Abstract
Probiotic bacteria provide unique opportunities to study the global responses and molecular mechanisms underlying the effects of gut-associated microorganisms in the human digestive tract. In this study, we show by comparative transcriptome analysis using DNA microarrays that the established probiotic Lactobacillus plantarum 299v specifically adapts its metabolic capacity in the human intestine for carbohydrate acquisition and expression of exopolysaccharide and proteinaceous cell surface compounds. This report constitutes the first application of global gene expression profiling of a commensal microorganism in the human gut. A core L. plantarum transcriptome expressed in the mammalian intestine was also determined through comparisons of L. plantarum 299v activities in humans to those found for L. plantarum WCFS1 in germ-free mice. These results identify the niche-specific adaptations of a dietary microorganism to the intestinal ecosystem and provide novel targets for molecular analysis of microbial–host interactions which affect human health.
Similar content being viewed by others
Main
Probiotic strains of Lactobacillus confer health benefits in the human gut and have a long history of safe use in high amounts (Floch et al., 2008). These organisms are therefore attractive models for quantifying the adaptations of gut-adapted commensal microorganisms to the conditions in the human digestive tract. Lactobacillus plantarum 299v is a commercially applied probiotic strain known to alleviate irritable bowel syndrome and reduce enteropathogen colonization in vivo (de Vries et al., 2006; Klarin et al., 2008). L. plantarum 299v and the highly related strain L. plantarum WCFS1 (Kleerebezem et al., 2003; Molenaar et al., 2005) are able to survive in the human gut for extended periods of time (Vesa et al., 2000; Molin, 2001). L. plantarum WCFS1 was also shown to differentially induce nuclear factor-κB-dependent pathways in the human intestine (van Baarlen et al., 2009) and specific cell products expressed by this strain were found to modulate anti-inflammatory responses in vivo (Grangette et al., 2005).
To determine the L. plantarum molecular properties expressed in the human intestine, which might be responsible for adaptation to the gut ecosystem and modulation of intestinal cell function, transcript profiling was performed on biopsy samples collected from healthy intestinal tissue during surgery of three colon cancer patients (see Supplementary Materials). For 8 consecutive days before surgery, the subjects consumed 100 ml of a fermented oatmeal drink containing 109 colony-forming units of L. plantarum 299v per ml. Biopsy samples were collected from normal tissue of the ileum (subject A) and colon (subjects A, B and C). A biopsy sample was also collected from a fourth subject (D) who had consumed a placebo oatmeal product without the probiotic culture. RNA was isolated from mucosal scrapings of the intestinal segments and hybridized to L. plantarum WCFS1 microarrays (Supplementary Materials). Microarray hybridization intensities for total mucosal RNA isolated from the biopsies of subjects A to C showed enrichment of L. plantarum in the mucosa compared with the placebo control (subject D) (data not shown). Real-time reverse transcriptase PCR on L. plantarum 16S rRNA from the biopsies of subjects who had consumed the probiotic drink confirmed the presence of elevated amounts of L. plantarum over a 40-fold range and ⩾160-fold more L. plantarum compared with the control subject (subject D) (Supplementary Materials and Supplementary Table S1). These data confirmed the enrichment of L. plantarum 299v RNA in the human mucosa and minimal interference of the indigenous gut microbiota, including Lactobacillus species, on the transcriptomes obtained for L. plantarum 299v.
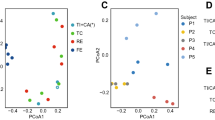
Global gene expression profiles of L. plantarum 299v were conserved in the ileum and colon within one individual and between the genetically unrelated human subjects (Figure 1). According to principal component analysis, these profiles were distinct from transcriptomes of L. plantarum WCFS1 in mono-associated mice fed a diet rich in plant polysaccharides (chow diet) or a ‘Western’ diet high in saturated and unsaturated fats (41% of the total calories) and sucrose (18% of the chow weight) (Marco et al., 2009) (Figure 1). These differences might have been at least partially due to the use of germ-free mice lacking an indigenous microbiota, which are likely to affect microbe–microbe interactions and nutrient availability in the gut. Moreover, global gene expression profiles of L. plantarum in the mice were strongly correlated to host diet (Figure 1) (Marco et al., 2009). According to the first component constituting the majority of the variance between the in vivo samples, the expression values in protein biosynthesis and hypothetical genes were least similar and therefore constituted the primary differences in L. plantarum responses between the three sample groups (data not shown).
Clustering of L. plantarum transcriptomes in the human intestine by principle component analysis. The in vivo profiles were normalized against transcriptomes obtained for L. plantarum WCFS1 grown in standard laboratory conditions (mid-exponential phase cells in de Man, Rogosa, and Sharpe (MRS) culture medium) to adjust for microarray platform-specific effects between the mice and human data sets. The first two components are shown, representing 53% and 20% of the variance, respectively.
To specify conserved responses by the ingested probiotic among the human subjects and the mono-associated mice, the in vivo transcriptomes were quantified against a diverse set of transcriptome patterns identified for in vitro laboratory cultures of L. plantarum WCFS1 (Supplementary Materials). The in vitro transcriptome profiles included L. plantarum responses to growth on rich (undefined) and chemically defined media containing known quantities of essential amino acids, vitamins and nucleotides. These media supported different growth rates, included exposure to environmental stresses and contained diverse individual carbohydrates or complex mixtures of plant polysaccharides. Remarkably, L. plantarum 299v upregulated genes in the human gut were largely restricted to only a few gene classes and this response was conserved for L. plantarum WCFS1 in mice. Induction of transport systems and cell envelope- and cell wall-localized genes constituted the conserved functional response in humans and mice on both dietary regimes relative to laboratory growth (Supplementary Figure S1). Amino acid biosynthesis and central intermediary metabolism were also upregulated in all human subjects (Supplementary Figure S1). In mice fed the Western diet, amino acid biosynthesis was induced, whereas genes associated with energy metabolism were significantly induced in the chow-fed mice (Marco et al., 2009) (Supplementary Figure S1).
Carbohydrate metabolism and amino acid biosynthesis pathways were significantly enriched in the core gut metagenome of the indigenous human intestinal microbiota (Turnbaugh and Gordon, 2009). Genes required for the uptake and degradation of carbohydrates by L. plantarum are distributed among several major functional gene classes of this organism and were collectively significantly induced in vivo (human subjects, P=1 × 10−6; mice fed with chow diet, P=0.0001; mice fed with the Western diet, P=0.0009, χ2-test) (Supplementary Figure S1 and Supplementary Table S2). Induction of both carbohydrate and amino acid biosynthesis gene classes of L. plantarum in the human gut confirms the central relevance of these cellular processes in microbial adaptation to the human intestinal ecosystem extending beyond the indigenous microbiota to a transient microbial resident entering the gut through foods and beverages.
Substantial overlaps in the expression patterns of L. plantarum carbohydrate metabolism genes in humans and mice were shown on hierarchical clustering of the gene expression profiles (Supplementary Table S2) and projections of these data at pathway levels (Figure 2). Genes encoding the transporters and enzymes for the utilization of maltose, cellobiose, lactose/galactose and melibiose were induced in humans and mice on either diet (Figure 2 and Supplementary Table S2). These disaccharides are constituents of plant-derived polysaccharides and the digestion of such compounds by L. plantarum in the mammalian gut might be expected for an organism known for its dominant roles in plant fermentations (de Vries et al., 2006). Furthermore, L. plantarum in humans and mice fed with the Western diet preferentially upregulated glycerol and glycerol-3-phosphate transporters, two α-mannosidases, three cell surface-localized glycosyltransferases, and seven ATP-binding cassette sugar transport systems for which the carbohydrate source is presently not known (Supplementary Table S2). Induction of the same sugar metabolism genes in humans and germ-free mice on the Western diet signify a common nutrient resource pool for L. plantarum in both mammals, which might be dependent on the nutritional landscape provided in the colonized human gut resembling that found in germ-free mice fed with the Western diet. The overlap in the expression of specific L. plantarum genes in humans and germ-free mice confirms the significance of animal models in studying gut-associated microorganisms, particularly when dietary specifications are included in the analyses.
L. plantarum carbohydrate and sugar alcohol metabolism in the intestines of humans and mice. The map is based on metabolic model of L. plantarum according to Simpheny (Teusink et al., 2006). The heat-map plot from left to right contains the following: ileum_subject A, colon_subject A, colon_subject B, colon_subject C, average of mice fed with chow diet and average of mice fed with the Western diet. The intensity gradient is based on a −1 (green) to 1 (red) ranked difference in expression.
L. plantarum cell surface properties were also significantly modified in vivo (Supplementary Figure S1). In humans, an average of 50 out of 133 genes in the cell envelope category encoding extracellular proteins and polysaccharide biosynthesis were induced (Supplementary Figure S2). In contrast, peptidoglycan, lipoteichoic acid and cell membrane biosynthesis pathways were either not affected or downregulated in vivo (data not shown). Among the upregulated genes, 23 were also induced in mice fed with the Western diet and a total of 9 genes were upregulated in humans and mice on either diet and constituted significant conservation in L. plantarum cell surface modifications in vivo (P=0.01, χ2-test) (Supplementary Figure S2). The L. plantarum response in all in vivo samples was dominated by cell-surface protein clusters (csc) predicted to be involved in carbohydrate acquisition (Siezen et al., 2006) and other genes with putative glycolytic or proteolytic functions (Boekhorst et al., 2006) (Supplementary Table S3). Two genes (lp_0800 and lp_2940) induced in the intestine of humans and germ-free mice on either diet were also identified as gut-inducible in conventionally raised mice (Bron et al., 2004; Marco et al., 2007) (Supplementary Table S3). The survival and persistence of a L. plantarum WCFS1 lp_2940 deletion mutant was severely compromised in the mouse digestive tract (Bron et al., 2007). Cell surface proteins such as Lp_2940 for which the functional properties are currently unknown are among a growing repertoire of Lactobacillus cell products providing specific contributions in the mammalian gut, which are not related to host diet (Konstantinov et al., 2008). These proteins are prime targets for unraveling the mechanisms by which probiotic bacteria affect human health through targeted interactions with intestinal epithelial and immune cells in ways that encompass the integrated contributions of the gut microbiota and host diet on the human body.
References
Boekhorst J, Wels M, Kleerebezem M, Siezen RJ . (2006). The predicted secretome of Lactobacillus plantarum WCFS1 sheds light on interactions with its environment. Microbiology 152: 3175–3183.
Bron PA, Grangette C, Mercenier A, de Vos WM, Kleerebezem M . (2004). Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J Bacteriol 186: 5721–5729.
Bron PA, Meijer M, Bongers RS, de Vos WM, Kleerebezem M . (2007). Dynamics of competitive population abundance of Lactobacillus plantarum ivi gene mutants in faecal samples after passage through the gastrointestinal tract of mice. J Appl Microbiol 103: 1424–1434.
de Vries MC, Vaughan EE, Kleerebezem M, de Vos WM . (2006). Lactobacillus plantarum- survival, functional and potential probiotic properties in the human intestinal tract. Int Dairy J 16: 1018–1028.
Floch MH, Walker WA, Guandalini S, Hibberd P, Gorbach S, Surawicz C et al. (2008). Recommendations for probiotic use—2008. J Clin Gastroenterol 42 (Suppl 2): S104–S108.
Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J et al. (2005). Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA 102: 10321–10326.
Klarin B, Molin G, Jeppsson B, Larsson A . (2008). Use of the probiotic Lactobacillus plantarum 299 to reduce pathogenic bacteria in the oropharynx of intubated patients: a randomised controlled open pilot study. Critical Care 12: R136.
Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R et al. (2003). Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA 100: 1990–1995.
Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F et al. (2008). S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci USA 105: 19474–19479.
Marco ML, Bongers RS, de Vos WM, Kleerebezem M . (2007). Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl Environ Microbiol 73: 124–132.
Marco ML, Peters TH, Bongers RS, Molenaar D, van Hemert S, Sonnenburg JL et al. (2009). Lifestyle of Lactobacillus plantarum in the mouse caecum. Environ Microbiol 11: 2747–2757.
Molenaar D, Bringel F, Schuren FH, de Vos WM, Siezen RJ, Kleerebezem M . (2005). Exploring Lactobacillus plantarum genome diversity by using microarrays. J Bacteriol 187: 6119–6127.
Molin G . (2001). Probiotics in foods not containing milk or milk constituents, with special reference to Lactobacillus plantarum 299v. Am J Clin Nutr 73: 380S–3385.
Siezen R, Boekhorst J, Muscariello L, Molenaar D, Renckens B, Kleerebezem M . (2006). Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics 7: 126.
Teusink B, Wiersma A, Molenaar D, Francke C, de Vos WM, Siezen RJ et al. (2006). Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J Biol Chem 281: 40041–40048.
Turnbaugh PJ, Gordon JI . (2009). The core gut microbiome, energy balance, and obesity. J Physiol 587: 4153–4158.
van Baarlen P, Troost FJ, van Hemert S, van der Meer C, de Vos WM, de Groot PJ et al. (2009). Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci USA 106: 2371–2376.
Vesa T, Pochart P, Marteau P . (2000). Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment Pharmacol Ther 14: 823–828.
Acknowledgements
We express our gratitude to the patients who participated in this study and thank all the staff at the University Hospital Malmö for their excellent technical assistance. We also kindly acknowledge Dr Jeffrey I Gordon and Dr Justin L Sonnenburg for their review of this paper and assistance with transcriptome analysis of L. plantarum WCFS1 in germ-free mice.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Marco, M., de Vries, M., Wels, M. et al. Convergence in probiotic Lactobacillus gut-adaptive responses in humans and mice. ISME J 4, 1481–1484 (2010). https://doi.org/10.1038/ismej.2010.61
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.61
Keywords
This article is cited by
-
Synbiotics: a New Route of Self-production and Applications to Human and Animal Health
Probiotics and Antimicrobial Proteins (2022)
-
Candidate probiotic Lactiplantibacillus plantarum HNU082 rapidly and convergently evolves within human, mice, and zebrafish gut but differentially influences the resident microbiome
Microbiome (2021)
-
Dietary Proteins Alter Fermentation Characteristics of Human Gut Microbiota In Vitro
Plant Foods for Human Nutrition (2020)
-
The gut microbiota in young and middle-aged rats showed different responses to chicken protein in their diet
BMC Microbiology (2016)
-
Gene expression of Lactobacillus plantarum and the commensal microbiota in the ileum of healthy and early SIV-infected rhesus macaques
Scientific Reports (2016)