Abstract
Nitrogen fixing and denitrifying bacteria, respectively, control bulk inputs and outputs of nitrogen in soils, thereby mediating nitrogen-based greenhouse gas emissions in an ecosystem. Molecular techniques were used to evaluate the relative abundances of nitrogen fixing, denitrifying and two numerically dominant ribotypes (based on the ⩾97% sequence similarity at the 16S rRNA gene) of bacteria in plots representing 10 agricultural and other land-use practices at the Kellogg biological station long-term ecological research site. Quantification of nitrogen-related functional genes (nitrite reductase, nirS; nitrous oxide reductase, nosZ; and nitrogenase, nifH) as well as two dominant 16S ribotypes (belonging to the phyla Acidobacteria, Thermomicrobia) allowed us to evaluate the hypothesis that microbial community differences are linked to greenhouse gas emissions under different land management practices. Our results suggest that the successional stages of the ecosystem are strongly linked to bacterial functional group abundance, and that the legacy of agricultural practices can be sustained over decades. We also link greenhouse gas emissions with specific compositional responses in the soil bacterial community and assess the use of denitrifying gene abundances as proxies for determining nitrous oxide emissions from soils.
Similar content being viewed by others
Introduction
The impact of agricultural practices on the environment has been studied extensively, leading to changes in land management policy worldwide (Tilman et al., 2002). Yet, surprisingly little is known about the interactions between agroecosystem management practices and the soil microbial community, which has a key role in nutrient transformation and chemical cycling (Staley and Reysenbach, 2002). The Kellogg biological station long-term ecological research (KBS-LTER) site has hosted numerous microbiological studies (Bruns et al., 1998, 1999; Broughton and Gross, 2000; Phillips et al., 2000a, 2000b; Buckley and Schmidt, 2001; Blackwood and Paul, 2003), but few studies have focused on quantitative analysis of bacterial community composition in relation to nitrogen turnover rates, specifically those related to greenhouse gas emissions. In addition, comparative quantitative analysis of specific functional or phylogenetic groups within the soil community is still limited. To date, genes encoding enzymes involved in nitrogen cycling have been targets of choice for studies focusing on functional groups (that is, guilds) of bacteria (Leininger et al., 2006; Henry et al., 2008). This focus is well founded as nitrogen is essential for plant growth, along with phosphate, carbon, hydrogen and oxygen. However, simultaneous comparison of the abundance of multiple N-cycle-related genes across multiple treatments and land-use types has not yet been conducted, especially when framed around an ecosystem-level process, such as, greenhouse gas emissions from soils.
The United States Department of Agriculture tracks emissions of multiple greenhouse gases related to agricultural activities and ranks them based on their global warming potential (GWP). Of all the sources of GWP in cropping systems, including CO2 and CH4, none are more poorly quantified than N2O production (Robertson and Grace, 2004). This represents a tremendous knowledge gap regarding the role of N2O in global warming, especially considering the fact that its GWP is 296-fold greater than that of CO2 and it is frequently the major source of GWP in agricultural systems (Robertson et al., 2000; Robertson and Grace, 2004; EIA, 2008). Poor quantification of N2O is to a large extent linked to the challenges of measuring N2O fluxes in the field, requiring numerous measurements with inherently high variability. This constraint has limited data collection, and, until refined, represents a rudimentary measurement of a globally important activity. The ability to construct more informative models, predictions and mitigation strategies related to greenhouse gas emissions depends on the development of new analytical approaches that are more efficient and accurate than currently available ones. As microbial populations control natural production and consumption of nitrous oxide, their abundance and activities represent a potential method for predicting gas emissions.
Using real-time quantitative PCR (qPCR), we examined the abundance of key soil microbial guilds and taxa, including nitrogen-fixing bacteria (through the nifH gene) and denitrifying bacteria (through the nirS and nosZ genes). Although nitrification represents a key step in the conversion of ammonia nitrogen into its gaseous forms (NO2 and NO3), this process is less relevant to N2O emissions. Further, nitrifier numbers are typically low in soils and are challenging to quantify using direct qPCR as it is applied to all other genes analyzed making nitrification measures beyond the scope of this study. Quantification of two numerically predominant operational taxonomic units (OTUs) belonging to the phyla Acidobacteria and Thermomicrobia obtained at KBS (Morales and Holben, 2009) was also performed for comparison with data obtained using function-based primers. These analyses were performed across 10 different treatments based on land-use types at the KBS-LTER. Analyses in other soils using 16S rRNA gene-based PCR denaturing gradient gel electrophoresis have shown that soil type may be the strongest selector of soil microbial community structure (Wakelin et al., 2008), but that study focused on large-scale rearrangements in community composition measured using a broad-scale technique. However, changes in functional or phylogenetic group abundance may go undetected when using such 16S rRNA gene-based approaches, as poor resolution between taxa due to gene conservation, nonspecific primers and other factors (for example, see Morales and Holben, 2009) can lead to mistaken conclusions about functional group abundance or population dynamics. To examine the extent to which this occurs in this system, the current study employed qPCR targets directly related to functional traits, as well as two 16S rRNA gene-based ribotypes, to compare the patterns observed based on the abundance of genes encoding key enzymatic activities with those observed using ‘OTU-based primers’.
Four general hypotheses were tested. The first stated that 16S-based taxon abundance estimates would be higher than those observed for the functional genes as the former can detect multiple phylogenetic subgroups that might each harbor multiple different bacterial functional groups (for example, denitrifiers and nitrogen fixers can both be found within the same genus). Second, we hypothesized that, due to the high number of leguminous and other nitrogen-fixing symbiotic plants in certain land-use treatments at the KBS-LTER site, nitrogenase reductase gene (nifH) numbers would be consistently higher in all treatments involving leguminous cover crops (that is, treatments 1–4 (T1–4), soybean; and T6, alfalfa). The third hypothesis predicted that abundance of denitrification bacterial genes (nosZ and nirS) would be relatively uniform across all treatments, given the widespread distribution of this metabolic activity across the breadth of bacterial phylogenetic groups. Finally, as the balance between input and output of nitrogen gas in soils is respectively controlled by nitrogen fixers and denitrifiers, we hypothesized that differences in bacterial gene abundances of these key nitrogen cycling genes between annual, perennial and successional sites would correspond to those observed for greenhouse gas emission rates for these sites.
Materials and methods
Study site and sample collection
Samples were collected from the KBS-LTER Row-Crop Agriculture site in mid-Michigan (for an overview of that project see http://lter.kbs.msu.edu/). Our study examined the bacterial community in the replicate plots of Treatment 1–8 (T 1–8) of the main experimental site, as well as two additional successional and forest sites (Table 1). Four of the eight main site treatments are annual crop rotations (T1–4), two are perennial (T5, poplars; and T6, alfalfa), and two are successional systems under native vegetation (T7, former agricultural site left fallow following spring plowing in 1989; and T8, never plowed or cultivated). The two forested sites comprised of a set of 40 to 60-year-old successional sites (former agricultural fields, SFR (successional forest) and DFR (a deciduous forest site that has never been cut)). All soils in these treatments are classified as fine loamy, mixed and mesic Typic Hapludalfs. Sampling was carried out on 2 May 2007 by collecting five randomly positioned, 0–20 cm soil cores using a soil probe from each of the treatment replicates as is standard at the LTER site (http://lter.kbs.msu.edu/protocols/112). Each set of five samples was sieved through 2 mm mesh and then mixed thoroughly in equal proportions, providing a single composite sample for each replicate treatment plot (that is, there were either three or six replicated plots per treatment, and one composited sample was developed and examined for each replicate plot). All soil samples were stored in Whirl-Pak bags on dry ice or at −70 °C immediately after sieving and mixing until processed for bacterial community DNA extraction. All samples were processed within 20 days of sampling.
DNA extraction
Total community DNA was extracted from 0.25 g of each soil sample, in triplicate, using the MoBio PowerSoil DNA Isolation Kit (MoBio, Solana Beach, CA, USA) according to the manufacturers instructions and using sterile MilliQ water in the final elution step. All DNA samples were stored at −20 °C until used in downstream analyses.
Real-time qPCR assays
Real-time qPCR was performed using an iCycler iQ thermocycler (Bio-Rad, Hercules, CA, USA) with an ABsolute QPCR SYBR green mix (AbGene, Epsom, UK) using primers and conditions previously described (nitrogenase reductase (nifH gene) (Rösch and Bothe, 2005; Yergeau et al., 2007); nitrite reductase (nirS gene) (Throback et al., 2004; Yergeau et al., 2007); nitrous oxide reductase (nosZ) (Henry et al., 2006); Thermomicrobia group 4 (OTU-specific based on ⩾97% sequence similarity at the 16S rRNA gene) (Morales and Holben, 2009); Acidobacteria group 6 (OTU-specific based on ⩾97% sequence similarity at the 16S rRNA gene) (Morales and Holben, 2009)) and are summarized in Appendix Table S1. Although no function- or 16S-based primer sets are necessarily comprehensive across the spectrum of microbial diversity, they have been widely used to good effect in comparative studies (for example, between treatments), as is the case in the current study. Known template standards were made from cloned PCR products amplified from whole-genome extracts of pure bacterial isolates (see Appendix Table S1), and each standard was sequenced to confirm target identity. Primer validation analyses were performed before use as previously described (Morales and Holben, 2009).
Variance in gene abundance measurements was determined between replicate plots of the same treatment by pooling three individual DNA preparations from each replicate plot in equimolar amounts to provide a representative sample for that replicate plot. A total of 5 ng of DNA was used to compare gene abundance in each plot. For comparing the effect of different treatments on the overall abundance of each gene, DNA preparations from all replicate plots and extractions within a treatment were combined in equimolar amounts to provide a representative sample for that treatment. All qPCR reactions for any single sample were run at least in triplicate, as described above.
Correct target amplification from soil DNA was confirmed by cloning PCR fragments from T1. Triplicate standard PCR reactions were performed separately as described above for each primer pair using total community DNA from T1, which represents the canonical treatment practice for the KBS-LTER site. The resultant PCR products were purified and cloned as previously described (Morales and Holben, 2009) to confirm that specific amplification of the corresponding target had occurred.
Statistical analysis
Relationships between microbial gene abundance, successional stage, greenhouse gas flux and other environmental parameters, were determined by principal components analysis (PCA) with data matrices composed of chemical and bacterial qPCR data (Supplementary Table S2) for T1–8, SFR and DFR of the KBS-LTER, collectively representing annual, perennial and successional sites. Chemical data were extracted from Robertson et al. (2000) and represented gas fluxes and their respective greenhouse warming potentials, aboveground net primary productivity, NO3-N, N mineralization potentials and soil carbon concentrations over an 8-year period. The chemical metadata set used was from 1991 to 1999 (as reported in Robertson et al., 2000) because this timeframe maximized the number of variables available for analysis, as several of the measurements were not continued beyond that point. However, more recent gas emission data through 2007 (presented in Supplementary Figure S6) strongly support the suggestion that the same soil processes and relationships persisted at KBS through our sampling time and beyond, as observed differences in these parameters are consistent across treatments and the relative relationships between treatments remain constant. Further, the classification of treatments as a sink or source of gases on the basis of current or past mean gas fluxes for any treatment remains the same. Data were organized with rows representing treatments and columns representing individual variables. Principal component scores were plotted by site, abbreviated as shown in the site description and in Table 1, for the first two principal components. Parameters driving the distribution of the PCA plots were determined by querying all variables against the first three principal components. Individual chemical factors were also independently queried to qPCR results on a per gene basis by conducting pairwise correlations.
To show co-trends between genetic and chemical variables, data were also analyzed using co-inertia analysis (CIA) (Dray et al., 2003), computed with the MADE4 package in the R statistical software environment (Culhane et al., 2005). CIA is a dimensional reduction procedure designed to measure the similarity of two sets of variables (measurements), as they are associated with a single set of cases. Due to very strong correlations (>0.99) between some chemistry measurements, we were able to unclutter the CIA plot by using CH4, N2O and Organic C as proxies for CH4-C, N2O-N and Organic C (kg), respectively.
Results
Bacterial gene abundance and diversity
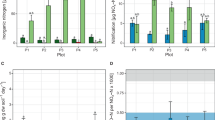
The efficiencies of real-time PCR assays for all targets averaged 92% (s.d.±4%), allowing for direct comparison of results for all targets. Statistically significant differences in gene target abundances were observed between replicate plots under the same treatment (T1) for all tested genes (Table 2). As anticipated, qPCR results obtained from pooling DNA extracted from each individual replicate plot and run as a representative sample for that treatment resulted in mean values within the standard deviation of the true replicates (Figure 1). Two 16S rRNA gene targets representing the numerically dominant bacterial groups Thermomicrobia and Acidobacteria based on sequences generated from this site (Morales et al., 2009) were the most abundant of all targets tested (Figure 1), supporting the first hypothesis regarding higher abundances based on phylogenetic targets (that is, ‘genus-level’ 16S rRNA genes) compared with functional gene (that is, enzyme coding) targets.
Specific detection of bacterial groups by quantitative PCR (qPCR). Values indicate gene copy numbers, as determined from 5 ng of DNA extracted from soil using a dilution curve with known standards. Error bars are one standard error of the mean (s.e.) of at least triplicate qPCR reactions (n⩾3). AB#6 and TM#4: operational taxonomic units based on ⩾97% sequence similarity to the 16S rRNA gene representing Acidobacteria group 6 and Thermomicrobia group 4, respectively. nifH, nitrogenase gene; nirS, nitrite reductase gene; nosZ, nitrous oxide reductase gene.
By contrast, the measured abundances of the nitrogenase gene (nifH) did not support our second hypothesis that predicted higher abundance in treatments containing crops with known symbiotic nitrogen fixation associations (for example, soybean rotations). Indeed, the abundance of nifH was generally higher in the successional treatments (SFR and DFR) than in traditional agricultural sites (Figure 1). Denitrifier abundance, as indicated by nirS and nosZ gene abundance, varied significantly between treatments, which did not support the third hypothesis predicting comparable denitrifier numbers between treatments.
Correlating bacterial gene abundance to environmental variables
Principal component analysis of annual ecosystem averages for key environmental parameters, global warming potential (GWP), bacterial qPCR results and a combined data set comprised of all variables was employed to assess relationships between bacterial gene abundance and process-level measurements. Strong clustering of samples based on aboveground plant cover type (that is, annual, perennial, successional) was observed (Figure 2), supporting the fourth hypothesis predicting differences in greenhouse gas emissions as being correlated to differences in the balance between nitrogen fixing and denitrifying bacteria. PCA based solely on qPCR results from bacterial gene targets accounted for the most variance within the first two components (∼89%). Lower combined principal component 1 and 2 scores were observed for annual ecosystem averages and GWP values (∼75%) and for the combined data set (∼68%) (Figure 2). All three PCA plots showed general clustering of sites based on land-use type, with the exception of T7 (the early-successional site which was previously a woodpile). Loadings for principal components 1 and 2 (that is, loading vectors) were plotted to show independent variable contributions to variance between the treatments (Figure 2). Successional sites were generally associated with higher organic carbon levels and nifH gene abundance. Perennial plant-based treatments, as well as the early native successional plot T7, exhibited increased nitrous oxide reducer abundance (nosZ), total carbon levels and abundance of Thermomicrobia and Acidobacteria. Aboveground annual productivity, nitrite reducers (nirS) and GWP were all strongly correlated with PC1, which was responsible for clustering of sites into annual, perennial and successional treatments, with annual sites showing the highest levels of nitrite reducers and GWP.
Principal component analysis (a) and factor loadings plot (b) for gene abundances (far right), ecosystem chemistry (center) and combined data sets (far left) summarized in Appendix S1 for different management systems at the KBS-LTER site (described in Table 1). The percentage of the variation in the samples described by the plotted principle components is indicated on the axis.
Although plotting the first two principal components accounted for much of the variance in the data, the third component significantly increased the percentage of variance accounted for. On the basis of the first three principal components for the qPCR data alone, ∼98% of the variance was accounted for, whereas the number decreased to ∼86% when only the annual ecosystem averages and GWP values were analyzed. The combined data set of all measured variables accounted for ∼82% of the variance within the first three components, with nosZ gene abundance being significantly and negatively correlated (−0.85, P⩽0.05) to the third component. Correlation values of individual variables with each principal component are summarized in Supplementary Table S3.
An alternative way of analyzing the data based on CIA showed a pattern similar to that in the PCA analysis (Appendix S4). The CIA gives an RV coefficient, a global measure of similarity between data sets based on a multivariate extension of the Pearson correlation coefficient, scaled from 0 (no similarity) to 1.0 (identical). The genetic and chemical variables show strong similarity with an RV=0.76. The significance of the similarity is underscored by a permutation test (Culhane et al., 2003), in which total co-inertia (the measure of co-variability of the two data sets) is computed after permutations to one of the two data sets (randomly chosen before each permutation). This shuffles the data, disassociating the soil sites from the genetic and chemical sample values. The test yielded 100 000 permutation-based values of total co-inertia. Under the null hypothesis that the data sets are independent, only five of these 100 000 values were as large or larger than that observed for the genetic and chemical data sets, for a P-value of 0.00005. The plots also provide a view of the relative strengths of relationships between genetic and chemical variables with respect to each other and to the different soil environments. Genetic and chemical variables show the strongest overall co-trend (shortest arrows) in the group of annuals (Appendix S4). This figure also indicates that, although chemical profiles in transition treatments (former agricultural sites left to undergo natural succession; namely T7 and SFR) closely resembled expected values for successional sites, genetic variables were slower to change, retaining their original signature for longer periods. This is to say, the original bacterial community signature associated to agricultural treatments is seemingly persistent (that is, apparent) after >40 years, but this cannot be unequivocally confirmed with our data as no temporal comparisons are available.
Pairwise correlations between all of the variables showed strong (>0.65) positive correlation between bacterial gene numbers and either greenhouse gas fluxes, GWP, annual productivity or carbon levels (Appendix S5). Nitrite reductase (nirS) gene abundance was positively correlated to greenhouse gas (N2O-N, NO3-N and CH4-C) emission values, GWP values and aboveground net primary productivity, while being negatively correlated to organic carbon. Nitrogenase (nifH) gene abundance was positively correlated to organic carbon levels. The abundance of the nifH gene shared a weak negative correlation (−0.358) with the abundance of the nirS gene (nitrite reductase) (Appendix S5). A second denitrification gene (nosZ), responsible for the reduction of nitrous oxide to dinitrogen, did not exhibit the same trend as nirS (Figure 1). The two target genes corresponding to the numerically dominant OTUs belonging to Thermomicrobia and Acidobacteria (Morales et al., 2009) were the most abundant of all targets measured (Figure 1). These genes also showed a strong positive correlation (Appendix S5) across all treatments, with the highest values found in single cultivar perennial treatment plots (Figure 1).
nirS–nosZ gene abundance as proxy for greenhouse gas (N2O) emissions
A simple regression analysis was conducted to compare direct measurements of nitrous oxide emissions from soils with the abundance of nirS gene targets minus nosZ gene targets (Figure 3), which showed a strong correlation (r2=0.74). Two discrete data clusters were observed related to direct N2O measurements, whereas a more incremental relationship was found between site successional stage and nirS–nosZ gene abundance (Figure 3).
Discussion
Multivariate analysis of annual ecosystem parameter averages, global warming potential and bacterial gene abundances in the current study support the hypothesis that the legacy of agriculture (that is, resilience of treatment effects) has a stronger influence on soil biogeochemistry than current environmental parameters (that is, real-time soil conditions), as previously suggested by other studies (Buckley and Schmidt, 2001, 2003). Strong correlations were obtained that suggest a key role for bacterial activities in controlling responses between agricultural practice or land-use regimes and greenhouse gas emissions. The data also support the interpretation of long-term repercussions at the microbial community level to certain land-use practices. However, given that our study does not include a temporal sampling sequence, these interpretations are based on hypothesized ecosystem successions, as observed on successional treatment plots at the KBS-LTER site after 40–60 years of cessation of agricultural management. Those plots represent more advanced successional stages of the current agricultural treatments, and can be used as references for comparisons.
Previous research at KBS showed differences in microbial community structure between treatments (for example, indicating effects on bacterial community composition based on 16S rRNA sequences (Buckley and Schmidt, 2001, 2003); denitrifiers (Cavigelli and Robertson, 2001; Stres et al., 2004); and ammonia oxidizers (Bruns et al., 1999)). In addition, treatment-based differences at KBS have been reported for greenhouse gas emission and have suggested possible mitigating properties of certain treatments (Robertson et al., 2000; Suwanwaree and Robertson, 2005). Related findings have also been reported for other systems, with plant species identity affecting denitrifying communities (Bremer et al., 2007), and vegetation type driving separation of community structure (Chim Chan et al., 2008).
We found carbon levels to be strongly correlated to the clustering of treatments based on cropping system, suggesting a strong role for carbon as a driver of bacterial community structure. Although higher emission values of the greenhouse gas nitrous oxide were positively correlated to traditional annual crop rotations, previous work has suggested that nitrate, as applied in fertilizer, does not select for denitrifiers (Tiedje, 1988). Instead, it has been proposed that denitrifiers are generally functioning as aerobic competitors for carbon, using their denitrification capabilities only under metabolically advantageous conditions (Tiedje, 1988). Although it appears that carbon has a major role in community structure in these treatments, the activity of an established community can be significantly altered by real-time events such as nitrogen deposition (Suwanwaree and Robertson, 2005). This leads us to suggest that community composition measurements (for example, DNA-based measurements of gene abundance in treatments) are good indicators of how treatment practices shape the community in the long run, whereas rRNA or mRNA measurements would be more useful to illustrate the response of the community to changing parameters in the short term (for example, diel cycles, rainfall, fertilizer application).
Although we included qPCR analysis of two predominant taxa (at approximately the sub-phylum or ‘genus’ level) based on 16S rRNA gene quantification to assess their ubiquity and abundance, data derived from those groups are hard to interpret in the context of biogeochemical cycling, given the lack of correlation between a specific 16S ribotype and its metabolic or catabolic capabilities. Where a direct link to a given biogeochemical reaction is desired, specific tracking of relevant functional genes is likely to be more productive. Thus, in the current study, we rely on quantification of functional genes for nitrogen cycling for correlation with process-level greenhouse gas emissions from KBS soils.
Contrary, perhaps, to common assumptions, the numbers of nitrogen fixers (as determined by nifH gene quantification) were found to be higher in forested or successional sites than in the agricultural fields, including those with regular soybean rotations. Although leguminous plants, which include beans, clover, alfalfa, lupine and peanuts, are among the best studied systems for nitrogen-fixing symbioses (Young and Haukka, 1996; van der Heijden et al., 2006; Nandasena et al., 2007; Houlton et al., 2008), other non-leguminous plants including grasses (Minamisawa et al., 2004; Coelho et al., 2008), pine (Izumi et al., 2006), wheat (Iniguez et al., 2004) and alder (Ridgway et al., 2004) have also been shown to posses significant populations of endophytic nitrogen-fixing bacteria. This suggests that the contribution of non-leguminous nitrogen-fixing plant symbioses and free-living nitrogen fixers is not just significant, but likely essential, in ecosystem development.
Two other nitrogen cycle-related genes encoding the denitrification enzymes nitrous oxide reductase (nosZ) and cytochrome cd1 nitrite reductase (nirS) did not exhibit similar trends in abundance across the system. Instead, what was observed was an apparent balance in the relative abundance of the two genes that can be used to predict greenhouse gas emissions and global warming potential (refer to Figures 3 and 4). To maximally relate our measurements to available metadata, we have used the same 1991–1999 data set as described by Robertson et al. (2000) for the KBS treatments. Whereas we note that gas emission data from 2000 to 2007 for both CH4 and N2O showed that emissions of these gases rose somewhat compared with the 1991–1999 timeframe (Robertson et al., unpublished data (http://lter.kbs.msu.edu/datatables/28)), the relationship of these parameters between treatments has not changed. As noted above and in Supplementary Figure S6, this suggests that the same soil processes and relationships between treatments have persisted through to our sampling time, and that classification of treatments as a sink or source of greenhouse gas emissions remains the same. As the KBS is an LTER site with same conditions carefully maintained for more than two decades, it is well supported that ecosystem processes should remain comparable within and between treatments.
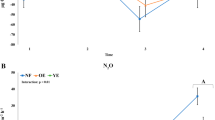
Generalized schematic model showing predicted changes in nitrogen flux, N-based greenhouse gas emissions and greenhouse warming potential in soils as a function of plant community differences and bacterial group abundance. Line thickness represents relative contribution of a given gene to nitrogen compound turnover rate based on data from this study and from a study by Robertson et al. (2000), and also summarized in Appendix S2.
In the schematic model that we have developed to explain this behavior (Figure 4), the rate of greenhouse gas emissions is controlled by the interplay between different guilds within the local bacterial community. In this initial study, we focused on nitrogen cycling and show how the prevalence of bacteria involved in key steps in the cycle are related to the overall outcome in terms of key environmental parameters, and also how differences in relative abundance of individual functional groups control, or at least relate to, whether a system is a net sink or a net source of greenhouse gases.
The data presented herein showed the presence of all targeted genes, and also illustrated how soil management practices have altered the relative abundance of two predominant ribotypes and several functional genes involved in different stages of the nitrogen cycle. These were linked, or at least correlated to, measured differences in greenhouse gas emissions and global warming potential resulting from various land-use practices. Although the general roles of nitrogen fixers and denitrifiers in the nitrogen cycle have been known for decades, the application of a quantitative approach based on functional gene abundance to provide predictive power regarding the fate of nitrogenous compounds in soils is novel.
Two major observations arise from these findings. The first is that monitoring bacterial community response variables, which can exhibit greater sensitivity to environmental change than other commonly used measures (Feris et al., 2009), represents a robust way to monitor geochemical dynamics. Second, the possibility, perhaps probability, that bacteria can respond rapidly to environmental change without major changes in community composition through altered patterns of gene expression suggests the importance of considering the contributions and responses of microbial populations in global biogeochemical cycles to such phenomena as climate change. The latter point further suggests that there is a role for both DNA- and RNA-based approaches in modern molecular microbial ecology depending on whether the investigator is looking at long-term drivers that shape community composition or at short-term response to perturbation and change, respectively.
The variability observed with our approach, as indicated by plot-to-plot replicate variability within a treatment, likely reflects an inherent property of small- or mid-scale heterogeneities in the soil environment that affect bacterial populations locally. Thorough sampling of study sites can compensate for such variability. This is readily achieved when using molecular methods by collecting multiple, small soil samples that can be pooled (composited), creating a representative sample for a site or treatment and thus its greenhouse gas production potential.
We note that our analyses focused on a single nitrite reductase gene, nirS. The copper-containing nitrite reductase (encoded by the nirK gene) was not analyzed in this study and might contribute to some of the variation not accounted for in our data. However, it has been shown that three-quarters of cultured denitrifying bacteria contain nirS rather than nirK (Zumft, 1997) and it has been found to predominate in most environments (Bothe et al., 2000). It is also important to note that these experiments targeted copy numbers of genes of interest, which represent the standing community and its potential for activity rather than an actual measure of real-time gene expression levels or the corresponding enzymatic activity. Ongoing methods development to directly measure actual gene expression levels will likely enhance the resolution and accuracy of studying microbial contributions or response to key environmental functions and activities. Although our data link key gene abundance data with measured greenhouse gas potential, changes in gene expression levels could transiently change a given soil or treatment from a greenhouse gas source to a sink. Thus, making more direct molecular measurements of flux in microbial community gene expression patterns (for example, through environmental transcriptomics) is indeed highly desirable for future work.
This study provides the first quantitative assessment of the effect of land management practice on multiple microbial community constituents at both the functional and phylogenetic level. We showed that microbial assemblages do not readily return to a native or baseline community state following agricultural disturbance, consistent with previous findings that soil nutrient levels require decades or more to recover after agriculture (Robertson et al., 1988, 1993; Drinkwater et al., 1998; Knops and Tilman, 2000). We also present the first quantitative study illustrating interactions between different bacterial activities and their role in controlling nitrogen flux as a response to ecosystem changes.
In conclusion, we note that this initial analysis linking bacterial gene abundance data to process-level greenhouse gas emission rates represents an early step in integrating key bacterial activities to larger-scale biogeochemical cycles. Additional research in this area will extend such capabilities and allow us to assess microbial contributions and responses to ecosystem, and even to global-scale ecological phenomena such as climate change. This is particularly important as it is widely acknowledged that microorganisms govern or at least contribute to global biogeochemical cycles, yet their roles and activities are generally not even considered in current large-scale models for climate change and other global phenomena.
References
Blackwood CB, Paul EA . (2003). Eubacterial community structure and population size within the soil light fraction, rhizosphere, and heavy fraction of several agricultural systems. Soil Biol Biochem 35: 1245–1255.
Bothe H, Jost G, Schloter M, Ward BB, Witzel K . (2000). Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol Rev 24: 673.
Bremer C, Braker G, Matthies D, Reuter A, Engels C, Conrad R . (2007). Impact of plant functional group, plant species, and sampling time on the composition of nirK-type denitrifier communities in soil. Appl Environ Microbiol 73: 6876–6884.
Broughton LC, Gross KL . (2000). Patterns of diversity in plant and soil microbial communities along a productivity gradient in a Michigan old-field. Oecologia 125: 420–427.
Bruns MA, Fries MR, Tiedje JM, Paul EA . (1998). Functional gene hybridization patterns of terrestrial ammonia-oxidizing bacteria. Microb Ecol 36: 293–302.
Bruns MA, Stephen JR, Kowalchuk GA, Prosser JI, Paul EA . (1999). Comparative diversity of ammonia-oxidizer 16S rRNA gene sequences in native, tilled, and successional soils. Appl Environ Microbiol 65: 2994–3000.
Buckley DH, Schmidt TM . (2001). The structure of microbial communities in soil and the lasting impact of cultivation. Microb Ecol 42: 11–21.
Buckley DH, Schmidt TM . (2003). Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ Microbiol 5: 441–452.
Cavigelli MA, Robertson GP . (2001). Role of denitrifier diversity in rates of nitrous oxide consumption in a terrestrial ecosystem. Soil Biol Biochem 33: 297–310.
Chim Chan O, Casper P, Sha LQ, Feng ZL, Fu Y, Yang XD et al. (2008). Vegetation cover of forest, shrub and pasture strongly influences soil bacterial community structure as revealed by 16S rRNA gene T-RFLP analysis. FEMS Microbiol Ecol 64: 449–458.
Culhane AC, Perrière G, Higgins DG . (2003). Cross-platform comparison and visualisation of gene expression data using co-inertia analysis. BMC Bioinformatics 4: 59.
Culhane AC, Thioulouse J, Perriere G, Higgins DG . (2005). MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics 21: 2789–2790.
Dray S, Chessel D, Thioulouse J . (2003). Co-inertia analysis and the linking of ecological data tables. Ecology 84: 3078–3089.
Drinkwater LE, Wagoner P, Sarrantonio M . (1998). Legume-based cropping systems have reduced carbon and nitrogen losses. Nature 396: 262–265.
EIA (2008). Emissions of Greenhouse Gases in the United States 2007 report number DOE/EIA-0573 (2007). In Corti J and Sweetnam GE (eds).
Feris KP, Ramsey PW, Gibbons SM, Frazar C, Rillig MC, Moore JN et al. (2009). Hyporheic microbial community development is a sensitive indicator of metal contamination. Environ Sci Technol 43: 6158–6163.
Henry S, Bru D, Stres B, Hallet S, Philippot L . (2006). Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16 s rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microbiol 72: 5181–5189.
Henry S, Texier S, Hallet S, Bru D, Dambreville C, Cheneby D et al. (2008). Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ Microbiol 10: 3082–3092.
Houlton BZ, Wang YP, Vitousek PM, Field CB . (2008). A unifying framework for dinitrogen fixation in the terrestrial biosphere. Nature 454: 327–330.
Iniguez AL, Dong Y, Triplett EW . (2004). Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Mol Plant Microbe Interact 17: 1078–1085.
Izumi H, Anderson IC, Alexander IJ, Killham K, Moore ERB . (2006). Diversity and expression of nitrogenase genes (nifH) from ectomycorrhizas of Corsican pine (Pinus nigra). Environ Microbiol 8: 2224–2230.
Knops JMH, Tilman D . (2000). Dynamics of soil nitrogen and carbon accumulation for 61 years after agricultural abandonment. Ecology 81: 88–98.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809.
Minamisawa K, Nishioka K, Miyaki T, Ye B, Miyamoto T, You M et al. (2004). Anaerobic nitrogen-fixing consortia consisting of Clostridia isolated from gramineous plants. Appl Environ Microbiol 70: 3096–3102.
Morales SE, Cosart TF, Johnson JV, Holben WE . (2009). Extensive phylogenetic analysis of a soil bacterial community illustrates extreme taxon evenness and the effects of amplicon length, degree of coverage and DNA fractionation on classification and ecological parameters. Appl Environ Microbiol 75: 668–675.
Morales SE, Holben WE . (2009). Empirical testing of 16S rRNA gene PCR primer pairs reveals variance in target specificity and efficacy not suggested by in silico analysis. Appl Environ Microbiol 75: 2677–2683.
Nandasena KG, O'Hara GW, Tiwari RP, Sezmis E, Howieson JG . (2007). In situ lateral transfer of symbiosis islands results in rapid evolution of diverse competitive strains of mesorhizobia suboptimal in symbiotic nitrogen fixation on the pasture legume Biserrula pelecinus L. Environ Microbiol 9: 2496–2511.
Phillips CJ, Harris D, Dollhopf SL, Gross KL, Prosser JI, Paul EA . (2000a). Effects of agronomic treatments on the structure and function of ammonia oxidizing communities. Appl Environ Microbiol 66: 5410–5418.
Phillips CJ, Paul EA, Prosser JI . (2000b). Quantitative analysis of ammonia oxidising bacteria using competitive PCR. FEMS Microbial Ecology. FEMS Microbiol Ecol 32: 167–175.
Ridgway KP, Marland LA, Harrison AF, Wright J, Young JPW, Fitter AH . (2004). Molecular diversity of Frankia in root nodules of Alnus incana grown with inoculum from polluted urban soils. FEMS Microbiol Ecol 50: 255–263.
Robertson G, Grace P . (2004). Greenhouse gas fluxes in tropical and temperate agriculture: the need for a full-cost accounting of global warming potentials. Environ Dev Sustain 6: 51–63.
Robertson GP, Crum JR, Ellis BG . (1993). The spatial variability of soil resources following long-term disturbance. Oecologia 96: 451–456.
Robertson GP, Huston MA, Evans FC, Tiedje JM . (1988). Spatial variability in a successional plant community: patterns of nitrogen availability. Ecology 69: 1517–1524.
Robertson GP, Paul EA, Harwood RR . (2000). Greenhouse gases in intensive agriculture: contributions of individual gases to the radiative forcing of the atmosphere. Science 289: 1922–1925.
Coelho MR, de Vos M, Carneiro NP, Marriel IE, Paiva E, Seldin L . (2008). Diversity of nifH gene pools in the rhizosphere of two cultivars of sorghum (Sorghum bicolor) treated with contrasting levels of nitrogen fertilizer. FEMS Microbiol Lett 279: 15–22.
Rösch C, Bothe H . (2005). Improved assessment of denitrifying, N2-fixing, and total-community bacteria by terminal restriction fragment length polymorphism analysis using multiple restriction enzymes. Appl Environm Microbiol 71: 2026–2035.
Staley JT, Reysenbach AL . (2002). Biodiversity of Microbial Life: Foundation of Earth's Biosphere. John Wiley & Sons, Inc.: New York.
Stres B, Mahne I, Avgustin G, Tiedje JM . (2004). Nitrous oxide reductase (nos Z) gene fragments differ between native and cultivated Michigan soils. Appl Environ Microbiol 70: 301–309.
Suwanwaree P, Robertson GP . (2005). Methane oxidation in forest, successional, and no-till agricultural ecosystems effects of nitrogen and soil disturbance. Soil Sci Soc Am J 69: 1722–1729.
Throback IN, Enwall K, Jarvis A, Hallin S . (2004). Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol 49: 401–417.
Tiedje JM . (1988). Ecology of denitrification and dissimilatory nitrate reduction to ammonium. Biol Anaerob Microorg 179: 244.
Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S . (2002). Agricultural sustainability and intensive production practices. Nature 418: 671–677.
van der Heijden MGA, Bakker R, Verwaal J, Scheublin TR, Rutten M, van Logtestijn R et al. (2006). Symbiotic bacteria as a determinant of plant community structure and plant productivity in dune grassland. FEMS Microbiol Ecol 56: 178–187.
Wakelin SA, Macdonald LM, Rogers SL, Gregg AL, Bolger TP, Baldock JA . (2008). Habitat selective factors influencing the structural composition and functional capacity of microbial communities in agricultural soils. Soil Biol Biochem 40: 803–813.
Yergeau E, Kang S, He Z, Zhou J, Kowalchuk GA . (2007). Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. ISME J 1: 163–179.
Young JPW, Haukka KE . (1996). Diversity and phylogeny of Rhizobia. New Phytol 133: 87–94.
Zumft WG . (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616.
Acknowledgements
This project was funded by the US Department of Agriculture–National Research Initiative (USDA–CSREES Grant No. 2004–03501). Soil samples for this project were graciously provided by the Kellogg biological station long-term ecological research project. We gratefully acknowledge the anonymous reviewers and Dr RM Callaway for helpful comments leading to and during revision of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Morales, S., Cosart, T. & Holben, W. Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J 4, 799–808 (2010). https://doi.org/10.1038/ismej.2010.8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.8
Keywords
This article is cited by
-
Soil water extract and bacteriome determine N2O emission potential in soils
Biology and Fertility of Soils (2023)
-
Effects of Land-Use Type and Flooding on the Soil Microbial Community and Functional Genes in Reservoir Riparian Zones
Microbial Ecology (2022)
-
Effect of mixed inhibitor application on N2O production pathways in paddy soil
Journal of Soils and Sediments (2022)
-
Examining the genomic features of human and plant-associated Burkholderia strains
Archives of Microbiology (2022)
-
How climate and soil properties affect the abundances of nitrogen-cycling genes in nitrogen-treated ecosystems: a meta-analysis
Plant and Soil (2022)