Abstract
As part of an ongoing survey of microbial community gene expression in the ocean, we sequenced and compared ∼38 Mbp of community transcriptomes and ∼157 Mbp of community genomes from four bacterioplankton samples, along a defined depth profile at Station ALOHA in North Pacific subtropical gyre (NPSG). Taxonomic analysis suggested that the samples were dominated by three taxa: Prochlorales, Consistiales and Cenarchaeales, which comprised 36–69% and 29–63% of the annotated sequences in the four DNA and four cDNA libraries, respectively. The relative abundance of these taxonomic groups was sometimes very different in the DNA and cDNA libraries, suggesting differential relative transcriptional activities per cell. For example, the 125 m sample genomic library was dominated by Pelagibacter (∼36% of sequence reads), which contributed fewer sequences to the community transcriptome (∼11%). Functional characterization of highly expressed genes suggested taxon-specific contributions to specific biogeochemical processes. Examples included Roseobacter relatives involved in aerobic anoxygenic phototrophy at 75 m, and an unexpected contribution of low abundance Crenarchaea to ammonia oxidation at 125 m. Read recruitment using reference microbial genomes indicated depth-specific partitioning of coexisting microbial populations, highlighted by a transcriptionally active high-light-like Prochlorococcus population in the bottom of the photic zone. Additionally, nutrient-uptake genes dominated Pelagibacter transcripts, with apparent enrichment for certain transporter types (for example, the C4-dicarboxylate transport system) over others (for example, phosphate transporters). In total, the data support the utility of coupled DNA and cDNA analyses for describing taxonomic and functional attributes of microbial communities in their natural habitats.
Similar content being viewed by others
Introduction
Marine microbial communities, centrally involved in the fluxes of matter and energy in the global oceans, are major drivers of global biogeochemical cycling (Karl and Lukas, 1996; Arrigo, 2005). Our knowledge of abundance, diversity and gene content of planktonic microbes has been fundamentally advanced over the past three decades, by both model organism-based studies (Giovannoni et al., 2005b; Coleman and Chisholm, 2007), as well as metagenomic surveys of natural microbial communities (DeLong et al., 2006; Rusch et al., 2007; Dinsdale et al., 2008; Ghai et al., 2010). In particular, metagenomic comparisons of distinct microbiomes (DeLong et al., 2006; Martin-Cuadrado et al., 2007; Dinsdale et al., 2008) have revealed habitat-dependent distribution of taxa and gene families, in part shaped by the biogeochemical dynamics characterizing each environment. Clearly, determining if and how such genomic variations are manifested at the level of gene expression and regulation, represents another important step towards understanding the interplay between microbes and their environments, as well as the metabolic strategies they use in distinct ecological niches.
Metatranscriptomics involve the direct sampling and sequencing of gene transcripts from natural microbial assemblages, allowing assessment of relative transcript abundance within the community, without requiring a priori knowledge of taxonomic and genomic compositions. We first carried out a pilot metatranscriptomic study at the Hawaii Ocean Time-series (HOT) Station ALOHA (Frias-Lopez et al., 2008), where community transcripts were analyzed in parallel with genomic sequences for a bacterioplankton assemblage at 75 m depth (within the mixed layer). One unexpected finding from that study was that many highly abundant transcripts (most of which were designated as hypothetical genes) were absent or in low abundance in the coupled DNA library, suggesting that they originated from low abundance microorganisms (or less frequently represented genes in hypervariable genomic regions). Subsequently, comparative analyses of surface water samples have shed light on the day/night and geographical differences in community gene expression (Poretsky et al., 2009; Hewson et al., 2010). More recently, to effectively enhance sequencing coverage across the functional transcript pool, Stewart et al. developed a universal ribosomal RNA (rRNA)-subtraction protocol that was shown to physically remove large amount of rRNA molecules from RNA samples, reducing rRNA transcript abundance by 40–58% (Stewart et al., 2010). The implications of these metatranscriptomic studies are clear, although the sequencing of microbial community transcripts has just begun and is far from comprehensive, it complements the metagenomic approach. Such studies are beginning to yield valuable information on genes that are actively expressed in naturally occurring microbial assemblages.
In this study we analyze coupled metatranscriptomic and metagenomic data from four bacterioplankton samples taken at Station ALOHA, along the stratified water column characterized by warm, nutrient-depleted surface waters underlain by a steep pycnocline and nutricline (Dore and Karl, 1996; Karl and Lukas, 1996). The goal was to assess in parallel microbial metabolic potential (in DNA) and transcriptional activity (in complementary DNA (cDNA)) along the vertical gradient. In addition to the recent use of these data sets to search and compare putatively novel RNA regulatory elements (small RNAs) highly abundant in these habitats (Shi et al., 2009), the results obtained in this study demonstrate that coupled metagenomic and metatranscriptomic analyses provide useful perspectives on microbial activity, biogeochemical potential and regulation in indigenous microbial populations.
Methods
Sample collection
Bacterioplankton samples (size fraction 0.22 μm–1.6 mm) from the photic zone (25 m, 75 m and 125 m) and the mesopelagic zone (500 m) were collected from the Hawaii Ocean Time-series (HOT) Station ALOHA site in March 2006, as described previously (Shi et al., 2009). See Supplementary Methods for further details on the seawater collection and RNA/DNA extraction.
Complementary DNA (cDNA) synthesis and sequencing
The synthesis of microbial community cDNA from small amounts of mixed-population microbial RNA was performed as previously described (Frias-Lopez et al., 2008). Briefly, ∼100 ng of total RNA was amplified using MessageAmp II (Ambion, Foster City, CA, USA) following the manufacturer's instructions and substituting the T7-BpmI-(dT)16VN oligo in place of the oligo(dT) supplied with the kit. The SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA) was used to convert amplified RNA to microgram quantities of cDNA, which was then digested with BmpI to remove poly(A) tails. Purified cDNA was then directly sequenced by pyrosequencing (GS20). See Supplementary Methods for further details.
Bioinformatic analyses
rRNA sequences were first identified by comparing the data sets with a combined 5S, 16S, 18S, 23S and 28S rRNA database derived from available microbial genomes and sequences from the ARB SILVA LSU and SSU databases (http://www.arb-silva.de). 16S rRNA reads were further selected and subjected to taxonomic classification. Non-rRNA sequences were compared with NCBI-nr, SEED and Global Ocean Sampling (GOS) protein clusters databases using BLASTX for functional gene analyses as previously described (Frias-Lopez et al., 2008; Shi et al., 2009). Two custom databases (one nucleotide and one amino acid) were constructed from then publicly available 2067 microbial genome sequences, and were used to recruit cDNA and DNA reads. See Supplementary Methods for further details.
Data deposit
The nucleotide sequences are available from the NCBI Sequence Read Archive under accession numbers SRA007802.3, SRA000263, SRA007804.3 and SRA007806.3 corresponding to cDNA sequences, and SRA007801.5, SRA000262, SRA007803.3 and SRA007805.4 corresponding to DNA sequences, for 25 m, 75 m, 125 m and 500 m samples, respectively.
Results and discussion
Bacterioplankton samples and pyrosequencing data sets
The four sampling depths represent discrete zones in the water column at Station ALOHA (22°45′ N, 158°W), which includes the middle of the mixed layer (25 m), the base of the mixed layer (75 m), the deep chlorophyll maximum (DCM, 125 m) at the top of the nutricline and the upper mesopelagic zone (500 m). Bacterioplankton samples were collected from each depth for RNA and DNA extraction and sequencing. As the sampling times for these four sets of seawater samples were different (25 m at 22 h local time, 75 m at 3 h, 125 m at 6 h and 500 m at 6 h), we expected that the observed gene expression patterns would reflect spatial geochemical gradients (Supplementary Figure S1), as well as temporal differences (discussed below).
A total of ∼38 Mbp and ∼157 Mbp of sequences were obtained for the four metatranscriptomic and four metagenomic data sets, respectively (Table 1). The number of cDNA reads per GS20 run is roughly a quarter of that of the DNA reads, likely due to incomplete removal of poly(A) tags added during RNA amplification step. Despite the lower yield of cDNA reads, we observed significant reproducibility and fidelity in transcript profiles of Prochlorococcus cultures (Frias-Lopez et al., 2008). Subsequent to the data sets reported in this study, significant improvements have been made in the cDNA preparing and sequencing protocols, using the GS-FLX platform (Stewart et al., 2010). Nevertheless, the earlier transcriptome data sets reported in this study provide new perspective on coupled metagenomic and metatranscriptomic data sets, and provide new information of transcriptional activity in parallel with community structure, gene abundance and genetic variation.
Taxonomic composition: ribosomal RNA (rRNA) sequence-based analyses in the DNA samples
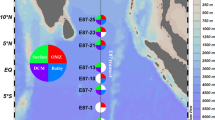
Roughly 0.3% of total DNA reads were designated as rRNA operon sequences (1188, 1117, 954 and 1029 reads for the 25 m, 75 m, 125 m and 500 m samples, respectively), including bacterial, archaeal, and eukaryotic small and large subunit rRNAs and intergenic spacer sequences. This sampling frequency was within the expected range based on the rRNA operon size (∼5000 bp), assuming average genome size of ∼2 Mbp for marine bacteria and archaea. To assess the taxonomic diversity within the four microbial communities, we classified these 16S rRNA gene sequences (Figure 1, upper panel), using the online Greengenes alignment and classification tools (http://greengenes.lbl.gov/cgi-bin/nph-classify.cgi) (DeSantis et al., 2006), which was reported to yield the highest accuracy for assigning taxonomy to short pyrosequencing reads compared with other methods such as RDP classifier or BLAST (Liu et al., 2008). Roughly 254–339 reads were classified for each of the four DNA samples; and these taxonomic assignments were further corroborated (Supplementary Figure S2; Pearson's correlation r>0.95 for all four depths), using a full set of ‘shotgun’ DNA library sequences (average read length 565 bp) from the same source DNA samples (Martinez et al., 2010).
Taxonomic classification based on 16S rRNA-bearing reads in DNA and cDNA data sets. Taxonomic assignments were binned at the order level, using the Hugenholtz taxonomy of Greengenes (see Supplementary Methods). 16S rRNA sequences that could not be classified were excluded from the analysis. Y-axis scale represents the percentage of the total classified 16S rRNA reads. Only taxa that represented ⩾1% of all classified reads are displayed. Also note here that, as no replicate data were available for each sample, error bars were absent and thus no statistical inference could be made from the figure.
Each of the four microbial communities was dominated by two or three major groups (Figure 1, upper panel). Consistiales (predominantly Pelagibacter) recruited ∼13–35% of the total classified 16S rRNA gene reads from all depths, supporting the high abundance of Pelagibacter populations throughout the water column (Eiler et al., 2009) and their under-representation in large-insert metagenomic libraries, at least for the populations residing shallower depths (Pham et al., 2008; Temperton et al., 2009). The other major groups included Prochlorales in the photic zone (∼17–51%), Cenarchaeales (∼22%) and the uncultured delta-proteobacterial group SVA0853 (∼9%) at 500 m, and Acidimicrobidae (∼2–8%) at all depths. This depth distribution was generally consistent with previous cultivation-independent surveys at this site, but variability (likely both biological and methodological) was apparent. For instance, a fosmid library-based survey (DeLong et al., 2006) reported a significant decrease in the relative abundance of Prochlorococcus populations at 75 m depth, potentially caused by cyanophage infection, as suggested by the large number of cyanophage sequences recovered in the same cellular size fraction. In contrast, in this survey large numbers of phage sequences were not detected, and Prochlorococcus relative abundance peaked at 75 m depth, regardless of DNA library type and sequencing method (pyrosequencing, Figure 1; fosmid clone library, Supplementary Table S1).
Taxonomic composition: protein-coding sequence-based analyses in the DNA samples
Another common approach to assess taxonomic composition from metagenomic data sets is to infer taxonomic origins from open reading frame (ORF) sequences (Huson et al., 2007). In this study, we observed both consistencies as well as some discrepancies when comparing the community composition derived from rRNA gene sequences (discussed above) to those derived from ORF sequences using MEGAN (Huson et al., 2007). As seen in Figure 1 and Supplementary Figure S3, Pelagibacter relative abundance decreased from ∼13–35% estimated from the 16S rRNA gene sequences, to ∼9–23% from the ORF sequences, and the uncultured delta-proteobacterium SVA0853 was completely missed in the latter. In contrast, Prochlorococcus-like sequences represented ∼39–71% of all annotated ORF sequences, much higher than that estimated from 16S rRNA gene sequences (∼17–51%). Higher representation of Prochlorococcus-like mRNA transcripts relative to their cell abundance was noted by Poretsky et al. in metatranscriptomic data sets from day and night samples from the same site, and was attributed to higher transcriptional activities of Prochlorococcus cells relative to coexisting heterotrophic microbes (Poretsky et al., 2009). However, it seems that differences in transcriptional activities may not be the explanation, as our DNA data sets showed the same trend of overrepresentation of Prochlorococcus-related ORF sequences. Assuming similar genome sizes, a more likely explanation is that the higher representation of Prochlorococcus-derived sequences reflects the uneven representation of taxa in current databases. That is, sequence annotation is biased in favor of taxa with more sequenced isolates, such as Prochlorococcus, than those with fewer or no sequenced isolates such as Pelagibacter and SVA0853-related delta-proteobacteria.
Taxonomic origin of transcripts in the cDNA samples
The simultaneous recovery of rRNA and mRNA transcripts from RNA samples provided a unique opportunity to use two different approaches to assess the contribution of different taxa to the community metabolic processes (as judged by transcript abundance). We performed taxonomic analyses with the 16S rRNA, as well as protein-coding mRNA transcript sequences exactly as described above for DNA samples (Figure 1, lower panel; Supplementary Figure S3, lower panel). Prochlorococcus populations inhabiting DCM layer (125 m), displayed highest transcriptional activity, relative to their DNA abundance at that depth. In contrast, Pelagibacter, the most numerically abundant heterotrophic bacteria in the open ocean, seemed to be relatively more abundant in cell numbers (DNA) but less active transcriptionally within DCM layer (mRNA). This was also evident in Pelagibacter genome-wide transcriptional activity analyses (see below). The DCM layer is characterized by two opposing resource gradients: light supplied from above and nutrients supplied from below, and thus co-existing photoautotrophic and heterotrophic microbes might alternate dominance at different times of a day or in different seasons of a year. Specifically, this apparently lower transcriptional activity of Pelagibacter may be influenced by the time of DCM sample collection: ∼6 h local time, when photosynthetic microorganisms such as Prochlorococcus may be relatively more active.
Finally, for the relatively under-studied mesopelagic zone (500 m), two observations are clear. Marine group I Crenarchaea and Pelagibacter compirsed a major fraction of microbial community in terms of both DNA and transcript abundance. Meanwhile, some groups in lower abundance such as Alteromonadales and Sphingomonadales, showed much higher transcript abundance relative to their DNA abundance, suggesting more active gene transcription per cell, in comparison with other taxa.
Global analysis of metabolic potential and functional activities
The majority of the non-rRNA cDNA reads (>50%), especially those derived from the 500 m sample (>70%), did not share any significant match against NCBI non-redundant (NCBI nr) and the SEED (Meyer et al., 2008) databases (Table 1). Not surprisingly, a significantly higher fraction of cDNA reads shared homology to sequences in the GOS peptide database, the largest marine-specific sequence database available (Yooseph et al., 2007). Furthermore, a large fraction of these cDNA sequences were not present in the coupled DNA libraries at the current sequencing depth (data not shown). These novel sequences likely represented actively expressed ORFs from low abundance microbial groups (alternatively, hyperdynamic genomic regions of well-known taxa), or non-coding regions that by definition are not translated into proteins but instead function as RNA molecules (Shi et al., 2009).
For sequences that were annotated as protein coding, we compared gene and transcript abundance in parallel, in order to investigate relative transcriptional activity in a normalized manner (see Supplementary Methods). Such normalization accounts for differences in community structure and gene content among samples, allowing detection of metabolic pathways and gene families in lower abundance but with relatively high transcriptional activity (see the example of crenarchaeal-mediated ammonia oxidation at 125 m below).
Known metabolic pathways
Several metabolic pathways exhibited high expression levels, as evidenced by a number of SEED subsystems that were found significantly enriched (at the 98% confidence level) in each transcript library, relative to the corresponding DNA library (Figure 2; Table 2). In the surface sample (25 m) collected at 22 h local time, the active expression of oxidative stress-related genes was likely a result of high UV doses during daytime. Aerobic respiration, expected to be enriched relative to photosynthesis at night, was reflected in the expression of cytochrome c oxidases and menaquinone–cytochrome c reductase complexes. The sample collected from DCM layer (125 m) at 6 h local time, exhibited high abundance of transcripts associated with carbon fixation and photosynthesis, compared with the other two photic zone samples (despite the relatively lower abundance of photosynthetic genes in the DNA, see Table 2). This is consistent with laboratory observations where Prochlorococcus carbon-fixation genes were maximally expressed at dawn, and photosynthetic gene transcription was elevated on the appearance of light (Zinser et al., 2009). Highly expressed subsystems in the mesopelagic sample (500 m) included peptidoglycan biosynthesis that may be involved in maintenance of cell wall integrity at greater depths, and ammonia assimilation that has a significant role in energy metabolism for mesopelagic Crenarchaea (Konneke et al., 2005).
Clustering of all cDNA and DNA data sets based on relative abundance of SEED subsystems. Only the most abundant subsystems that together recruited 95% of all reads are displayed. Hierarchical clustering of four DNA and four cDNA samples were performed with euclidean distance and single-linkage method using MATLAB. Color scale represents the proportion of reads assigned to SEED categories relative to the total library size in each sample. Blue to red color indicates low to high representation of SEED categories.
Not surprisingly, light-harvesting cellular subsystems were among the most highly expressed in the photic zone. The differentiated clustering of photic zone DNA and cDNA samples observed (Figure 2; Supplementary Figure 5) may be partly attributable to sampling times, given the commonality of diel rhythms among photosynthetic microbes (Zinser et al., 2009). As expected, the metabolic signatures of mesopelagic communities reflected a completely different transcriptional signature, including energy sources, cellular structures and catabolic and anabolic biochemical pathways.
GOS protein families
The recent GOS expedition (Rusch et al., 2007; Yooseph et al., 2007) has greatly expanded our knowledge of open ocean-derived protein families. Among all protein families identified based on sequence similarity clustering, 3995 protein clusters consisted of only GOS sequences, 1700 of which have no detectable homology to previously known protein families (Yooseph et al., 2007). Many of these GOS-only protein clusters of unknown functions were detected in our transcript libraries, some in high abundance (Figure 3a), underscoring ecologically relevant functions associated with these novel/hypothetical protein families. Meanwhile, analysis of protein families with known or predicted functions highlighted genes that are highly expressed and therefore likely have active roles in maintaining ecosystem functions at each habitat (Figure 3b).
Community-level gene expression profiles based on the GOS protein family database. Cluster-based expression ratio was defined as representation of each GOS cluster in the cDNA library normalized by its representation in the DNA library. GOS clusters that recruited only cDNA reads were arbitrarily set a value of 1 copy of DNA read, to avoid a denominator of 0. (a) GOS clusters were ranked by their cluster-based expression ratios for four depths; (b) The most highly expressed GOS clusters with known or predicted functions were highlighted for each depth.
Nitrogen metabolism protein families
A suite of nitrogen metabolism genes (ammonium transporter, amt; dissimilatory nitrite reductase, nirK; urea transporter, urt; ammonia monooxygenase subunits, amoABC) was among the most highly expressed of GOS protein families detected (Figure 3b). An essential macronutrient, nitrogen availability and turnover limits biological production in many open ocean regions, including NPSG (Van Mooy and Devol, 2008). Ammonia/ammonium is a key reduced nitrogen compound that can either be incorporated into carbon skeleton via the glutamine synthetase (GS; glnA)/glutamate synthase (GOGAT; glsF) cycle, or can serve as energy source fueling autotrophic metabolism (Konneke et al., 2005). Thus, the transport of ammonia/ammonium is vital for planktonic microbes living in the nutrient deplete surface waters and energy constrained deep waters in an open ocean setting. Urea is another potentially important nitrogen source in the ocean, and is used by marine cyanobacteria (Moore et al., 2002). The more oxidized forms of nitrogen, nitrite and nitrate require more metabolic energy to use, but can serve as alternative nitrogen sources because of their much higher concentrations in deep euphotic zone and mesoplegic zone below the nitracline.
To assess the prevalent nitrogen-using pathways in the genomes of the most abundant planktonic microbial populations, we compared the observed frequency (normalized to gene length and data set size) of several essential nitrogen metabolism genes with that of the 16S rRNA gene of Prochlorococcus and marine group I Crenarchaea. The observed frequency of Prochlorococcus-related amt, glnA, urt, urease genes is equivalent to that of Prochlorococcus 16S rRNA gene (Supplementary Figure S4A, left panel), suggesting that ammonium and urea assimilation is preserved in naturally occurring Prochlorococcus populations. In contrast, the assimilatory nitrite reductase gene (nirA) was present in only a small fraction of Prochlorococcus cells (c.a., 7%, 8% and 15% at 25 m, 75 m and 125 m, respectively), consistent with expectation based on genomic and physiological studies of Prochlorococcus isolates (Moore et al., 2002; Rocap et al., 2003). Furthermore, the transcripts of these nitrogen metabolism genes (except nirA) were also detected in our metatranscriptomic data sets (Supplementary Figure S4A, right panel), suggesting active deployment of these nitrogen metabolism pathways by Prochlorococcus cells in situ. The amt gene was the most actively transcribed, likely an adaptive mechanism to efficiently scavenge low-concentration ammonium as the most preferred nitrogen source. The dramatic decrease in the relative transcriptional activity of amt gene at 125 m, however, was not expected. It is possible that the apparently higher primary production at 125 m (DCM) has caused accumulation of ammonium via active nutrient regeneration processes. In fact, ammonium maxima near the DCM layer are common in stratified oligotrophic waters (Brzezinski, 1988). As a result, the presumably elevated ammonium concentration may result in downregulation of the amt gene expression, as observed in many cyanobacterial isolates.
Marine group I Crenarchaea exist in high abundance in mesopelagic zone, where distinct forms and concentrations of nitrogen species (for example, nitrate, nitrite and urea) are present. Nitrosopumilus maritimus, an isolate of related Crenarchaea from marine aquarium, has been shown definitively to grow chemolithoautotrophically on ammonia (Konneke et al., 2005). Further genomic analyses of marine group I Crenarchaea have provided insights into the metabolism of other forms of nitrogen compounds (Hallam et al., 2006; Walker et al., 2010). In this study, our data showed that amt, amoABC and glnA genes were prevalent and expressed in planktonic crenarchaeal populations, whereas urea-usage genes, while present and expressed, appeared in lower abundance (Supplementary Figure S4B, left panel). Clearly, despite the apparent lack of such genes in the N. maritimus genome (Walker et al., 2010), a fraction of planktonic crenarchaeal populations encode genes for using urea as nutrient or energy source. The normalized expression levels of Crenarchaea-related amt and amoABC genes (especially amoC gene) was among the highest in our data sets (orders of magnitude higher than most other protein-coding genes) (Figure 3b). Interestingly, the anomalously high amoC gene expression seems to be common, as it is also observed in bacterial nitrifiers (Berube et al., 2007), for as-yet unknown reasons. Consistent with a quantitative PCR-based study (Church et al., 2010), the amoABC transcripts were detected in high abundance at 125 m depth despite the small planktonic crenarchaeal population size (Supplementary Figure S4B, right panel). Together with previous report of remarkably high substrate affinity and kinetics of crenarcheal amo genes (Martens-Habbena et al., 2009), these data further support a role for marine Crenarchaea in nitrification in the ocean via active ammonia oxidation.
Nitrite, an end product of archaeal ammonia oxidation, could exert toxic effects to cells if accumulated, and an upper primary nitrite maximum is often observed near DCM layer (125 m in this study) in the open ocean (Dore and Karl, 1996). Consistent with the hypothesis that dissimilatory nitrite reductase (nirK) in ammonia-oxidizing microbes is involved in nitrite detoxification (Casciotti and Ward, 2001; Hallam et al., 2006), nirK was found highly expressed at 125 m (Supplementary Figure S4B, right panel). Finally, nitrate reductase genes (narH and narG) and transcripts were frequently detected in the 500 m data sets, and seemed to be most similar to homologs found in Candidatus Kuenenia stuttgartiensis, a planctomycete, suggesting that planktonic Crenarchaea may not participate in the first step of nitrate respiration.
Photoheterotrophy
We detected in the photic-zone active expression of genes involved in photoheterotrophy, including those encoding proteorhodopsins. Proteorhodopsin (PR) is a photoprotein that functions as light-driven proton pump, generating biochemical energy via proton motive force (Béjà et al., 2000). PR photosystems have been detected in a large percentage (up to 80% at the DNA level) of ocean surface-dwelling bacteria and archaea (DeLong and Béjà, 2010), and were suggested to be horizontally transferred among phylogenetically divergent microbial taxa (Frigaard et al., 2006; McCarren and DeLong, 2007). Laboratory-based experiments have suggested that PR photosystem increases cellular fitness to bacterial cells under adverse growth conditions (González et al., 2008; Gómez-Consarnau et al., 2007, 2010).
Our depth profile data allow us to directly assess the in situ abundance and taxonomic origins of PR gene and transcripts. Abundance of PR transcripts decreased dramatically from euphotic zone to 500 m (in which only 4 cDNA reads shared homology to known PR genes) (Supplementary Figure S5A). Although PR DNA and cDNA reads seemed to be originated from a diverse range of taxa, the majority shared homology to known PR genes from SAR11-like organisms (Supplementary Figure S5B). Notably, PR genes were found most highly expressed in the 75 m sample (collected at 22 h), followed by the 25 m and 125 m samples (collected at 3 h and 6 h, respectively) (Supplementary Figure S5A; also see the Pelagibacter genome-wide gene expression analysis below), suggesting that PR genes may be constitutively expressed in the photic zone. Laboratory studies of PR-containing isolates, as well as a recently reported microcosm experiment have reported inconsistent observations, some suggesting constitutive PR expression (Giovannoni et al., 2005a; Riedel et al., 2010), whereas others suggesting light regulation of PR expression (Gómez-Consarnau et al., 2007; Lami et al., 2009; Poretsky et al., 2009). Higher-resolution metatranscriptomic studies are necessary to provide further insight into light effects on PR gene expression in different taxa, and in different oceanographic provinces.
Evidence for another form of phototrophy mediated by aerobic anoxygenic phototrophic (AAP) bacteria was also observed. Recent studies suggest that AAPs constitute a considerable fraction of marine planktonic community, and may contribute significantly to the carbon cycle in the ocean via facultative photoheterotrophy (Kolber et al., 2001; Béjà et al., 2002). Living in an oligotrophic environment, oceanic AAPs likely are capable of efficiently controlling the expression of their photosynthetic apparatus, supplementing heterotrophic metabolism with light-dependent energy harvest. In this depth profile, AAPs were most abundant in 25 m and 75 m samples based on observed gene frequencies of bacteriochlorophyll biosynthesis genes (bchXYZ), light-harvesting complex I genes (pufAB) and the reaction center genes (pufLM). The majority of these photosynthetic genes were closely related to Roseobacter-like AAP sequences, particularly a BAC clone insert retrieved from the Red Sea (eBACred25D05; accession number: AY671989) (Oz et al., 2005). GOS protein clusters associated with these AAP genes were found highly expressed in the 75 m sample (Figure 3b), and most of this AAP gene expression originated from the puf operon (Supplementary Figure S6). Collectively, the data indicate photosynthetically active population of AAPs, at 75 m in particular.
Reference genome-centric analyses
We used a total of 2067 genomic references (including finished and draft genomes), to recruit DNA and cDNA reads at high stringency, based on BLASTN comparison (see Supplementary Methods). About 29%, 40%, 15% and 7% of total DNA reads, and 30%, 24%, 26%, and 18% of total cDNA reads were recruited to the reference genomic data for 25 m, 75 m, 125 m and 500 m sample, respectively. Notably, the percentage of recruited cDNA reads for each sample was significantly higher than that of cDNA reads that could be assigned to NCBI-nr protein database (Table 1), a result of cDNA recruitment to expressed non-coding regions on the genomes. For instance, about 1539 reads in the 25 m sample were recruited to an intergenic region of Prochlorococcus strain MIT 9215 genome, corresponding to the Group_2 small RNA previously reported by Shi et al. (2009).
The relative representation of genomes/genome fragments is shown in a three-way comparison plot to illustrate the similarities and differences of communities dwelling in specific habitats (Figure 4). For this analysis, the 75 m and 125 m samples were pooled together, as they shared the most similar profile at cDNA level, and were relatively similar at the DNA level (Figure 2). All genomes recruiting >50 DNA reads are also listed in Supplementary Table S2. In this study, general separation of photic zone populations with mesopelagic populations was observed, with a few exceptions that were found more evenly distributed along the depth, including the ubiquitous Pelagibacter, and the alphaproteobacterium Erythrobacter sp. SD-21, a Mn(II) oxidizing bacterium that has been isolated from many diverse marine environments including surface and deep oceans (Francis et al., 2001).
Three-way comparison of representation of genomes and genome fragments (fully sequenced fosmids) in DNA and cDNA data sets. The 75 m and 125 m data sets were combined as they were the most similar. Each dot represents a genome (fragment), and its proximity to a vertex reflects the enrichment of the corresponding genome (fragment) in the respective sample. Only genomes recruited >0.1% of total reads are displayed. Station ALOHA fosmids represent fosmid sequences that were reported by (DeLong et al., 2006). See Supplementary Methods for detail.
Such genome recruitment analysis provides direct measurement of vertical distribution of ecologically coherent populations (represented by reference genomes) in nature, such as high-light and low-light adapted Prochlorococcus ‘ecotypes’ (Moore and Chisholm, 1999). Notably, despite an expected significant increase of low-light adapted Prochlorococcus populations (mostly eNATL2A) at 125 m, where light intensity dramatically decreased compared with shallower depths, >80% of the Prochlorococcus-like reads at 125 m were most similar to sequences of high-light adapted isolates (mostly eMIT9312) (Supplementary Table S2). Although possibly a result of physical homogenization of the water column due to deep mixing in the winter (Malmstrom et al., 2010), these high-light-like Prochlorococcus cells displayed elevated transcriptional activity at 125 m (Supplementary Table S2), suggesting that they were unlikely sinking dead cells. Zinser et al. (2006) showed that in deeper waters (below 75 m) at the western North Atlantic site, a significant fraction of Prochlorococcus population cannot be detected by qPCR probes designed to capture currently known ecotypes, suggesting significant deep populations of Prochlorococcus yet to be identified and characterized. Results of this study suggest the presence of a high-light-like Prochlorococcus population that may be well adapted to the lower euphotic zone, under low-light conditions.
Population transcriptomic analysis of Pelagibacter
As the most abundant heterotrophic bacterial group throughout the ocean water column, Pelagibacter (member of the alphaproteobacteria SAR11 clade) provides a useful model example for how culture-based and metagenomic/metatranscriptomic data can be integrated to study the ecophysiology of wild populations. Subsets of DNA and cDNA reads from all four depths were mapped onto the reference genome of the open ocean Pelagibacter isolate HTCC7211 (see Supplementary Methods). The expression level of annotated protein-coding genes provided clues on the prevailing metabolic activities of Pelagibacter populations at each depth (Figure 5; Supplementary Table S3). Overall, the expression profile of protein-coding genes confirmed the observation based on the rRNA profile (Figure 1) that Pelagibacter cells at 125 m were less transcriptionally active at the time of sampling, compared with their counterparts at 25 m and 75 m. Indeed, ribosomal proteins were among the most highly expressed genes in 25 m and 75 m samples, and most ORFs showed lower expression levels in the 125 m sample.
Genome-wide expression profiles of Pelagibacter-related populations, in all four depths. X-axis shows the arbitrary numbering of ORFs along the genome of Pelagibacter strain HTCC7211. Y-axis scale represents normalized cDNA to DNA ratio (normalized expression level; see Supplementary Methods) for each ORF. Each colored circle in the stem plot represents a given ORF at a given depth.
Nutrient-uptake genes of Pelagibacter, particularly those encoding periplasmic solute-binding proteins of ATP-binding cassette (ABC) families, represented the most abundant class of transcripts (Figure 5). The disproportionally high abundance of transporter genes in Pelagibacter genomes is believed to contribute to their capability of efficiently using a broad variety of substrates (Giovannoni et al., 2005b). In this study we observed high transcriptional levels of solute-binding proteins families 1, 3 and 7 (Figure 5), which involve in the uptake of sugars, polar amino acids and organic polyanions, respectively (Tam and Saier, 1993). Polyamines (for example, spermidine/putrescine), trace elements (for example, selenium) and possible osmolytes (for example, glycine betaine) also seemed to be actively transported. In addition, a few transporter families other than the ABC superfamily were also expressed, including Na+/solute symporter (Ssf family) and tripartite ATP-independent periplasmic (TRAP) dicarboxylate transporter genes for the uptake of mannitol and/or C4-dicarboxylates, which relies on proton motive force rather than ATP hydrolysis. Notably, different expression levels among the four depths were discernible for these transporter genes, potentially a result of substrate availability and preference for Pelagibacter populations residing different depths.
Sowell et al. have observed in Pelagibacter metaproteomes collected from the Sargasso Sea surface water a dominant signal of periplasmic transport proteins for substrates such as phosphate, amino acids, phosphonate and spermidine/putrescine (Sowell et al., 2008). The overall consistent observation that nutrient-uptake transporters were most highly expressed both at transcriptional level (this study) and translational level (Sowell et al., 2008), corroborates the oligotrophic nature of both oceanic sites. However, significant differences in peptide versus transcript expression levels were also apparent among certain categories of transporters. For example, we did not detect gene expression for phosphate and phosphonate transporter genes (pstS and phnD) related to Pelagibacter in our data sets. In fact, no phnD-related sequences were detected in the DNA reads recruited to the Pelagibacter HTCC7211 genome, suggesting phnD gene is absent in most Pelagibacter cells at Station ALOHA. This observation contrasts sharply with that of Sowell et al., reflecting the significant biogeochemical difference between the two oceanic sites (for example, phosphate concentrations at BATS are much lower than that at Station ALOHA (Wu et al., 2000)). The effect of geography-dependent phosphorus limitation seems to be reflected in the gene content of native Prochlorococcus cells (Martiny et al., 2009), as well as other picoplankton populations (Martinez et al., 2010).
HTCC7211-specific genes
It has been well established that genomic plasticity of microbes, reflected by variations in gene content of closely related strains, may facilitate microbial adaptation to their natural habitats (Coleman et al., 2006; Cuadros-Orellana et al., 2007). We compared the genome sequences of two Pelagibacter coastal isolates (strains HTCC1062 and HTCC1002) and the open ocean isolate (HTCC7211, used as reference genome in the genome-centric analysis above), and asked which HTCC7211-specific genes might be highly expressed and thus functionally important in the open ocean environment.
There are 296 HTCC7211-specific genes (see Supplementary Methods), 154 detected in at least one of our metatranscriptomic data sets (Supplementary Figure S7). Two ORFs encoding ABC-type periplasmic solute-binding proteins seemed to be specific to open ocean-dwelling Pelagibacter, and were highly expressed. One ORF encodes a selenium-binding protein, which may contribute to the synthesis of selenoproteins (Zhang and Gladyshev, 2008). The other ORF encodes an extracellular solute-binding protein family 1, which is associated with the uptake of malto-oligosaccharides, multiple sugars, alpha-glycerol phosphate and iron (Tam and Saier, 1993). In addition, the C4-dicarboxylate transport (Dct) system, which relies on extracytoplasmic solute-binding receptors with high specificity and affinity, seemed to be important in oceanic Pelagibacter populations. Not only were four dct operons present in the strain HTCC7211 (as opposed to apparently only one copy in coastal strains HTCC1062 and HTCC1002), but the three HTCC7211-specific dctP paralogues (encoding a periplasmic C4-dicarboxylate-binding protein) were also expressed (Supplementary Figure S7). Dct transporters are secondary carriers that use an electrochemical H+ gradient as the driving force for transport rather than ATP hydrolysis, and allow the uptake of mannitol and/or C4-dicarboxylates like succinate, fumarate and malate, pointing to such organic compounds as important carbon and energy source for oceanic Pelagibacter.
Caveats and challenges
Given the complex, nonlinear relationship between gene expression, protein expression and biochemical function, the transcript profiles need to be carefully interpreted in the context of other supporting data. Transcript abundance will not always correlate directly with cognate protein levels, and the kinetics that couple gene expression to phenotypic manifestation varies among different transcript classes (Steunou et al., 2008). Nonetheless, reasonably good correlation between transcriptomes and proteomes, especially for transcripts and peptides in higher abundance, has been observed in several model organisms (Eymann et al., 2002; Corbin et al., 2003; Scherl et al., 2005). In the Pelagibacter genome-centric analyses reported in this study (Figure 5), we observed considerable overlap between highly abundant transcripts and the most represented peptides previously reported in a SAR11-centric metaproteomic study (Sowell et al., 2008). This general consistency between the population transcriptomes and proteomes of the most abundant and ubiquitous heterotrophic bacteria clade in the open ocean, supports the use of metatranscriptomics to assess inventories of functionally relevant gene families based on their expression levels.
The work reported in this study, along with previous studies (Hewson et al., 2009; Poretsky et al., 2009), also illustrates several challenges for future metatranscriptomic studies. First, because of great diversity found in most natural systems and predominant transcriptional signal of the genes involved in central metabolism and protein synthesis machinery, sequencing depth needs to be greatly expanded (Gifford et al., 2010; Stewart et al., 2010). Our data showed that for one pyrosequencing run on an open ocean bacterioplankton sample, about 66–74% of sequences with a taxonomic assignment belonged to the top two most abundant taxonomic groups (Supplementary Figure S3, lower panel). Thus, the majority of the diversity of the transcript pool was represented by low abundance reads with little statistical confidence, albeit these may well contain important information. As a good demonstration, two technical replicate metatranscriptomic data sets were found to only share 37% of the NCBI-nr reference entries, suggesting the rarefaction curve of functional diversity is far from leveling off (Stewart et al., 2010).
Another challenge is associated with the frequent observation that hypothetical genes are among the most highly expressed genes in the genomes examined (Supplementary Figure S8). Such hypothetical genes are potentially of great relevance to the ecology of host populations in their native environment, but understanding and characterizing unknown functions in these hypothetical ORFs represents a continuing challenge. It has recently been reported that about two thirds of the gene families with unknown functions likely represent very divergent branches of known and well-characterized families (Jaroszewski et al., 2009). Expression patterns in the environment, combined with structure and sequence homology search, provides a starting point for formulating and testing hypotheses about the biological functions of these uncharacterized ORFs. As an example, four highly expressed hypothetical genes, putatively originating from marine crenarchaeal genomes, were annotated to contain putative polycystic kidney disease domain (PF00801). Polycystic kidney disease domains are mostly present on the cell surface, and are involved in protein–protein interactions. Particularly, polycystic kidney disease domains were found predominant in archaeal surface layer proteins that were thought to protect the cell from extreme environments (Jing et al., 2002), or in exported proteins of marine heterotrophic bacteria that may be involved in the binding and degradation of extracellular polymers (carbohydrate and protein) (Zhao et al., 2008). The taxonomic origins and prevalence in community transcriptomes of these polycystic kidney disease domain-containing hypothetical genes now render them reasonable targets for future functional characterizations in planktonic marine Crenarchaea.
Conclusions and future directions
Through analysis of four coupled metagenomic and metatranscriptomic data sets, we have demonstrated that microbial community transcriptomes can be profiled (for abundant microbial populations, at a reasonable coverage), and compared in the context of genomic compositions and ambient environmental conditions. Our results provide insight into: (1) sequence characteristics, such as the uniqueness and vast diversity, of microbial community transcriptomes in the open ocean ecosystem; (2) specific metabolic processes that characterize each of the four habitats investigated; (3) highly expressed gene families, and their putative taxonomic breakdown; and (4) population variability and physiological signals from abundant taxa of the microbial assemblages, inferred via reference genome-centric analyses. Given the great complexity found in the transcriptome of even small genomes (Guell et al., 2009), it must be assumed that we are at present just scratching the surface of the dynamic, complex transcriptional network orchestrated by microbe–microbe and microbe–environment interactions. Future metagenomic and metatranscriptomic surveys at more highly resolved spatial and temporal dimensions will help to provide a more comprehensive picture of microbial functional diversity in natural settings. Subtractive methods, similar to rRNA substraction, might also be explored to remove more abundant transcripts, to better access the low abundance transcript pool at higher statistical significance. Additionally, the application of coupled metagenomics and metatranscriptomics in experimental settings should facilitate more controlled assessment of microbial community responses to environmental changes, and allow simultaneous study of microbial community physiology, population and community dynamics.
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Arrigo KR . (2005). Marine microorganisms and global nutrient cycles. Nature 437: 349–355.
Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP et al. (2000). Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science 289: 1902–1906.
Béjà O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T et al. (2002). Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415: 630–633.
Berube PM, Samudrala R, Stahl DA . (2007). Transcription of all amoC copies is associated with recovery of Nitrosomonas europaea from ammonia starvation. J Bacteriol 189: 3935–3944.
Brzezinski MA . (1988). Vertical-distribution of ammonium in stratified oligotrophic waters. Limnol Oceanogr 33: 1176–1182.
Casciotti KL, Ward BB . (2001). Dissimilatory nitrite reductase genes from autotrophic ammonia-oxidizing bacteria. Appl Environ Microbiol 67: 2213–2221.
Church MJ, Wai B, Karl DM, DeLong EF . (2010). Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol 12: 679–688.
Coleman ML, Chisholm SW . (2007). Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol 15: 398–407.
Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF et al. (2006). Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770.
Corbin RW, Paliy O, Yang F, Shabanowitz J, Platt M, Lyons CE et al. (2003). Toward a protein profile of Escherichia coli: Comparison to its transcription profile. Proc Natl Acad Sci USA 100: 9232–9237.
Cuadros-Orellana S, Martin-Cuadrado AB, Legault B, D’Auria G, Zhaxybayeva O, Papke RT et al. (2007). Genomic plasticity in prokaryotes: the case of the square haloarchaeon. ISME J 1: 235–245.
DeLong EF, Béjà O . (2010). The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol 8: e1000359.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM et al. (2008). Functional metagenomic profiling of nine biomes. Nature 452: 629–632.
Dore JE, Karl DM . (1996). Nitrite distributions and dynamics at Station ALOHA. Deep Sea Res Part II Top Stud Oceanogr 43: 385–402.
Eiler A, Hayakawa DH, Church MJ, Karl DM, Rappe MS . (2009). Dynamics of the SAR11 bacterioplankton lineage in relation to environmental conditions in the oligotrophic North Pacific subtropical gyre. Environ Microbiol 11: 2291–2300.
Eymann C, Homuth G, Scharf C, Hecker M . (2002). Bacillus subtilis functional genomics: Global characterization of the stringent response by proteome and transcriptome analysis. J Bacteriol 184: 2500–2520.
Francis CA, Co EM, Tebo BM . (2001). Enzymatic manganese(II) oxidation by a marine alpha-proteobacterium. Appl Environ Microbiol 67: 4024–4029.
Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW et al. (2008). Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA 105: 3805–3810.
Frigaard NU, Martinez A, Mincer TJ, DeLong EF . (2006). Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 439: 847–850.
Ghai R, Martin-Cuadrado AB, Molto AG, Heredia IG, Cabrera R, Martin J et al. (2010). Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J 4: 1154–1166.
Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA . (2010). Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J; e-pub ahead of print 16 September 2010.
Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL et al. (2005a). Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438: 82–85.
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005b). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245.
Gómez-Consarnau L, Akram N, Lindell K, Pedersen A, Neutze R, Milton DL et al. (2010). Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol 8: e1000358.
Gómez-Consarnau L, Gonzalez JM, Coll-Llado M, Gourdon P, Pascher T, Neutze R et al. (2007). Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445: 210–213.
González JM, Fernandez-Gomez B, Fernandez-Guerra A, Gomez-Consarnau L, Sanchez O, Coll-Llado M et al. (2008). Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp MED152 (Flavobacteria). Proc Natl Acad Sci USA 105: 8724–8729.
Guell M, van Noort V, Yus E, Chen W-H, Leigh-Bell J, Michalodimitrakis K et al. (2009). Transcriptome Complexity in a Genome-Reduced Bacterium. Science 326: 1268–1271.
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM et al. (2006). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaea. PLoS Biol 4: 520–536.
Hewson I, Poretsky RS, Dyhrman ST, Zielinski B, White AE, Tripp HJ et al. (2009). Microbial community gene expression within colonies of the diazotroph, Trichodesmium, from the Southwest Pacific Ocean. ISME J 3: 1286–1300.
Hewson I, Rachel SP, Tripp HJ, Joseph PM, Jonathan PZ . (2010). Spatial patterns and light-driven variation of microbial population gene expression in surface waters of the oligotrophic open ocean. Environ Microbiol 12: 1940–1956.
Huson DH, Auch AF, Qi J, Schuster SC . (2007). MEGAN analysis of metagenomic data. Genome Res 17: 377–386.
Jaroszewski L, Li ZW, Krishna SS, Bakolitsa C, Wooley J, Deacon AM et al. (2009). Exploration of Uncharted Regions of the Protein Universe. PLoS Biol 7: e1000205.
Jing H, Takagi J, Liu JH, Lindgren S, Zhang RG, Joachimiak A et al. (2002). Archaeal surface layer proteins contain beta propeller, PKD, and beta helix domains and are related to metazoan cell surface proteins. Structure 10: 1453–1464.
Karl DM, Lukas R . (1996). The Hawaii Ocean Time-series (HOT) program: Background, rationale and field implementation. Deep Sea Res Part II Top Stud Oceanogr 43: 129–156.
Kolber ZS, Plumley FG, Lang AS, Beatty JT, Blankenship RE, VanDover CL et al. (2001). Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292: 2492–2495.
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Lami R, Cottrell MT, Campbell BJ, Kirchman DL . (2009). Light-dependent growth and proteorhodopsin expression by Flavobacteria and SAR11 in experiments with Delaware coastal waters. Environ Microbiol 11: 3201–3209.
Liu ZZ, DeSantis TZ, Andersen GL, Knight R . (2008). Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 36: e120.
Malmstrom RR, Coe A, Kettler GC, Martiny AC, Frias-Lopez J, Zinser ER et al. (2010). Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J 4: 1252–1264.
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA . (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461: 976–979.
Martin-Cuadrado AB, Lopez-Garcia P, Alba JC, Moreira D, Monticelli L, Strittmatter A et al. (2007). Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. Plos One 2: e914.
Martinez A, Tyson GW, DeLong EF . (2010). Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol 12: 222–238.
Martiny AC, Huang Y, Li WZ . (2009). Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol 11: 1340–1347.
McCarren J, DeLong EF . (2007). Proteorhodopsin photosystem gene clusters exhibit co-evolutionary trends and shared ancestry among diverse marine microbial phyla. Environ Microbiol 9: 846–858.
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M et al. (2008). The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9: 386.
Moore LR, Chisholm SW . (1999). Photophysiology of the marine cyanobacterium Prochlorococcus: Ecotypic differences among cultured isolates. Limnol Oceanogr 44: 628–638.
Moore LR, Post AF, Rocap G, Chisholm SW . (2002). Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol Oceanogr 47: 989–996.
Oz A, Sabehi G, Koblizek M, Massana R, Beja O . (2005). Roseobacter-like bacteria in Red and Mediterranean Sea aerobic anoxygenic photosynthetic populations. Appl Environ Microbiol 71: 344–353.
Pham VD, Konstantinidis KT, Palden T, DeLong EF . (2008). Phylogenetic analyses of ribosomal DNA-containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre. Environ Microbiol 10: 2313–2330.
Poretsky RS, Hewson I, Sun SL, Allen AE, Zehr JP, Moran MA . (2009). Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol 11: 1358–1375.
Riedel T, Tomasch J, Buchholz I, Jacobs J, Kollenberg M, Gerdts G et al. (2010). Constitutive expression of the proteorhodopsin gene by a flavobacterium strain representative of the proteorhodopsin-producing microbial community in the North Sea. Appl Environ Microbiol 76: 3187–3197.
Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA et al. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424: 1042–1047.
Rodriguez-Brito B, Rohwer F, Edwards RA . (2006). An application of statistics to comparative metagenomics. BMC Bioinformatics 7: 162.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5: 398–431.
Scherl A, FranÁois P, Bento M, Deshusses JM, Charbonnier Y, Converset V et al. (2005). Correlation of proteomic and transcriptomic profiles of Staphylococcus aureus during the post-exponential phase of growth. J Microbiol Methods 60: 247–257.
Shi Y, Tyson GW, DeLong EF . (2009). Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature 459: 266–269.
Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF et al. (2008). Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J 3: 93–105.
Steunou AS, Jensen SI, Brecht E, Becraft ED, Bateson MM, Kilian O et al. (2008). Regulation of nif gene expression and the energetics of N-2 fixation over the diel cycle in a hot spring microbial mat. ISME J 2: 364–378.
Stewart FJ, Ottesen EA, DeLong EF . (2010). Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J 4: 896–907.
Tam R, Saier MH . (1993). Structural, Functional, and Evolutionary Relationships among Extracellular Solute-Binding Receptors of Bacteria. Microbiol Rev 57: 320–346.
Temperton B, Field D, Oliver A, Tiwari B, Muhling M, Joint I et al. (2009). Bias in assessments of marine microbial biodiversity in fosmid libraries as evaluated by pyrosequencing. ISME J 3: 792–796.
Van Mooy BAS, Devol AH . (2008). Assessing nutrient limitation of Prochlorococcus in the North Pacific subtropical gyre by using an RNA capture method. Limnol Oceanogr 53: 78–88.
Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ et al. (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA 107: 8818–8823.
Wu JF, Sunda W, Boyle EA, Karl DM . (2000). Phosphate depletion in the western North Atlantic Ocean. Science 289: 759–762.
Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K et al. (2007). The sorcerer II global ocean sampling expedition: Expanding the universe of protein families. PLoS Biol 5: 432–466.
Zhang Y, Gladyshev VN . (2008). Trends in selenium utilization in marine microbial world revealed through the analysis of the Global Ocean Sampling (GOS) Project. PLoS Genet 4: e1000095.
Zhao G-Y, Chen X-L, Zhao H-L, Xie B-B, Zhou B-C, Zhang Y-Z . (2008). Hydrolysis of insoluble collagen by deseasin MCP-01 from deep-sea Pseudoalteromonas sp. SM9913. J Biol Chem 283: 36100–36107.
Zinser ER, Coe A, Johnson ZI, Martiny AC, Fuller NJ, Scanlan DJ et al. (2006). Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl Environ Microbiol 72: 723–732.
Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, Coleman ML et al. (2009). Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS ONE 4: e5135.
Acknowledgements
We thank the captain and crew of the R/V Kilo Moana for facilitating sample collection. We also thank Stephan Schuster for collaboration on pyrosequencing. We are grateful to the J Craig Venter Institute, and the Gordon and Betty Moore Foundation for the microbial genome sequences. This work was supported by a grant from the Gordon and Betty Moore Foundation (EFD), the Office of Science (BER) US Department of Energy (EFD), and NSF Science and Technology Center Award EF0424599. This article is a contribution from the National Science Foundation Science and Technology Center for Microbial Oceanography: Research and Education (C-MORE).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Shi, Y., Tyson, G., Eppley, J. et al. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J 5, 999–1013 (2011). https://doi.org/10.1038/ismej.2010.189
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.189
Keywords
This article is cited by
-
Transcriptomic Insights into Archaeal Nitrification in the Amundsen Sea Polynya, Antarctica
Journal of Microbiology (2023)
-
Growth-stage-related shifts in diatom endometabolome composition set the stage for bacterial heterotrophy
ISME Communications (2022)
-
The contribution of nirK gene-containing thaumarchaea to denitrification and N2O production across coastal sediment and terrestrial ecosystems
Journal of Soils and Sediments (2022)
-
Global ecotypes in the ubiquitous marine clade SAR86
The ISME Journal (2020)
-
Species specific gene expression dynamics during harmful algal blooms
Scientific Reports (2020)