia-lyase, methylaspartate ammonia-lyase and methylaspartate mutase (reviewed in Sañudo-Wilhelmy et al. (2014)). Cobalamin is related to heme, chlorophyll and the F420 coenzyme of methanogens, and contains a tetrapyrrole corrin ring system surrounding a central cobalt atom. The fifth coordinated position of Co3+ is occupied by a dimethyl-benzamidazole nucleotide loop and the sixth catalytic upper ligand position is either occupied by a methyl group (i.e., methylcobalamin) or with a deoxyadenosine (i.e., adenosylcobalamin).
Because of the absolute requirement of cobalamin coenzymes by many marine bacterioplankton and most cultured phytoplankton, the supply of B vitamins (including vitamins B1, B7 and B12) have been implicated in controlling bloom distributions and species successions in the ocean water column (Sañudo-Wilhelmy et al., 2006; Bertrand et al., 2007; Gobler et al., 2007). In particular, widespread nutritional auxotrophy results in a complete dependence by many marine organisms on an exogenous source of cobalamin. Auxotrophic phytoplankton are thought to establish symbiotic relationships with bacterial partners to obtain this vitamin (Croft et al., 2005; Grant et al., 2014), providing a source of organic carbon to the prokaryotic symbionts in return. Another strategy is to develop costly alternative pathways (e.g., cobalamin-independent methionine synthase) or to scavenge organic nutrients directly from the environment, which has been suggested for the ubiquitous Pelagibacter ubique of the SAR11 clade (Tripp et al., 2008) and for a recently discovered class of uncultured marine Euryarchaeota (Iverson et al., 2012).
Cobalamin biosynthesis represents one of the most complex and ancient metabolic pathways known (Raux et al., 1999), and this vitamin has been described as ‘nature’s most beautiful cofactor’ (Stubbe, 1994). Owing to its elaborate structure, only a limited complement of Bacteria and Archaea are known to produce cobalamin. The ‘Black Queen Hypothesis’ (i.e., in the card game ‘Hearts’, players avoid holding the queen of spades; Morris et al., 2012) has been invoked to explain why cobalamin producers maintain biosynthesis of cobalamin for potential community benefit, despite the high metabolic cost for those few taxa responsible for its production (Giovannoni, 2012). Based on genome analysis, known dominant marine bacterial taxa implicated in cobalamin production include select members of the Cyanobacteria, Alphaproteobacteria, Gammaproteobacteria and Bacteroidetes (Sañudo-Wilhelmy et al., 2014). These characterized cobalamin producers synthesize cobalamin by either aerobic or anaerobic pathways that involve 20 enzymatic steps from a uroporphyrinogen III precursor, sharing several homologous enzymes, but with oxygen-requiring or oxygen-sensitive steps, depending on the pathway (Moore et al., 2013). The aerobic pathway involves a late-insertion of the cobalt ion into the already contracted tetrapyrrole structure, whereas the early insertion pathway can operate under anoxic conditions and incorporates cobalt prior to ring contraction.
Although genomic analyses have implicated Archaea in cobalamin production (Rodionov et al., 2003), examples have been limited to halophilic Crenarchaeaota of hypersaline ponds and marine methanogens from the Euryarchaeota. To our knowledge, no study has reported a potential cobalamin synthesis role for the Thaumarchaeota. This is surprising because Thaumarchaeota are widespread in aquatic and terrestrial environments (Francis et al., 2005; Leininger et al., 2006; Prosser and Nicol, 2008) and dominate marine water columns (Wuchter et al., 2006). Known Thaumarchaeota possess autotrophic and oligotrophic metabolisms that are fueled by ammonia oxidation, and may be stimulated by organic molecules such as pyruvate (Tourna et al., 2011; Stahl and de la Torre, 2012). Implicating cobalamin synthesis in thaumarchaeotal metabolism has important global biogeochemical and ecological implications, especially for the marine environment, where Thaumarchaeota are estimated to account for ∼20% of all prokaryotic cells (Karner et al., 2001).
Based on our observation of cobalamin synthesis genes within available thaumarchaeotal genomes (Hallam et al., 2006; Spang et al., 2010; Walker et al., 2010; Blainey et al., 2011; Kim et al., 2011; Lebedeva et al., 2013; Lloyd et al., 2013; Luo et al., 2014), combined with the ubiquity and abundance of these organisms, we hypothesized that Thaumarchaeota represent numerically relevant marine cobalamin producers. We also predicted that cobalamin synthesis gene distributions would demonstrate biogeographical patterns, with correlations to measured physical and chemical characteristics. Leveraging available marine metagenomic libraries, we demonstrate depth-specific, latitudinal and temporal gradients of thaumarchaeotal cobalamin synthesis genes. We also observed niche partitioning of phylum-specific cobalamin synthesis, which reinforces that vitamin synthesis is a coordinated and costly keystone ecosystem function provided by individual, rather than collective, community members. Our results implicate Thaumarchaeota among dominant global cobalamin producers and add vitamin B12 production to the roles recognized for these abundant and enigmatic marine microbial community members.
Materials and methods
Genome pathway analysis
The Integrated Microbial Genomes system (Markowitz et al., 2014) was used to detect matches to all enzymes within the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2008) reference pathway map for ‘porphyrin and chlorophyll metabolism’ (map00860), containing both the aerobic and anaerobic cobalamin biosynthesis pathways. The presence of each enzyme was assessed within all available archaeal genomes, and these data were retrieved to generate a presence/absence matrix. The proportion of archaeal genomes possessing each enzyme within the pathway was used to compute an archaeal ‘conservation score’. This score was then mapped onto a pathway heatmap using the KEGG Mapper tool (http://www.genome.jp/kegg/mapper.html).
Metagenome datasets and processing
We retrieved 430 aquatic metagenomes from 28 studies from the Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis (CAMERA) portal on 8 January 2014 (see Supplementary Table S1 for a full list of accession numbers). Included studies spanned all available marine and freshwater metagenomes, and excluded data from engineered environments or metagenomes related to host-associated, virus-, or bacteria-specific studies. Given that polar marine Thaumarchaeota were recently implicated in urea-fueled nitrification (Alonso-Sáez et al., 2012), we also included the specific Arctic metagenome used in this previous study (NCBI SRA accession number ERP001178).
Pipeline for detection and taxonomic classification of cobalamin biosynthesis genes
Metagenome profile Hidden Markov Models (profile-HMMs) were used for genes of the cobalamin biosynthesis pathway that were retrieved from the TIGRfam (Haft et al., 2003) and Pfam (Bateman et al., 2004) databases, as based on the KEGG pathway (map00860). A total of 11 genes (cobA_cysG C-terminal domain (TIGR01469), cobI_cbiL (TIGR01467), cobJ_cbiH (TIGR01466), cobM_cbiF (TIGR01465), cbiT_cobL (TIGR02469), cbiE_cobL (TIGR02467), cbiC_cobH (PF02570.10), cbiA_cobB (TIGR00379), cbiB_cobD (TIGR00380), cbiP_cobQ (TIGR00313), cobS (TIGR00317)) were selected as cobalamin pathway markers for further analysis due to their broad distribution throughout the cobalamin biosynthesis pathway. For each of these 11 genes, the corresponding HMM was used to scan for homologs in all sequence reads of each metagenome, which were initially processed into open reading frames using FragGeneScan (Rho et al., 2010). The program hmmsearch within HMMER version 3.1b1 (hmmer.org) was used with default parameters and an E-value threshold of 1 × 10−6. The use of profile-HMMs based on protein family alignments eliminated bias that would have otherwise been introduced by homology searches based on single sequence queries. Tools and taxonomy indices from the Krona package (Ondov et al., 2011) were used to assign taxonomy to recovered hits based on their top BLAST matches in the NCBI refseq database (version 60) with an E-value threshold of 1 × 10−6.
Per-taxon cobalamin gene contribution
Taxonomic frequency profiles were generated by dividing the number of cobalamin synthesis hits per taxon by the total number of hits for a given gene category. Removing samples with low overall cobalamin synthesis gene frequencies (i.e., n<10) reduced potential statistical bias resulting from low sample sizes. To generate an overall measure of per-taxon cobalamin gene contributions for each sample, we combined data for the 11 cobalamin marker genes as follows. For a given dataset, D, the per-taxon cobalamin (cob/cbi) gene contribution (Dt) was computed as:

Metadata, correlation and cluster analysis
Correlation analyses of numerical metadata from each metagenome compared individual physical and chemical characteristics with per-taxon cobalamin gene contributions (i.e., Dt from above). We analyzed metadata correlations across all individual metagenome datasets and also for pooled sample datasets for larger studies, with many samples characterized by common study-specific metadata. Standard Pearson correlations were used to calculate correlation coefficients (r). Maps were generated with the maps package in R (http://cran.r-project.org/web/packages/maps/index.html), with metagenome samples plotted by their associated latitude and longitude metadata. Hierarchical clustering was performed using the Ward method with Manhattan distances in R.
Phylogenetic marker gene analysis
The full analysis described above was repeated using the Pfam profile-HMMs of three universally conserved phylogenetic marker genes (Ribosomal_S12_S23 (PF00164), Ribosomal_L18e (PF00828), and Ribosomal_S9 (PF00380)) that were selected from a previous publication (Lang et al., 2013).
Results
Thaumarchaeotal cobalamin synthesis
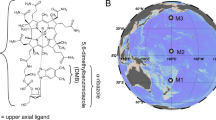
Genomic analysis of the ammonia-oxidizing archaeon Nitrosopumilus maritimus SCM1 and related thaumarchaeotal representatives indicates a genetic capacity for cobalamin biosynthesis. Using N. maritimus SCM1 as a reference, we identified a complete biosynthetic pathway and separate gene clusters encoding distinct stages of cobalamin biosynthesis (Figure 1, Supplementary Figures S1 and S2). Although both the aerobic and anaerobic pathways for cobalamin synthesis share a majority of their enzymes, several steps and associated enzymes are specific to each (Figure 1a). N. maritimus and related Thaumarchaeota (Figures 1b and c) lack genes encoding enzymes of the aerobic pathway (CobG, CobF, CobNST) and possess genes encoding enzymes specific to the anaerobic biosynthesis pathway (CbiX, CbiG, CbiD). This is supported by the presence and absence of marker genes as defined by the KEGG (Supplementary Figures S1 and S2). N. maritimus and other Thaumarchaeota were also found to encode cobalamin-dependent enzymes such as methionine synthase (Nmar_1267), ribonucleotide reductase (Nmar_1627), and methylmalonyl CoA mutase (B12 binding domain, Nmar_0958), as well as probable cobalt transporters (Nmar_0878).
Identification of the cobalamin synthesis pathway and associated gene clusters in available thaumarchaeotal genomes. The pathway in (a) is adapted from the studies by Moore et al. (2013) and Raux et al. (1999). Horizontal arrows indicate homology between aerobic and anaerobic pathway enzymes. Grey enzyme names were not detected in thaumarchaeotal genomes. *CbiJ is also not present in other known archaeal cobalamin producers and therefore should not be considered an essential gene for this pathway in Thaumarchaeota. (b) A six-member cobalamin synthesis gene cluster with conserved synteny across several thaumarchaeotal genomes. The cluster encodes enzymes (highlighted by blue boxes in (a)) in the upper pathway, and includes three enzymes (bolded) specific to the anaerobic pathway. (c) A four-member gene cluster possessing enzymes (highlighted by green boxes in (a)) corresponding to the final steps of cobalamin synthesis. The apparent missing cobD gene in AR1 may be a genome annotation error given that it appears to be in the genome but may be truncated (data not shown).
As further support for cobalamin biosynthetic potential, we identified dual cob/cbi gene clusters conserved across all known ammonia-oxidizing thaumarchaeotal genomes. The two relevant gene clusters are present in the genomes of Candidatus Nitrosoarchaeum limnia, Candidatus Nitrosopumilus salaria, Candidatus Nitrososphaera gargensis, and Candidatus Nitrosopumilus koreensis AR1 (Figures 1b and c), in addition to Cenarchaeum symbiosum and Candidatus Nitrosotenuis uzonensis (Supplementary Figure S2 and data not shown, respectively). With the possible exception of Can. N. gargensis, these gene clusters exhibit highly conserved synteny (Figures 1b and c, Supplementary Figure S2), indicative of selection for coordinated regulation. Furthermore, these gene clusters encode separate stages of cobalamin biosynthesis, with the first stage encoding six enzymes for the early pathway converting uroporphyrinogen III to cobyrinic acid a,c-diamide (Figures 1a and b) and the second stage encoding four enzymes for the later conversion of cobyrinic acid a,c-diamide to vitamin B12 (Figures 1a and c).
Our results also show that many Archaea possess a similar complement of cobalamin biosynthetic genes, which cluster together and co-occur genomically (Supplementary Figures S2 and S3). This, along with the observed sequence and gene order conservation, suggests widespread functional conservation of the cobalamin biosynthetic pathway among the Archaea.
Environmental distributions of cobalamin synthesis genes
We searched for cobalamin biosynthesis genes within 430 metagenomes derived from 28 studies, representing a diverse range of marine, freshwater and hypersaline environments. Eleven cobalamin synthesis genes were selected based on their representation of both the aerobic and anaerobic pathway (Figure 1a), broad distribution throughout the cobalamin pathway and consistency with a core majority of genes used previously to identify cobalamin producers (Sañudo-Wilhelmy et al., 2014).
Our screen identified 187 223 total cobalamin biosynthetic genes across all metagenomes, 95 911 of which matched the 11 marker genes (Figure 2a). Taxonomic assignments classified these 95 911 genes into three main phyla (Figure 2b): Proteobacteria (P), Cyanobacteria (C), and Thaumarchaeota (T). At the species level, Thaumarchaeota dominated among prevalent organisms (Table 1), comprising three of the four most frequent contributors of cob/cbi genes overall. Thaumarchaeota were also significantly overrepresented within the context of cobalamin synthesis (contributing 16.1% of cob/cbi genes) relative to their community abundance (contributing 2.5% of ribosomal marker genes). These results indicate that Thaumarchaeota are abundant global cobalamin producers through the anaerobic biosynthetic pathway, and may be a keystone species for cobalamin production.
Survey of cobalamin synthesis genes across aquatic metagenomes. (a) The full set of cob/cbi genes and subset of eleven representative marker genes (black) used in the screen and their abundance (total number of detected genes). (b) Taxonomic breakdown of all detected cob/cbi genes across 430 aquatic metagenomes. (c) Distributions of thaumarchaeotal, cyanobacterial and proteobacterial cobalamin gene contributions (% of total cob/cbi genes) for 28 different studies. (d) Clustering and heatmap visualization of taxonomic composition of cob/cbi genes for the 430 metagenomes. The color scale reflects the proportion of cob/cbi genes in each sample affiliated with a particular taxonomic group. More information on each sample can be found in Supplementary Table S1. (e) Global distribution of cobalamin synthesis genes. To emphasize Thaumarchaeota-rich metagenomes, data points with higher thaumarchaeotal contributions have been slightly enlarged and overlayed preferentially in cases where there are overlapping data points. The Thaumarchaeota-rich Arctic metagenome described in the text is indicated by an asterisk.
In addition to evaluating all metagenome datasets together, we partitioned studies into their individual metagenome samples (430 in total), each of which was associated with distinct environmental parameters. Environment-specific metagenomes varied widely in taxonomic composition of cobalamin genes, within and between studies (Figure 2c). Hierarchical clustering of cobalamin taxonomic profiles of all metagenomes demonstrated visual ecotype partitioning (Figure 2d), including distinct T, P and C environments, or mixed environments (e.g., P+C or P+T). These recurring ecotypes show broad geographic distribution and non-uniform latitudinal gradients (Figure 2e).
Thaumarchaeota ranged from undetected to contributing as much as ∼70–80% of the cobalamin genes in various samples from the Microbial Initiative in Low Oxygen areas off Concepcion and Oregon (MI_LOCO; http://mi-loco.coas.oregonstate.edu), Western Channel Observatory Microbial Metagenomic Study (http://camera.crbs.ucsd.edu/projects/details.php?id=CAM_PROJ_WesternChannelOMM; Gilbert et al., 2010) and Antarctica metagenome (http://camera.crbs.ucsd.edu/projects/details.php?id=CAM_PROJ_AntarcticaAquatic) studies (Figure 2c, Supplementary Table S1). In addition, Thaumarchaeota were the dominant source of cob/cbi genes in several metagenomes included in this analysis (Supplementary Table S1), including the Arctic metagenome (66%, Figure 3b), deep-ocean bathypelagic habitat metagenome (50%), the station ALOHA (500 m depth) metagenome (67%) and individual sites from the Global Ocean Surveys dataset. Several metagenome studies included few samples (e.g., a single sample for the Arctic metagenome), and may therefore not be representative of their respective environments.
Phylum and metadata correlations. (a) Correlation matrix for all species-metadata correlations with r>0.25. Correlations were measured between numerical metadata from all 430 metagenome samples and the per-taxon proportion of cobalamin genes. Correlations between taxonomic proportions were also measured to identify potential species–species associations. (b) Left: proportion of cobalamin synthesis genes vs depth for three phyla (computed using all 430 metagenome samples). Middle: proportion of cobalamin synthesis genes vs different time points from the 2008–2009 L4 station time series. Right: per-gene taxonomic proportions of cobalamin genes for an Arctic metagenome. Thaumarchaeota (red), Cyanobacteria (green), Proteobacteria (blue), Unclassified (dark grey), Bacteroidetes/Chlorobi (purple), Actinobacteria (black), Firmicutes (orange), and Other (light gray). Bar widths are proportional to the square root of the sample sizes. (c) Phyla-vs-Phyla contributions of cobalamin synthesis genes across all 430 metagenome samples. All correlations exhibit a negative linear relationship, except for Thaumarchaeota-vs-Cyanobacteria, which exhibits an either-or relationship, suggestive of competitive or niche exclusion. (d) Correlation analysis for the MI_LOCO metagenome. The strongest correlating variables with thaumarchaeotal cob/cbi gene contribution are shown on the left, and plots for the top two positively (nitrate concentration and sample depth) and negatively correlated (ammonium and bacterial cell concentration) variables are shown on the right.
Thaumarchaeotal cobalamin genes correlate with environmental variables
The observed variation (Figure 2c) and clustering (Figures 2d and e) of distinct and recurring cobalamin ecotypes (Figures 2d and e) indicate that specific environmental variables drive the dynamics of cobalamin production. We performed correlation analyses across and within all metagenome datasets, and identified specific factors, including depth, nutrient levels, temperature and seasonality, that correlate with the taxonomic composition of cobalamin synthesis genes (Figure 3). The proportion of thaumarchaeotal cobalamin genes correlated most strongly (r=0.55) with sample depth (Figures 3a and b), consistent with observations of thaumarchaeotal dominance in deep water column metagenome studies (Figure 3 and Supplementary Figure S4). The correlation with depth was stronger across 63 samples from the MI_LOCO study (r=0.73, Figure 3d), which is likely attributed to higher within-study methodological consistency.
Further analysis of the MI_LOCO dataset revealed additional correlating variables that are consistent with known aspects of thaumarchaeotal ecology. Levels of thaumarchaeotal cobalamin synthesis genes exhibited strong positive correlations with numerous factors (Figure 3d), including nitrate (r=0.75), sample depth (r=0.73), phosphate (r=0.61) and dissolved organic nitrogen (r=0.60). The strongest negative correlations with thaumarchaeotal cob/cbi genes included bacterial cell number (r=−0.53) and ammonium concentration (r=−0.40). Low ammonium mixing ratios are consistent with a role for marine Thaumarchaeota as high affinity ammonia oxidizers (Martens-Habbena et al., 2009; Horak et al., 2013; Nakagawa and Stahl, 2013).
Taxonomic cobalamin gene contributions also varied temporally. This was most apparent in a single metagenomic study from the Western English Channel time-series experiments sampled from station L4 (Figure 3b). The average proportion of thaumarchaeotal cobalamin synthesis genes was low (∼3%) for the August and April time points, but increased up to 73% in January, suggesting that Thaumarchaeota are dominant contributors to cobalamin production in cold or winter-associated marine waters.
Additional phyla-vs-phyla correlations revealed a strong anticorrelation between thaumarchaeotal and cyanobacterial cobalamin synthesis genes (Figure 3a and see either-or effect in Figure 3c). This was evident in additional analyses that revealed no apparent T+C-rich ecotypes (Figure 2d). Similar patterns were observed for three phylogenetic marker genes (Supplementary Figure S5), suggesting a broader niche partitioning or exclusion between these two major marine clades. Unlike Thaumarchaeota, cyanobacterial cobalamin synthesis gene abundances correlated positively with temperature (Figure 3a), consistent with warm water cyanobacterial blooms, and negatively with depth (Figure 3a). We also identified a positive correlation between cobalamin gene proportions of Thaumarchaeota and Nitrospinae, a nitrite-oxidizing bacterial phylum. This likely reflects a functional association between these two groups, linked by nitrification.
Discussion
Available genomes and metagenomic studies provide strong support that Thaumarchaeota are globally important vitamin B12 producers. Although cobalamin synthesis is distributed throughout both Archaea and Bacteria, only a restricted subset of microorganisms encodes this genetic capacity. Bacterial cobalamin synthesis is associated with select members of Cyanobacteria, Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes (Sañudo-Wilhelmy et al., 2014), which are consistent with the dominant phyla observed in the metagenomic datasets analyzed here (Figure 2b). Within the Archaea, only methanogenic Euryarchaeota and extremophilic crenarchaeal representatives were recognized as cobalamin producers prior to this study. For example, methanogens are known to produce and require cobalamin for acetoclastic and hydrogenotrophic methanogenesis (reviewed in DiMarco et al. (1990)). In addition, cobamides are produced and required by Halobacterium (Woodson et al., 2003) and known sulfur-metabolizing Crenarchaeaota (Kräutler et al., 1988). Given that Thaumarchaeaota are estimated to represent the most abundant Archaea on the planet, and are among the most abundant prokaryotes in the ocean (Karner et al., 2001), the capacity to synthesize vitamin cofactors within this phylum implies global biogeochemical, metabolic and ecological significance.
Genetic capacity for cobalamin synthesis is very tightly conserved. For example, several genes from the multi-step cobalamin synthesis pathway are considered reliable indicators for the entire pathway. If a genome encodes cbiA/cobB, cbiC/cobH or cobT, then the entire cobalamin synthesis pathway is likely encoded and expressed in that microorganism (Bertrand et al., 2011). Indeed, all available thaumarchaeotal genomes encode this capacity, and demonstrate conserved clusters of cobalamin synthesis genes (Figure 1 and Supplementary Figures S2 and S3). Providing physiological support for cobalamin synthesis by the Thaumarchaeota, existing pure cultures of ammonia-oxidizing archaea (including, N. maritimus SCM1 and Nitrosospaera viennensis) are grown in medium prepared without vitamin supplementation (Martens-Habbena and Stahl, 2011; Tourna et al., 2011). Given that the gene for the low efficiency B12-independent methionine synthesis pathway (metE) is absent in the genome of N. maritimus SCM1, growth in the absence of vitamin supplementation provides strong supporting evidence for thaumarchaeotal cobalamin synthesis. Direct measurements of gene expression and cobalamin production by pure cultures of N. maritimus SCM1 and additional Thaumarchaeota would confirm thaumarchaeotal cobalamin synthesis and provide additional information on the structure, regulation and fate of this vitamin when produced by marine Thaumarchaeota. Although all known ammonia-oxidizing archaea possess the genetic capacity for cobalamin synthesis, the absence of homologous genes in Caldiarchaeaum subterranean (Supplementary Figure S2), a deep branching archaeon related to the Thaumarchaeota (Nunoura et al., 2011), suggests that cobalamin synthesis is not universally distributed among related Archaea.
Cobalamin gene analysis of available aquatic metagenomic datasets provides compelling support for niche specificity and biogeochemical controls on cobalamin production. Taxonomic affiliations indicate that although proteobacterial cobalamin synthesis genes can co-occur with either Cyanobacteria or Thaumarchaeota, these two non-proteobacterial cobalamin producers display strong mutual exclusion (Figures 2 and 3). This trend was also observed when repeating the analysis using ribosomal phylogenetic markers (Supplementary Figure S5), suggesting broad niche partitioning between these major marine groups. In addition to light sensitivity of Thaumarchaeota (described below), a possibility is that high cyanobacterial photosynthetic production of molecular oxygen interferes with the anaerobic thaumarchaeotal cobalamin biosynthetic pathway, which possesses an oxygen-sensitive intermediate (cobalt-precorrin-5B; Moore et al., 2013). In addition to niche partitioning of thaumarchaeotal and cyanobacterial cobalamin synthesis genes, metadata correlations revealed a positive correlation with marine nitrite-oxidizing Nitrospinae, which is consistent with the an ammonia-oxidizing role for most marine Thaumarchaeota. Nitrospina-like bacteria have also been implicated as the primary nitrite oxidizers in oxygen minimum upwelling zones (Levipan et al., 2014), which is consistent with high thaumarchaeotal abundance in these marine environments (Supplementary Figure S4).
The metagenomic data are also consistent with observed geographical and temporal patterns of thaumarchaeotal abundances. In particular, the urea-fueled polar and halocline abundances of Thaumarchaeota (Alonso-Sáez et al., 2012) were reflected here in polar cobalamin synthesis gene abundances in the same datasets. Winter peak abundances of Thaumarchaeota (Wuchter et al., 2006; Herfort et al., 2007; Galand et al., 2010; Pitcher et al., 2011) are consistent with our observation of a winter thaumarchaeotal cobalamin gene maximum at station L4. In addition, previous research has reported high thaumarchaeotal abundance in oxygen minimum zones (Pitcher et al., 2011; Peng et al., 2013), which reflects our observation of abundant thaumarchaeotal cobalamin genes in the MI_LOCO metagenomes. The longstanding observation of abundant Thaumarchaeota (then ‘mesophilic Crenarchaeota’) in deep marine water columns (Karner et al., 2001; Sintes et al., 2013) is also supported strongly by the metagenomic data survey conducted in this study. Importantly, polar environments, aphotic water columns, oxygen minimum zones, and winter time points share low light conditions, which would be a likely explanation for the observed anticorrelation between thaumarchaeotal and cyanobacterial cobalamin synthesis and ribosomal genes. The biogeographical cobalamin synthesis gene distributions observed in this study are consistent with observations of light-dependent decreases in marine thaumarchaeotal abundance below sea ice (Alonso-Sáez et al., 2012) and known photoinhibition of thaumarchaeotal ammonia oxidation (Merbt et al., 2012). Other factors proposed to explain aquatic thaumarchaeotal distributions include nutrient availability, temperature, dissolved oxygen and organic substrates (Peng et al., 2013).
Observations of vitamin B12 maxima in ocean waters below the marine photic zone are consistent with an important role for Thaumarchaeota in marine cobalamin production, given their high proportional abundance with depth. Previous observations of increased cobalamin concentrations in ocean mesopelagic zones (Sañudo-Wilhelmy et al., 2012) have led to suggestions that vitamin-producing microbial plankton below the photic zone are important contributors to marine biogeochemistry, in addition to essential suppliers of growth factors for phytoplankton and microbial communities in the photic zone (Sañudo-Wilhelmy et al., 2014). Although ocean upwelling zones might be considered a mechanism for supplying cobalamin to surface waters, vitamin B12 concentrations did not correlate with upwelling index off the coast of Baja California, Mexico (Panzeca et al., 2009). With respect to high latitude environments, cobalt supplementation of North Atlantic marine samples led to increased vitamin B12 production (Panzeca et al., 2008). Our metagenomic analysis of high latitude samples implies that Thaumarchaeota were responsible for vitamin B12 production in this experiment, and implies that thaumarchaeotal productivity may be limited by cobalt concentrations. Importantly, vitamin B12 was shown to influence phytoplankton growth and community composition in samples taken from the subarctic Gulf of Alaska (Koch et al., 2011), which is consistent with controls on phytoplankton growth by vitamin B12 in freshwater environments as well (Daisley, 1969; Cavari and Grossowicz, 1977). Together, these results provide early evidence that cobalamin production by Thaumarchaeota may directly influence phytoplanktonic communities in polar surface waters, freshwater and possibly in marine surface waters via mixing with ocean water column vitamin pools.
Conclusion
Thaumarchaeotal vitamin production in most of the global ocean would have enormous potential impacts on microbial communities and food web dynamics. Here we provide evidence that Thaumarchaeota are among the most abundant global cobalamin vitamin producers. All known ammonia-oxidizing members of the Thaumarchaeota possess the genetic capacity to produce cobalamin, and data from multiple metagenomic studies reveal niche partitioning of this keystone community function. Thaumarchaeotal cobalamin synthesis genes demonstrate spatial and temporal biogeographical distributions. Combining pure culture experimental work and field studies will help assess the proportion of marine cobalamin pools derived from archaeal synthesis, identify the role of archaeal cobalamin in marine biogeochemistry, and characterize links between global thaumarchaeotal cobalamin production and microbial community composition and succession.
References
Alonso-Sáez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL et al. (2012). Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci USA 109: 17989–17994.
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S et al. (2004). The Pfam protein families database. Nucleic Acids Res 32: D138–D141.
Bertrand EM, Saito MA, Rose JM, Riesselman CR, Lohan MC, Noble AE et al. (2007). Vitamin B12 and iron colimitation of phytoplankton growth in the Ross Sea. Limnol Oceanogr 52: 1079–1093.
Bertrand EM, Saito MA, Jeon YJ, Neilan BA . (2011). Vitamin B12 biosynthesis gene diversity in the Ross Sea: the identification of a new group of putative polar B12 biosynthesizers. Environ Microbiol 13: 1285–1298.
Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR . (2011). Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6: e16626.
Cavari B, Grossowicz N . (1977). Seasonal distribution of vitamin B12 in Lake Kinneret. Appl Environ Microbiol 34: 120–124.
Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG . (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93.
Daisley K . (1969). Monthly survey of vitamin B12 concentrations in some waters of the English Lake District. Limnol Oceanogr 14: 224–228.
DiMarco AA, Bobik TA, Wolfe RS . (1990). Unusual coenzymes of methanogenesis. Ann Rev Biochem 59: 355–394.
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB . (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102: 14683–14688.
Galand PE, Gutiérrez-Provecho C, Massana R, Gasol JM, Casamayor EO . (2010). Inter-annual recurrence of archaeal assemblages in the coastal NW Mediterranean Sea (Blanes Bay Microbial Observatory). Limnol Oceanogr 55: 2117–2125.
Gilbert JA, Meyer F, Schriml L, Joint I, Mühling M, Field D . (2010). Metagenomes and metatranscriptomes from the L4 long-term coastal monitoring station in the Western English Channel. Stand Genomic Sci 3: 183–193.
Giovannoni SJ . (2012). Vitamins in the sea. Proc Natl Acad Sci USA 109: 13888–13889.
Gobler CJ, Norman C, Panzeca C, Taylor GT, Sañudo-Wilhelmy SA . (2007). Effect of B-vitamins (B1, B12) and inorganic nutrients on algal bloom dynamics in a coastal ecosystem. Aquat Microb Ecol 49: 181–194.
Grant MA, Kazamia E, Cicuta P, Smith AG . (2014). Direct exchange of vitamin B12 is demonstrated by modelling the growth dynamics of algal–bacterial cocultures. ISME J 8: 1418–1427.
Haft DH, Selengut JD, White O . (2003). The TIGRFAMs database of protein families. Nucleic Acids Res 31: 371–373.
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM et al. (2006). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4: e95.
Herfort L, Schouten S, Abbas B, Veldhuis MJ, Coolen MJ, Wuchter C et al. (2007). Variations in spatial and temporal distribution of Archaea in the North Sea in relation to environmental variables. FEMS Microbiol Ecol 62: 242–257.
Horak RE, Qin W, Schauer AJ, Armbrust EV, Ingalls AE, Moffett JW et al. (2013). Ammonia oxidation kinetics and temperature sensitivity of a natural marine community dominated by Archaea. ISME J 2013: 2023–2033.
Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV . (2012). Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science 335: 587–590.
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484.
Karner MB, DeLong EF, Karl DM . (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510.
Kim BK, Jung MY, Yu DS, Park SJ, Oh TK, Rhee SK et al. (2011). Genome sequence of an ammonia-oxidizing soil archaeon, "Candidatus Nitrosoarchaeum koreensis" MY1. J Bacteriol 193: 5539.
Koch F, Alejandra Marcoval M, Panzeca C, Bruland KW, Sañudo-Wilhelmy SA, Gobler CJ . (2011). The effect of vitamin B12 on phytoplankton growth and community structure in the Gulf of Alaska. Limnol Oceanogr 56: 1023–1034.
Kräutler B, Kohler HPE, Stupperich E . (1988). 5′-Methylbenzimidazolyl-cobamides are the corrinoids from some sulfate-reducing and sulfur-metabolizing bacteria. Eur J Biochem 176: 461–469.
Lang JM, Darling AE, Eisen JA . (2013). Phylogeny of bacterial and archaeal genomes using conserved genes: supertrees and supermatrices. PLoS One 8: e62510.
Lebedeva EV, Hatzenpichler R, Pelletier E, Schuster N, Hauzmayer S, Bulaev A et al. (2013). Enrichment and genome sequence of the group I.1a ammonia-oxidizing archaeon “Ca. Nitrosotenuis uzonensis” representing a clade globally distributed in thermal habitats. PLoS One 8: e80835.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol G et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809.
Levipan HA, Molina V, Fernandez C . (2014). Nitro spina-like bacteria are the main drivers of nitrite oxidation in the seasonal upwelling area of the Eastern South Pacific (Central Chile ∼36°S). Environ Microbiol Rep doi:10.1111/1758-2229.12158.
Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD et al. (2013). Predominant archaea in marine sediments degrade detrital proteins. Nature 496: 215–218.
Luo H, Tolar BB, Swan BK, Zhang CL, Stepanauskas R, Moran MA et al. (2014). Single-cell genomics shedding light on marine Thaumarchaeota diversification. ISME J 8: 732–736.
Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M et al. (2014). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42: D560–D567.
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA . (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461: 976–979.
Martens-Habbena W, Stahl DA . (2011). Nitrogen metabolism and kinetics of ammonia-oxidizing archaea. Methods Enzymol 496: 465–487.
Merbt SN, Stahl DA, Casamayor EO, Martí E, Nicol GW, Prosser JI . (2012). Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett 327: 41–46.
Moore SJ, Lawrence AD, Biedendieck R, Deery E, Frank S, Howard MJ et al. (2013). Elucidation of the anaerobic pathway for the corrin component of cobalamin (vitamin B12). Proc Natl Acad Sci USA 110: 14906–14911.
Morris JJ, Lenski RE, Zinser ER . (2012). The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3: e00036–00012.
Nakagawa T, Stahl DA . (2013). Transcriptional response of the archaeal ammonia oxidizer Nitrosopumilus maritimus to low and environmentally relevant ammonia concentrations. Appl Environ Microbiol 79: 6911–6916.
Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H et al. (2011). Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res 39: 3204–3223.
Ondov BD, Bergman NH, Phillippy AM . (2011). Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12: 385.
Panzeca C, Beck AJ, Leblanc K, Taylor GT, Hutchins DA, Sañudo-Wilhelmy SA . (2008). Potential cobalt limitation of vitamin B12 synthesis in the North Atlantic Ocean. Glob Biogeochem Cycles 22: GB2029.
Panzeca C, Beck AJ, Tovar-Sanchez A, Segovia-Zavala J, Taylor GT, Gobler CJ et al. (2009). Distributions of dissolved vitamin B12 and Co in coastal and open-ocean environments. Estuar Coast Shelf Sci 85: 223–230.
Peng X, Jayakumar A, Ward BB . (2013). Community composition of ammonia-oxidizing archaea from surface and anoxic depths of oceanic oxygen minimum zones. Front Microbiol 4: 177.
Pitcher A, Villanueva L, Hopmans EC, Schouten S, Reichart G-J, Damsté JSS . (2011). Niche segregation of ammonia-oxidizing archaea and anammox bacteria in the Arabian Sea oxygen minimum zone. ISME J 5: 1896–1904.
Prosser JI, Nicol GW . (2008). Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10: 2931–2941.
Raux E, Schubert HL, Roper JM, Wilson KS, Warren MJ . (1999). Vitamin B12: insights into biosynthesis's mount improbable. Bioorg Chem 27: 100–118.
Rho M, Tang H, Ye Y . (2010). FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res 38: e191–e191.
Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS . (2003). Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem 278: 41148–41159.
Sañudo-Wilhelmy SA, Cutter LS, Durazo R, Smail EA, Gómez-Consarnau L, Webb EA et al. (2012). Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci USA 109: 14041–14045.
Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA . (2014). The role of B vitamins in marine biogeochemistry. Ann Rev Mar Sci 6: 339–367.
Sañudo-Wilhelmy S, Gobler C, Okbamichael M, Taylor G . (2006). Regulation of phytoplankton dynamics by vitamin B12 . Geophys Res Lett 33: L04604.
Sintes E, Bergauer K, De Corte D, Yokokawa T, Herndl GJ . (2013). Archaeal amoA gene diversity points to distinct biogeography of ammonia-oxidizing Crenarchaeota in the ocean. Environ Microbiol 15: 1647–1658.
Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E et al. (2010). Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol 18: 331–340.
Stahl DA, de la Torre JR . (2012). Physiology and diversity of ammonia-oxidizing archaea. Ann Rev Microbiol 66: 83–101.
Stubbe J . (1994). Binding site revealed of nature's most beautiful cofactor. Science 266: 1663–1664.
Tourna M, Stieglmeiera M, Spanga A, Könneke M, Schintlmeisterc A, Uricha T et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA 108: 8420–8425.
Tripp HJ, Kitner JB, Schwalbach MS, Dacey JW, Wilhelm LJ, Giovannoni SJ . (2008). SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452: 741–744.
Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ et al. (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA 107: 8818–8823.
Woodson JD, Peck RF, Krebs MP, Escalante-Semerena JC . (2003). The cobY gene of the archaeon Halobacterium sp. strain NRC-1 is required for de novo cobamide synthesis. J Bacteriol 185: 311–316.
Wuchter C, Abbas B, Coolen MJ, Herfort L, van Bleijswijk J, Timmers P et al. (2006). Archaeal nitrification in the ocean. Proc Natl Acad Sci USA 103: 12317–12322.
Acknowledgements
We thank Mirko Vucicevich for technical assistance. This research was supported by the National Science and Engineering Research Council of Canada (NSERC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
This work is licensed under a Creative Commons Attribution 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/
About this article
Cite this article
Doxey, A., Kurtz, D., Lynch, M. et al. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J 9, 461–471 (2015). https://doi.org/10.1038/ismej.2014.142
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.142
This article is cited by
-
Vitamin B12 is not shared by all marine prototrophic bacteria with their environment
The ISME Journal (2023)
-
Phylotype resolved spatial variation and association patterns of planktonic Thaumarchaeota in eastern Chinese marginal seas
Marine Life Science & Technology (2023)
-
Inactivation of the MSTN gene expression changes the composition and function of the gut microbiome in sheep
BMC Microbiology (2022)
-
Availability of vitamin B12 and its lower ligand intermediate α-ribazole impact prokaryotic and protist communities in oceanic systems
The ISME Journal (2022)
-
Viral community analysis in a marine oxygen minimum zone indicates increased potential for viral manipulation of microbial physiological state
The ISME Journal (2022)