Abstract
To obtain evidence for blood pressure (BP) trait association, we conducted an association study of selected candidate gene variants. In Japan, a total of 19 426 individuals underwent testing for genetic associations with systolic BP (SBP)/diastolic BP (DBP) and 9271 individuals (3460 cases and 5811 controls) underwent testing for genetic associations with dichotomous hypertension. Association with seven notable candidate genes was tested, namely, ACE, ADD1, ADRB2, AGT, CYP11B2, GNB3 and NOS3, followed by a joint meta-analysis involving previously reported multi-study populations, including >20 000 individuals (for SBP/DBP) and >17 000 individuals (for hypertension). BP trait associations at two loci (AGT rs699 and CYP11B2 rs1799998) were consistently replicated in the Japanese association study and joint meta-analysis involving the populations described above. Hypertension association reached genome-wide significance for the two variants, specifically, P=7.3 × 10−10 for AGT rs699 and P=3.9 × 10−8 for CYP11B2 rs1799998. In our study panels, the most significant association was found for CYP11B2 rs1799998 with all three BP traits: P=1.5 × 10−5 for SBP, P=1.8 × 10−5 for DBP and P=2.3 × 10−5 for hypertension. A suggestive association with SBP (P=0.042), DBP (P=0.01) and hypertension (P=1.4 × 10−5) was also detected for ACE rs4340 (a proxy for ACE D/I polymorphism) in the joint meta-analysis. Our data provide evidence for true BP trait associations with two candidate gene variants. These variants were not identified in the previous genome-wide association studies, presumably because they did not reach a given threshold in the discovery stage. Thus, certain variants in genes with clinical and physiological relevance are likely to account for a portion of BP variance in the general population and are worth following up via a target gene approach.
Similar content being viewed by others
Introduction
Despite the high heritability estimate (30–50%) of blood pressure (BP) and hypertension,1, 2, 3 to date, associations among candidate genes and these traits have rarely been convincingly demonstrated. With recent technological advances, genome-wide association (GWA) studies, which interrogate a large proportion of the common genetic variants (that is, by genotyping hundreds of thousands of common single-nucleotide polymorphisms (SNPs)) throughout the genome, have allowed the investigation of genetic susceptibility to these traits without a priori hypotheses regarding the underlying pathophysiology.3 In the first wave of GWA studies,4, 5, 6, 7, 8, 9 few studies appeared to identify loci associated with BP traits at a genome-wide significance level (P<5 × 10−8) when adjusted for multiple testing. In 2009, two large-scale meta-analyses of GWA studies from the Global BP Genetics (Global BPgen) and Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) consortia identified a total of 13 independent loci significantly associated with BP.10, 11 Subsequently, meta-analyses of GWA studies in East Asians12 and in a combined population of Global BPgen and CHARGE13 further identified a total of 21 additional loci. Most of these associations are in or near genes that had not been suspected of involvement in BP regulation. The sample size of genome-wide exploration in these meta-analyses (n=19608–69395)10, 11, 12, 13 was much larger than that of the previous GWA studies of BP traits,4, 5, 6, 7, 8, 9 indicating that very large samples are required to identify molecular variants with modest genetic effects on BP variation in the general population. In addition, continuous BP traits are implied to have greater power than dichotomous hypertension traits, at least for the genetic loci commonly affecting both types of traits; that is, the chances of obtaining significant results are higher with a continuous trait, either systolic (SBP) or diastolic (DBP) BP.3
Therefore, we conducted a large-scale association study of eight variants from seven notable candidate genes, namely, ACE (angiotensin-converting enzyme), ADD1 (adducin 1), ADRB2 (beta-2 adrenergic receptor), AGT (angiotensinogen), CYP11B2 (aldosterone synthase), GNB3 (G protein, beta-3 subunit), and NOS3 (nitric oxide synthase 3), in the Japanese (n=21851 in total). The individual contribution of each of these genes to BP variation and/or hypertension in the general population has not been thoroughly elucidated, although a number of studies have repeatedly investigated them as candidate genes.14, 15, 16, 17, 18, 19, 20 Moreover, we performed meta-analyses of the current data on the Japanese and the data from previous studies to verify consistency and to increase the statistical power of SNP trait associations.
Methods
Study population
Detailed characteristics of the subjects analyzed in each study panel are described in the Supplementary Information and Table 1. Our genetic studies for BP and hypertension were originally organized as part of an ongoing GWA study for cardiometabolic disorders among Japanese subjects in the Cardiometabolic Genome Epidemiology (CAGE) network.21 Three CAGE study panels were used for the association analysis with continuous BP traits, SBP and DBP, and dichotomous hypertension. The CAGE study panel 1 comprised 1526 Japanese subjects who were genotyped for a GWA scan and included 403 cases and 452 controls. The CAGE study panel 2 involved 5331 Japanese subjects (hereafter known as the Amagasaki cohort) consecutively enrolled in a population-based setting as described elsewhere22 and 2425 subjects primarily recruited for the case–control study; in total, 1750 cases and 1776 controls were selected for case–control comparison from panel 2 (see the footnote to Table 1). The CAGE study panel 3 also comprised 12 569 subjects (hereafter called the Fukuoka cohort) who were randomly selected from the general Japanese population.23 From panel 3, 1307 cases and 3583 control subjects were further chosen for case–control comparison. All participants in these studies provided written informed consent, and the local ethics committees approved the protocols.
BP levels were classified according to the Japanese Society of Hypertension Guidelines for the Management of Hypertension (JSH2009) as described elsewhere.24 Cases (except participants in the Fukuoka cohort) were enrolled from clinical practices or during annual medical checkups at medical institutions and university hospitals in accordance with uniformly defined criteria. These criteria included (i) SBP⩾160 mm Hg and/or DBP⩾100 mm Hg for untreated subjects (Grade II and III hypertension in JSH2009); (ii) currently receiving chronic antihypertensive treatment(s); (iii) no secondary form of hypertension as evaluated by an extensive workup, including serum creatinine and electrolytes, chest radiography, electrocardiogram, urinalysis, and other hematological screening tests; and (iv) age of hypertension onset⩽60 years. Normotensive controls were defined as follows: (i) SBP<130 mm Hg and DBP<85 mm Hg without antihypertensive treatments (normal BP in JSH2009) and (ii) age⩾50 years.
SNP genotyping
In panel 1, genotyping was performed with the Infinium HumanHap550 BeadArray system (Illumina, San Diego, CA, USA), interrogating 555352 SNPs as previously described.21 The lambda value for the genomic control was 1.00 to 1.02, indicating the absence of systematic confounding, such as population stratification, in our GWA study samples (panel 1). With the exception of four variants (which were on the HumanHap550 BeadArray) in panel 1, samples were genotyped using the TaqMan assay for eight SNPs from seven unique candidate genes (Table 2).14, 15, 16, 17, 18, 19, 20 These genes included AGT (rs699, M235T), ADD1 (rs4961, G460W), ADRB2 (rs1042713, R16G; rs1042714, E27Q), NOS3 (rs1799983, D298E), CYP11B2 (rs1799998, −344C/T), GNB3 (rs5443, C825T) and ACE (rs4341,25 a proxy SNP for D/I polymorphism). The genotypic distribution of all tested SNPs was in Hardy–Weinberg equilibrium (P>10−3). We obtained successful genotyping call rates of >99% for the entire characterized sample.
Statistical Analyses
SNP association analysis
The SNPs were tested for associations with BP and hypertension using linear regression analysis and the Cochran–Armitage trend test, respectively. In the linear regression models, we adjusted continuous SBP and DBP for age, age squared, and body mass index stratified by sex (all panels) and sample enrollment site (panel 1 only). For individuals receiving antihypertensive therapies, BP was imputed by adding 15 mm Hg and 10 mm Hg to SBP and DBP, respectively.10, 21, 26 One-tailed P<0.006≈0.05/8 (two-tailed P<0.012) was considered significant with Bonferroni’s correction for multiple testing; for an association to be considered significant, it had to involve the same risk allele as that previously reported and was assessed accordingly with the one-tailed test. For both the quantitative trait analysis and case–control study, the genetic effects estimated in each of the Japanese panels were combined with the inverse variance method (Supplementary Information). We used PLINK, the R software, and the rmeta and meta packages to test for the associations.
Meta-analysis
First, the previous multi-ethnic meta-analysis studies of each candidate gene variant were identified in the PubMed database and included in the current meta-analysis. When >1 meta-analysis studies were identified per variant, only the latest and most comprehensive one was used. Next, we identified additional studies published after the chosen meta-analysis studies by extended computer-based searches of PubMed. In terms of BP association, meta-analyses had not been published, except for the AGT and NOS3 variants.17, 20 In these searches, we used the following search criteria: (‘gene name’ or ‘gene symbol (official and alias)’) and ‘polymorphism’ and (‘hypertension’ or ‘blood pressure’). The retrieved studies were read in their entirety to assess their appropriateness for inclusion in the meta-analysis; eligible studies were included if they fulfilled the following criteria: (i) evaluated one of the eight variants using either a case–control or cross-sectional design; (ii) contained non-overlapping data; (iii) showed the relevant data to calculate the effect size and its variance; and (iv) involved a sample size >1000 (for either case–control or quantitative trait analysis). We included the fourth criterion to reduce possible fluctuation in genetic effects due to small sample sizes, that is, publication bias, which seemed to be particularly pertinent to studies that were published after the meta-analysis report(s) of a given variant.
The main endpoints were the influence of candidate gene variants on BP as continuous traits (SBP and DBP) and the risk of hypertension as a categorical variable. The strength of association with hypertension was measured using an odds ratio (OR) and its 95% confidence interval (CI). Heterogeneity among the study results was formally assessed using Cochran’s Q-statistic or Woolf’s test, with P<0.05 being considered to be significant.
Results
Replication of eight variants from seven candidate genes in Japanese
From the three CAGE study panels, a total of 19 426 individuals and 9271 individuals (3460 cases and 5811 controls) were subjected to quantitative trait analysis and a case–control study, respectively (Table 1). Among the eight variants tested in the Japanese, rs1799998 in CYP11B2 had the most significant association with all three BP traits: β=0.91 mm Hg, P=1.5 × 10−5 for SBP; β=0.53 mm Hg, P=1.8 × 10−5 for DBP; and OR=1.15 (95% CI, 1.08–1.23), P=2.3 × 10−5 for hypertension (Table 2). A significant association (two-tailed P <0.012) was also detected for rs699 in AGT with SBP (β=0.78 mm Hg, P=0.002) and DBP (β=0.49 mm Hg, P=0.001), whereas a borderline association was replicated for this variant with hypertension (OR=1.09 (95% CI, 1.01–1.19), P=0.033). For the two SNPs mentioned above, the major alleles—T of rs1799998 and C of rs699—were associated with elevated levels of SBP and DBP and increased risks of hypertension in accordance with the previous meta-analysis findings.17, 18
Upon examining the association results separately by panel, we found that the direction and strength of the associations with BP traits were almost concordant for the two SNPs; that is, CYP11B2 rs1799998 and AGT rs699 (Supplementary Table 1). In addition, we found significant associations of ADRB2 rs1042713 and ACE rs4341 with SBP (P=0.004 for rs4341), DBP (P=0.005 for rs1042713 and P=0.0118 for rs4341), and/or hypertension (P=0.008 for rs1042713 and P=0.002 for rs4341) in the CAGE study panel 3, in directions that were consistent with those of previous studies;16, 27, 28 however, these associations were not replicable in panel 1 and panel 2 (Supplementary Table 1).
Joint meta-analysis for seven candidate genes
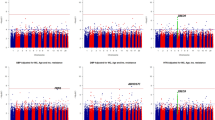
In the PubMed database, we identified multi-ethnic meta-analyses of BP association for two (of eight tested) SNPs and those of hypertension association for eight SNPs (Supplementary Tables 2 and 3).14, 15, 16, 17, 18, 19, 20 Compared with Japanese samples genotyped in this study, the relative increase in joint meta-analysis samples appeared to be modest (that is, 5–40%) at all loci except AGT rs699 for SBP/DBP association, whereas this increase was prominent (that is, 105–586%) for hypertension association, thereby providing the joint meta-analysis, in particular, for case–control study (5757–17399 cases vs. 11240–23338 controls) with reasonable power. For SBP and DBP, there were significant associations (two-tailed P<0.012, putative risk-allele homozygote vs. non-risk-allele homozygote) with AGT rs699 (P=0.001 for SBP), CYP11B2 rs1799998 (P=2.0 × 10−6 for SBP; P=3.1 × 10−6 for DBP), and ACE rs4340 (P=0.01 for DBP) (Supplementary Table 2). For hypertension, conversely, reproducible association was detected at AGT rs699 (P=7.3 × 10−10), CYP11B2 rs1799998 (P=3.9 × 10−8), and ACE rs4340 (P=1.4 × 10−5) (Figures 1 and 2 and Supplementary Table 3). In this study, heterogeneity analyses in multi-study populations identified some heterogeneity at ACE rs4340 (Phetero=0.019, DD vs. II).
Forest plots showing the association with BP traits (SBP, DBP and hypertension) in the studies included in the present meta-analysis (individually and in meta-analysis) for the two SNPs robustly associated (P<5 × 10−8) with hypertension. See also Supplementary Tables 2 and 3. For dichotomous hypertension, the effect size (OR and 95% CI) is depicted for risk/risk vs. non-risk/non-risk homozygous subjects
Forest plots showing the association with hypertension in the studies included in the present meta-analysis (individually and in meta-analysis) for the five SNPs tested for hypertension association, except AGT M235T, CYP11B2 -344T/C and GNB3 C825T. The corresponding data (the effect size for risk/risk vs. non-risk/non-risk homozygous subjects) are not available for GNB3 C825T.
Discussion
We examined evidence for the association of three BP traits (SBP, DBP and hypertension) with seven notable candidate genes assumed to have etiological roles in BP regulation. We conducted a large-scale replication study by testing eight previously reported variants in a total of >21 000 Japanese individuals from the CAGE network followed by joint meta-analyses involving an additional sample of 876–17492 individuals for BP association and 10 133–54764 individuals (cases plus controls) for hypertension association. Notably, BP trait associations at two loci (AGT and CYP11B2) were consistently replicated in both a Japanese association study in the CAGE network and the joint meta-analysis of multi-study populations. In addition, a suggestive association with DBP and hypertension was detected for ACE rs4340 in the joint meta-analysis. For the first time, our data demonstrate that an association with hypertension of two candidate variants, namely, AGT rs699 and CYP11B2 rs1799998, reached genome-wide significance (P<5.0 × 10−8), thereby confirming the validity of the target gene approach, which has been an issue of debate, given the complex nature of hypertension genetics.
The necessity of large study samples (roughly >10 000 individuals) has been recognized in identifying genes associated with BP traits following the great success of consortium-based GWA studies.10, 11, 12 Until those studies were conducted, people had raised general concerns about the utility of genetic association studies for BP traits. Effect sizes of each risk allele have turned out to be modest for the variants previously identified in the GWA studies using common SNPs;10, 11, 12, 29 1 mm Hg for SBP and 0.5 mm Hg for DBP, on average, and 5–20% increase in dichotomous disease risk. On the basis of these findings, it appears that solid evidence of genetic association with BP traits is attainable when conducting large-scale association studies. The present study did enable us accordingly to determine association signals at several candidate SNP loci.
Components of the renin–angiotensin system (RAS) were most repeatedly investigated as candidate genes for essential hypertension. In particular, suggestive or significant association was detected in certain studies for the angiotensinogen M235T (AGT rs699),17, 30 angiotensin I-converting enzyme D/I (a proxy for ACE rs4340),27, 28 and aldosterone synthase C-344T (CYP11B2 rs1799998)18, 31 polymorphisms, although the results remained largely inconclusive.32, 33, 34, 35 Besides reproducible associations in the present study (Figure 1), a recent, large-scale association study of hypertension drug target genes in populations of European descent (involving a total of 86 588 individuals)36 confirmed BP trait associations with two candidate genes—AGT and ACE—in the RAS. Using the GWA scan data without focusing on particular variants selected a priori, the authors reported that rs2004776 in AGT (P=3.8 × 10−6 for SBP, P=5.0 × 10−8 for DBP, and P=3.7 × 10−7 for hypertension) and rs4305 in ACE (P=3.0 × 10−5 for SBP, P=4.6 × 10−4 for DBP, and P=6.0 × 10−5 for hypertension) showed prominent association signals in each gene.36 Two pairs of SNPs—rs2004776 and rs699 in AGT (r2=0.362 in HapMap JPT/CHB; r2=0.49 in HapMap CEU) and rs4305 and rs4341 in ACE (r2=0.80 in HapMap JPT/CHB; r2=0.48 in HapMap CEU)—exhibit moderate to high levels of linkage disequilibrium in East Asian and European populations, suggesting a potential common link between the studies. Another locus in the RAS demonstrating significant BP trait association in the present study is CYP11B2 rs1799998. This SNP (T to C substitution in the 5′-distal promoter region of the gene at position –344) disrupts a putative recognition site for the transcriptional factor, steroidogenic factor 1, and is considered to have functional relevance to gene expression,37 despite conflicting reports about its association with hypertension. One previous meta-analysis involving 5343 cases and 5882 controls reported a significant association with hypertension, OR=0.834 (95% CI, 0.76–0.914) for CC vs. TT, P<0.0001.18 By further increasing the sample size, hypertension association of CYP11B2 rs1799998 reached genome-wide significance with an almost equivalent effect size (OR=0.81 for non-risk alleles, P=3.9 × 10−8), and BP association was also found to be highly significant (P=2.0 × 10−6 for SBP and P=3.1 × 10−6 for DBP) in our meta-analysis (Supplementary Tables 2 and 3).
The strength of the current study lies in its extensive, quantitative trait analysis conducted in two general Japanese populations, specifically, the Amagasaki (n=5331, CAGE panel 2) and Fukuoka (n=12569, CAGE panel 3) cohorts, in addition to the GWA study samples (n=1526, CAGE panel 1), which have been validated for the absence of systematic confounding such as population stratification (see Methods) with genome-wide SNP genotyping data.21 These cohorts are among the largest reported to date for quantitative trait analysis of BP (Supplementary Table 2) and hypertension case–control studies (Supplementary Table 3). Nevertheless, given the relatively modest effect sizes for BP/hypertension susceptibility loci, statistical power may not be sufficient to robustly confirm or refute the BP trait associations in the present study (Figure 2 and Supplementary Table 4). Moreover, several limitations of the use of continuous BP traits in the association study must be noted. For instance, we applied treatment adjustments to impute expected baseline effects; that is, the individuals receiving antihypertensive therapies were imputed to have 15 mm Hg higher SBP and 10 mm Hg higher DBP than the observed measurements. Although this imputation has been shown to improve statistical power as a whole,26 it could lead to over- or underadjustment at the individual level, depending on the treatment intensity. The confounding effects of non-genetic factors (for example, obesity, sex, age and lifestyle factors such as diet) may also result in inter-individual variation in BP measurements and hamper the identification of true association signals to a substantial extent,21 although determining the effects of these factors is beyond the scope of the present study.
Apart from a targeted, candidate gene approach,36 neither of the two loci (AGT and CYP11B2) has been reported to show genome-wide significant association with BP traits via GWA scans. Because AGT rs699 and CYP11B2 rs1799998 themselves and/or their proxies are included in the list of SNPs that are assayed by most of the GWA scan platforms, it is unlikely that these SNPs have failed to be tested for BP trait associations in the previous GWA studies.4, 5, 6, 7, 8, 9, 10, 11, 12, 13 Rather, it is probable that the associations were not followed up in the multi-staged screening approach, principally because their statistical significance in the discovery stage did not attain a given threshold (for example, P<1 × 10−5). This value may not indicate the lack of modest association at these loci, as we reported in the present study (Figure 3). During the preparation of our manuscript, a multi-staged GWA study of hypertension reported a new susceptibility locus (rs3918226) in the promoter region of NOS3 in populations of European descent;38 this SNP does not appear to be polymorphic in East Asians (according to HapMap JPT/CHB data), although further examination is warranted.
Relationship of two genome-wide significant loci (two loci on the left hand) and those previously reported21 (seven loci on the right hand, which are presented here for the purpose of comparability of effect sizes) for SBP, DBP and hypertension. Bars at each locus show the effects of each variant on continuous SBP and DBP and on the OR for dichotomous hypertension compared with normotension. For comparability, SBP and DBP effects are shown on the s.d. scale (SBP s.d.=22.2 mm Hg, DBP s.d.=12.7 mm Hg). In addition, for comparability, the OR for hypertension is scaled to have the same average level as BP over the nine loci. Whiskers indicate standard error. Alleles are coded as shown in Table 2 and in the previous report.21 See Supplementary Tables 2 and 3 for the association with BP traits at AGT rs699 (M235T) and CYP11B2 rs1799998 (−344T/C). A full color version of this figure is available at the Hypertension Research journal online.
In the present study, we chose seven gene variants based on the existing biological knowledge of the genetics of hypertension. In addition to studying such ‘biologically plausible candidate genes’, the target gene approach should be used to test suggestive associations with ‘positional candidate genes’, such as those genes (for example, ADD2 and M6PR) that we previously reported in a genome-wide exploratory approach,5 which was, by current standards, modest with regard to sample size.
In summary, our data provide evidence for true BP trait associations with two candidate gene variants that were not identified in previous GWA studies.4, 5, 6, 7, 8, 9, 10, 11, 12, 13 Although the effect sizes could be rather modest, certain variants in genes with clinical and physiological relevance are likely to account for a proportion of the BP variance in the general population and are worth further investigation via the target gene approach to complement GWA scans.
References
Mitchell GF, DeStefano AL, Larson MG, Benjamin EJ, Chen MH, Vasan RS, Vita JA, Levy D . Heritability and a genome-wide linkage scan for arterial stiffness, wave reflection, and mean arterial pressure: the Framingham heart study. Circulation 2005; 112: 194–199.
Pilia G, Chen WM, Scuteri A, Orrú M, Albai G, Dei M, Lai S, Usala G, Lai M, Loi P, Mameli C, Vacca L, Deiana M, Olla N, Masala M, Cao A, Najjar SS, Terracciano A, Nedorezov T, Sharov A, Zonderman AB, Abecasis GR, Costa P, Lakatta E, Schlessinger D . Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet 2006; 2: e132.
Ehret GB . Genome-wide association studies: contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep 2010; 12: 17–25.
Yang HC, Liang YJ, Wu YL, Chung CM, Chiang KM, Ho HY, Ting CT, Lin TH, Sheu SH, Tsai WC, Chen JH, Leu HB, Yin WH, Chiu TY, Chen CI, Fann CS, Wu JY, Lin TN, Lin SJ, Chen YT, Chen JW, Pan WH . Genome-wide association study of young-onset hypertension in the Han Chinese population of Taiwan. PLoS One 2009; 4: e5459.
Kato N, Miyata T, Tabara Y, Katsuya T, Yanai K, Hanada H, Kamide K, Nakura J, Kohara K, Takeuchi F, Mano H, Yasunami M, Kimura A, Kita Y, Ueshima H, Nakayama T, Soma M, Hata A, Fujioka A, Kawano Y, Nakao K, Sekine A, Yoshida T, Nakamura Y, Saruta T, Ogihara T, Sugano S, Miki T, Tomoike H . High-density association study and nomination of susceptibility genes for hypertension in the Japanese National Project. Hum Mol Genet 2008; 17: 617–627.
Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, Rotimi C . A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet 2009; 5: e1000564.
Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, Vasan RS, Mitchell GF . Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet 2007; 8: S3.
Org E, Eyheramendy S, Juhanson P, Gieger C, Lichtner P, Klopp N, Veldre G, Döring A, Viigimaa M, Sõber S, Tomberg K, Eckstein G, KORA Kelgo P, Rebane T, Shaw-Hawkins S, Howard P, Onipinla A, Dobson RJ, Newhouse SJ, Brown M, Dominiczak A, Connell J, Samani N, Farrall M, BRIGHT-Caulfield MJ, Munroe PB, Illig T, Wichmann HE, Meitinger T, Laan M . Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet 2009; 18: 2288–2296.
Wang Y, O'Connell JR, McArdle PF, Wade JB, Dorff SE, Shah SJ, Shi X, Pan L, Rampersaud E, Shen H, Kim JD, Subramanya AR, Steinle NI, Parsa A, Ober CC, Welling PA, Chakravarti A, Weder AB, Cooper RS, Mitchell BD, Shuldiner AR, Chang YP . Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci USA 2009; 106: 226–231.
Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL Wellcome Trust Case Control Consortium Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Döring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O'Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O'Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvänen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dörr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Völker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Völzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB . Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet 2009; 41: 666–676.
Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Köttgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O'Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM . Genome-wide association study of blood pressure and hypertension. Nat Genet 2009; 41: 677–687.
Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen CH, Zhang Y, Yamamoto K, Katsuya T, Yokota M, Kim YJ, Ong RT, Nabika T, Gu D, Chang LC, Kokubo Y, Huang W, Ohnaka K, Yamori Y, Nakashima E, Jaquish CE, Lee JY, Seielstad M, Isono M, Hixson JE, Chen YT, Miki T, Zhou X, Sugiyama T, Jeon JP, Liu JJ, Takayanagi R, Kim SS, Aung T, Sung YJ, Zhang X, Wong TY, Han BG, Kobayashi S, Ogihara T, Zhu D, Iwai N, Wu JY, Teo YY, Tai ES, Cho YS, He J . Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet 2011; 43: 531–538.
International Consortium for Blood Pressure Genome-Wide Association Studies Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011; 478: 103–109.
Staessen JA, Wang JG, Ginocchio G, Petrov V, Saavedra AP, Soubrier F, Vlietinck R, Fagard R . The deletion/insertion polymorphism of the angiotensin converting enzyme gene and cardiovascular-renal risk. J Hypertens 1997; 15: 1579–1592.
Niu WQ, Zhang Y, Ji KD, Gao PJ, Zhu DL . Lack of association between alpha-adducin G460W polymorphism and hypertension: evidence from a case-control study and a meta-analysis. J Hum Hypertens 2010; 24: 467–474.
Lou Y, Liu J, Huang Y, Liu J, Wang Z, Liu Y, Li Z, Li Y, Xie Y, Wen S . A46G and C79G polymorphisms in the β2-adrenergic receptor gene (ADRB2) and essential hypertension risk: a meta-analysis. Hypertens Res 2010; 33: 1114–1123.
Sethi AA, Nordestgaard BG, Tybjaerg-Hansen A . Angiotensinogen gene polymorphism, plasma angiotensinogen, and risk of hypertension and ischemic heart disease: a meta-analysis. Arterioscler Thromb Vasc Biol 2003; 23: 1269–1275.
Sookoian S, Gianotti TF, González CD, Pirola CJ . Association of the C-344T aldosterone synthase gene variant with essential hypertension: a meta-analysis. J Hypertens 2007; 25: 5–13.
Bagos PG, Elefsinioti AL, Nikolopoulos GK, Hamodrakas SJ . The GNB3 C825T polymorphism and essential hypertension: a meta-analysis of 34 studies including 14,094 cases and 17,760 controls. J Hypertens 2007; 25: 487–500.
Pereira TV, Rudnicki M, Cheung BM, Baum L, Yamada Y, Oliveira PS, Pereira AC, Krieger JE . Three endothelial nitric oxide (NOS3) gene polymorphisms in hypertensive and normotensive individuals: meta-analysis of 53 studies reveals evidence of publication bias. J Hypertens 2007; 25: 1763–1774.
Takeuchi F, Isono M, Katsuya T, Yamamoto K, Yokota M, Sugiyama T, Nabika T, Fujioka A, Ohnaka K, Asano H, Yamori Y, Yamaguchi S, Kobayashi S, Takayanagi R, Ogihara T, Kato N . Blood pressure and hypertension are associated with 7 loci in the Japanese population. Circulation 2010; 121: 2302–2309.
Tsuchihashi-Makaya M, Serizawa M, Yanai K, Katsuya T, Takeuchi F, Fujioka A, Yamori Y, Ogihara T, Kato N . Gene-environmental interaction regarding alcohol-metabolizing enzymes in the Japanese general population. Hypertens Res 2009; 32: 207–213.
Nanri A, Yoshida D, Yamaji T, Mizoue T, Takayanagi R, Kono S . Dietary patterns and C-reactive protein in Japanese men and women. Am J Clin Nutr 2008; 87: 1488–1496.
Ogihara T, Kikuchi K, Matsuoka H, Fujita T, Higaki J, Horiuchi M, Imai Y, Imaizumi T, Ito S, Iwao H, Kario K, Kawano Y, Kim-Mitsuyama S, Kimura G, Matsubara H, Matsuura H, Naruse M, Saito I, Shimada K, Shimamoto K, Suzuki H, Takishita S, Tanahashi N, Tsuchihashi T, Uchiyama M, Ueda S, Ueshima H, Umemura S, Ishimitsu T, Rakugi H . The Japanese society of hypertension guidelines for the management of hypertension (JSH 2009). Hypertens Res 2009; 32: 3–107.
Tanaka C, Kamide K, Takiuchi S, Miwa Y, Yoshii M, Kawano Y, Miyata T . An alternative fast and convenient genotyping method for the screening of angiotensin converting enzyme gene polymorphisms. Hypertens Res 2003; 26: 301–306.
Tobin MD, Sheehan NA, Scurrah KJ, Burton PR . Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med 2005; 24: 2911–2935.
O'Donnell CJ, Lindpaintner K, Larson MG, Rao VS, Ordovas JM, Schaefer EJ, Myers RH, Levy D . Evidence for association and genetic linkage of the angiotensin-converting enzyme locus with hypertension and blood pressure in men but not women in the Framingham Heart Study. Circulation 1998; 97: 1766–1772.
Higaki J, Baba S, Katsuya T, Sato N, Ishikawa K, Mannami T, Ogata J, Ogihara T . Deletion allele of angiotensin-converting enzyme gene increases risk of essential hypertension in Japanese men: the Suita Study. Circulation 2000; 101: 2060–2065.
Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, Cha SH, Kim JW, Han BG, Min H, Ahn Y, Park MS, Han HR, Jang HY, Cho EY, Lee JE, Cho NH, Shin C, Park T, Park JW, Lee JK, Cardon L, Clarke G, McCarthy MI, Lee JY, Lee JK, Oh B, Kim HL . A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet 2009; 41: 527–534.
van Rijn MJ, Bos MJ, Isaacs A, Yazdanpanah M, Arias-Vásquez A, Stricker BH, Klungel OH, Oostra BA, Koudstaal PJ, Witteman JC, Hofman A, Breteler MM, van Duijn CM . Polymorphisms of the renin-angiotensin system are associated with blood pressure, atherosclerosis and cerebral white matter pathology. J Neurol Neurosurg Psychiatry 2007; 78: 1083–1087.
Yamada Y, Ando F, Shimokata H . Association of gene polymorphisms with blood pressure and the prevalence of hypertension in community-dwelling Japanese individuals. Int J Mol Med 2007; 19: 675–683.
Hahntow IN, Mairuhu G, van Valkengoed IG, Koopmans RP, Michel MC . Are ‘functionally related polymorphisms’ of renin-angiotensin-aldosterone system gene polymorphisms associated with hypertension? BMC Cardiovasc Disord 2010; 10: 23.
Gjesing AP, Sparsø T, Borch-Johnsen K, Jørgensen T, Pedersen O, Hansen T, Olsen NV . No consistent effect of ADRB2 haplotypes on obesity, hypertension and quantitative traits of body fatness and blood pressure among 6,514 adult Danes. PLoS One 2009; 4: e7206.
Niu W, Qi Y, Hou S, Zhai X, Zhou W, Qiu C . Haplotype-based association of the renin-angiotensin-aldosterone system genes polymorphisms with essential hypertension among Han Chinese: the Fangshan study. J Hypertens 2009; 27: 1384–1391.
Kingah PL, Luu HN, Volcik KA, Morrison AC, Nettleton JA, Boerwinkle E . Association of NOS3 Glu298Asp SNP with hypertension and possible effect modification of dietary fat intake in the ARIC study. Hypertens Res 2010; 33: 165–169.
Johnson AD, Newton-Cheh C, Chasman DI, Ehret GB, Johnson T, Rose L, Rice K, Verwoert GC, Launer LJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, Caulfield M, van Duijn CM, Ridker PM, Munroe PB, Levy D . On behalf of the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium, Global Bpgen Consortium, and Women's Genome Health Study. Association of hypertension drug target genes with blood pressure and hypertension in 86 588 individuals. Hypertension 2011; 57: 903–910.
Bassett MH, Zhang Y, Clyne C, White PC, Rainey WE . Differential regulation of aldosterone synthase and 11beta-hydroxylase transcription by steroidogenic factor-1. J Mol Endocrinol 2002; 28: 125–135.
Salvi E, Kutalik Z, Glorioso N, Benaglio P, Frau F, Kuznetsova T, Arima H, Hoggart C, Tichet J, Nikitin YP, Conti C, Seidlerova J, Tikhonoff V, Stolarz-Skrzypek K, Johnson T, Devos N, Zagato L, Guarrera S, Zaninello R, Calabria A, Stancanelli B, Troffa C, Thijs L, Rizzi F, Simonova G, Lupoli S, Argiolas G, Braga D, D'Alessio MC, Ortu MF, Ricceri F, Mercurio M, Descombes P, Marconi M, Chalmers J, Harrap S, Filipovsky J, Bochud M, Iacoviello L, Ellis J, Stanton AV, Laan M, Padmanabhan S, Dominiczak AF, Samani NJ, Melander O, Jeunemaitre X, Manunta P, Shabo A, Vineis P, Cappuccio FP, Caulfield MJ, Matullo G, Rivolta C, Munroe PB, Barlassina C, Staessen JA, Beckmann JS, Cusi D . Genomewide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension 2011; 59: 248–255.
Acknowledgements
We thank all the people who have continuously supported the Hospital-based Cohort Study at the National Center for Global Health and Medicine, the Amagasaki Study and the Kyushu University Fukuoka Cohort Study. We also thank Dr Yukio Yamori, Dr Akihiro Fujioka, Dr Shotai Kobayashi, Dr Suminori Kono, Dr Masato Isono, Mr Masahiro Ogasawara, Mr Kazutoyo Ienaga, Dr Chikanori Makibayashi and the many physicians of the Amagasaki Medical Association for their assistance in collecting the DNA samples and the accompanying clinical information and in genotyping the SNPs. This work was supported by grants for the Core Research for Evolutional Science and Technology (CREST) from the Japan Science Technology Agency; the Program for Promotion of Fundamental Studies in Health Sciences, National Institute of Biomedical Innovation Organization (NIBIO); the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT); the Grant of National Center for Global Health and Medicine; and the Ministry of Health, Labor and Welfare.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Hypertension Research website
Supplementary information
Rights and permissions
About this article
Cite this article
Takeuchi, F., Yamamoto, K., Katsuya, T. et al. Reevaluation of the association of seven candidate genes with blood pressure and hypertension: a replication study and meta-analysis with a larger sample size. Hypertens Res 35, 825–831 (2012). https://doi.org/10.1038/hr.2012.43
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hr.2012.43
Keywords
This article is cited by
-
Association between eNOS rs1799983 polymorphism and hypertension: a meta-analysis involving 14,185 cases and 13,407 controls
BMC Cardiovascular Disorders (2021)
-
Natural selection and local adaptation of blood pressure regulation and their perspectives on precision medicine in hypertension
Hereditas (2019)
-
Genetic variations in IL1A and IL1RN are associated with the risk of preeclampsia in Chinese Han population
Scientific Reports (2014)
-
Association between single-nucleotide polymorphisms in six hypertensive candidate genes and hypertension among northern Han Chinese individuals
Hypertension Research (2014)
-
Multivariate meta-analysis of the association of G-protein beta 3 gene (GNB3) haplotypes with cardiovascular phenotypes
Molecular Biology Reports (2014)