Abstract
Many flowering plant species exhibit a variety of distinct sexual morphs, the two most common cases being the co-occurrence of females and males (dioecy) or the co-occurrence of hermaphrodites and females (gynodioecy). In this study, we compared DNA sequence variability of the three genomes (nuclear, mitochondrial and chloroplastic) of a gynodioecious species, Silene nutans, with that of a closely related dioecious species, Silene otites. In the light of theoretical models, we expect cytoplasmic diversity to differ between the two species due to the selective dynamics that acts on cytoplasmic genomes in gynodioecious species: under an epidemic scenario, the gynodioecious species is expected to exhibit lower cytoplasmic diversity than the dioecious species, while the opposite is expected in the case of balancing selection maintaining sterility cytoplasms in the gynodioecious species. We found no difference between the species for nuclear gene diversity, but, for the cytoplasmic loci, the gynodioecious S. nutans had more haplotypes, and higher nucleotide diversity, than the dioecious relative, S. otites, even though the latter has a relatively high rate of mitochondrial synonymous substitutions, and therefore presumably a higher mutation rate. Therefore, as the mitochondrial mutation rate cannot account for the higher cytoplasmic diversity found in S. nutans, our findings support the hypothesis that gynodioecy in S. nutans has been maintained by balancing selection rather than by epidemic-like dynamics.
Similar content being viewed by others
Introduction
Mating systems are major factors affecting species’ genetic and genomic diversity (Charlesworth and Wright, 2001; Glémin et al., 2006). Mating system differences are particularly striking in flowering plants, including a variety of sexual polymorphisms, that is, the co-occurrence of morphologically distinct sex phenotypes (reviewed by Barrett (2010)). Among these are dioecy, the co-occurrence of females and males within a given species, and gynodioecy, females co-occurring with hermaphrodites (Darwin, 1877; Renner and Ricklefs, 1995). Gynodioecy has been considered either as a stable mating system or as a transient state during the evolution of dioecy. The maintenance of gynodioecy has long been considered an evolutionary puzzle. It often involves a genomic conflict between the nuclear and cytoplasmic genomes, which differ in their mode of transmission (Lewis, 1941; Cosmides and Tooby, 1981; Saumitou-Laprade et al., 1994). Specifically, female (that is, male-sterile) individuals in gynodioecious species result from factors in the maternally inherited mitochondrial genome (called cytoplasmic male sterility or CMS factors). Hermaphroditic individuals can result either when male-sterility factors are absent, or from the presence of bi-parentally transmitted nuclear restorer factors that counteract the action of the male-sterility factors and allow normal pollen development (reviewed by Chase (2007) and Delph et al. (2007)). Hermaphrodites in gynodioecious species reproduce via both their female and male functions, while females reproduce only via female functions, so females might be expected to be at a selective disadvantage and quickly be eliminated, resulting in a monomorphic hermaphroditic population (Valdeyron et al., 1973). Two classes of theoretical models have been proposed to account for the maintenance of sterility factors in populations.
In the first class of models, females must have a selective advantage in female functions (that is, higher seed fitness of females than hermaphrodites, due to resource reallocation to female function or avoidance of inbreeding depression). This female advantage combined with a cost of restorer alleles, at least when they are associated with cytoplasms different from the one they restore, can allow the maintenance of a nuclear-cytoplasmic polymorphism. This is a form of balancing selection involving negative frequency-dependent selection (Charlesworth, 1981; Gouyon et al., 1991; Dufay et al., 2007). Under such assumptions, CMS factors are advantageous only when restorer alleles are rare (when they are mainly carried by females), while restorer alleles are selected for only when CMS factors are frequent.
The second class of models posits gene flow between a set of interconnected populations (a metapopulation), causing recurrent invasions of CMS factors, which results in transient male sterility in the populations, again through a female fertility advantage. The increase in frequency of CMS factors within a local population provides a selective advantage for restorer factors, which may invade from other populations, and ultimately become fixed in the local population, leading to loss of its sexual polymorphism until a new CMS invades. Under this class of models, the maintenance of gynodioecy results from epidemic-like dynamics (Frank, 1989; Couvet et al., 1998).
The two classes of models make opposite predictions for cytoplasmic diversity. Epidemic dynamics should reduce nucleotide diversity, because new sterilizing cytoplasms repeatedly sweep through local populations, leading to homogenization of the cytoplasmic genotype within and across populations (Ingvarsson and Taylor, 2002). In contrast, the balancing selection involved in the stable nucleo-cytoplasmic polymorphism model should lead to higher nucleotide diversity of the mitochondrial genome in gynodioecious species compared with hermaphroditic or dioecious species, because non-recombining haplotypes are maintained, potentially over long periods of time, and can accumulate different mutations (Hudson and Kaplan, 1988; Charlesworth, 2002; Städler and Delph, 2002; Touzet and Delph, 2009).
These assumptions have been tested in the genus Silene, which includes a diversity of mating systems, including hermaphroditic, gynodioecious and dioecious species (for example, Desfeux et al., 1996; Jürgens et al., 2002) and thus allows the use of comparative tests of whether balancing selection or epidemic dynamics have predominantly affected the evolutionary dynamics of gynodioecy. However, previous studies comparing cytoplasmic diversity among Silene species with different reproductive systems have led to contradictory conclusions. Ingvarsson and Taylor (2002) showed that sequence variation at chloroplast loci within the gynodioecious species Silene vulgaris is low relative to that in Silene latifolia, a closely related dioecious species, whereas the two species did not differ in diversity at a nuclear gene studied, tending to support epidemic dynamics. Conversely, Städler and Delph (2002) studying the nucleotide diversity of a mitochondrial gene in gynodioecious S. acaulis, described a large number of divergent haplotypes, which they attributed to the signature of balancing selection. Moreover, Houliston and Olson (2006) showed also high mitochondrial gene diversity in S. vulgaris contradicting Ingvarsson and Taylor’s conclusion. Finally, a comparative study of mitochondrial gene diversity on a sample of three gynodioecious (S. acaulis, S. vulgaris and S. nutans) and seven non-gynodioecious Silene species showed that mitochondrial gene diversity was high in gynodioecious species when compared with non-gynodioecious ones, favouring again the ‘balancing selection’ model (Touzet and Delph, 2009). One major problem, unresolved by previous studies, is that the difference of mitochondrial diversity between species can be explained not only by the mating system but also by the mitochondrial mutation rate, which has been found to be extremely variable among genes and among species in the Silene genus (Barr et al., 2007; Mower et al., 2007; Sloan et al., 2008; Sloan et al., 2009). It is thus necessary in comparative studies to control this effect to disentangle the confounding effects of balancing selection and an elevated mitochondrial mutation rate. In the current study, we therefore compared two closely related Silene species belonging to the same subgenus, the gynodioecious S. nutans, with nucleo-cytoplasmic gynodioecy (Garraud et al., 2011), and the dioecious S. otites. To assess the most likely evolutionary scenario involved in the maintenance of gynodioecy in S. nutans, we compared diversity in the two species, using loci sampled from all three genomes, mitochondrial, chloroplastic and nuclear. Nuclear genes help us to control for possible demographic differences between the two species, such as recent bottlenecks reducing diversity. We then used HKA tests (Hudson et al., 1987) to control for mutation rate differences, and also used chloroplast loci (after testing for molecular clock rate differences for the chloroplast genome) as a way to test whether the observed differences could be due to mitochondrial mutation rate variation. Owing to their predominant uniparental inheritance, linkage disequilibrium (LD) is expected between the chloroplast genome and the targets of selection in the mitochondrial genome, and therefore both cytoplasmic genomes should exhibit the same signature of selection (whether epidemics or balancing selection). However, paternal leakage has been documented in other Silene species, disrupting complete LD between the cytoplasmic genomes (McCauley et al., 2005), so we also tested for recombination between and within the mitochondrial and chloroplast genomes of both species.
Materials and methods
Species and plant material
S. nutans (Caryophyllaceae) is a diploid, long-lived perennial rosette plant growing in dry, open grass communities of hillsides. It is a gynomonoecious–gynodioecious (gynodioecious, but with some individuals having flowers of both sex types) self-compatible species (Desfeux et al., 1996; Dufay et al., 2010). It has a wide distribution range, extending from North-Western Europe to Siberia and the Caucasus (Hegi, 1979; Van Rossum et al., 1996; Van Rossum et al., 1999). S. otites (Caryophyllaceae) is a dioecious perennial plant common in low-altitude rocks and arid slopes (Desfeux et al., 1996). It is distributed across Europe, extending from the centre of Spain, eastwards to Lithuania and Bulgaria (Flora Europaea).
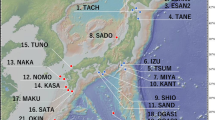
We sampled a single individual per population of both species, in a paired sampling scheme with geographically ‘co-located’ accessions, on a wide geographic scale (Figure 1). We obtained a total of 47 accessions per species, and sequenced 20–37 accessions per gene/species. The S. nutans plants were collected from natural populations (Table 1), whereas those of S. otites were obtained from the herbarium of the Meise Botanical Garden, Belgium (F. Van Rossum), except for four populations for which seeds were grown in the greenhouse (Supplementary Table 1). We used one plant of the dioecious species S. latifolia as an outgroup.
Molecular analyses
To assess mitochondrial diversity, we sequenced two genes, coding for cytochrome b (cob) and for the first sub-unit of cytochrome oxidase (cox1). There have been no known transfer of either of these genes to the nuclear genome among angiosperms, that is, they are exclusively mitochondrial (Gray et al., 1999; Adams et al., 2002; Touzet and Delph, 2009). Four nuclear autosomal genes were also sequenced, the ATP-binding-cassette transporter gene (ABCtrp), the gene coding for the α sub-unit of the eukaryotic elongation factor-1 (ELF), the α tubulin gene (ATUB) and X4, putatively coding for fructose-2,6-bisphosphatase protein (Atanassov et al., 2001; Marais et al., 2011). Note that X4 is not sex-linked in S. otites (Mrackova et al., 2008). Finally, we sequenced four chloroplast fragments: three intergenic spacer sequences trnG-trnS (GS), trnL-trnF (LF) and psbA-trnH (psbA), and the fragment of the matK gene, that is believed to code for a maturase based on structural similarities to other such gene (Neuhaus and Link, 1987; Mohr et al., 1993; Hilu et al., 2003) and is the only maturase of higher plant plastids (Vogel et al., 1997).
Total genomic DNA was extracted and purified from leaves using the NucleoSpin 96 Plant kit (Macherey-Nagel, Düren, Germany). PCR reactions were performed using 40 cycles of 30 s at 94 °C, 45 s at annealing temperature (Supplementary Table 2) and 1 min at 72 °C, with an initial step of 1 min at 94 °C and a final step of 10 min at 72 °C. Each mitochondrial gene was amplified with two pairs of primers, generating overlapping fragments (Supplementary Table 2).
PCR products were purified using Millipore MultiScreen-PCRμ96 filter plates (PCR filter plates) (Millipore Corporation, Billerica, MA, USA). Using the Big Dye Terminatorv3.1 Cycle Sequencing Kit and an ABI 3130 (Applied Biosystems, Carlsbad, CA, USA), we directly sequenced both strands of the purified PCR products except for the two nuclear genes ATUB and ELF; these two genes were cloned using the TA Cloning Kit with pCR 2.1 vector (Invitrogen, Carlsbad, CA, USA). Positive colonies were then screened for presence of the appropriate-sized insert by direct PCR, using the conditions described by the manufacturer, with the primers M13-F (5′-CACGACGTTGTAAAACGAC-3′) and M13-R (5′-GGATAACAATTTCACACAGG-3′). When a haplotype was found only once, it was confirmed by sequencing from an independent PCR reaction. All sequences were deposited in EMBL (accessions KC211324 to KC211517).
Statistical analyses
Sequences were aligned manually using Bioedit version 7.0.5.3 (Hall, 1999).
Plant mitochondrial transcripts are known to undergo post-transcriptional C–U editing at non-synonymous sites (Gray and Covello, 1993; Maier et al., 1996; Brennicke et al., 1999). Such editing may result in C–T DNA polymorphism not being reflected as a polymorphism in the mRNA. Consequently, while the site would be predicted to be non-synonymous from the DNA, with editing, the mutation would not alter the amino-acid sequence. Edited sites were predicted using the online resource PREP-Mt (http://www.prep-mt.net; Mower, 2005), with a cutoff value of 0.2.
We estimated nucleotide diversity both as π, the average number of nucleotide differences per site between a pair of randomly chosen sequences (Nei, 1987), and as Watterson’s θw (Watterson, 1975). We also estimated the average numbers of nucleotide substitutions per site, K, between the species studied and the outgroup S. latifolia and Ks, the value for synonymous site. To compare the numbers of haplotypes and numbers of segregating sites of nuclear and cytoplasmic sequences between the two species, one-sided paired Wilcoxon signed-rank tests were performed using R version 2.11.1. The minimum numbers of recombination events Rm, were estimated by the four-gametes test of Hudson and Kaplan (1985) and LD between cytoplasmic polymorphic sites was estimated by |D’| (Lewontin, 1964). All parameters were estimated with DnaSP version 5 (Librado and Rozas, 2009). A permutation procedure was used to test whether LD observed within genomes (between polymorphic sites located within either the chloroplastic or the mitochondrial genomes) was significantly different from that observed between genomes (between polymorphic sites, one located on the chloroplast and the other in the mitochondrion).
Mitochondrial synonymous substitution rates vary greatly between different Silene species, potentially confounding mutation rate differences affecting diversity with diversity differences due to different selection regimes. We took account of potential mutation rate differences in two different ways. First, we compared synonymous divergence from the outgroup S. latifolia of the mitochondrial genes with that of the chloroplast genes (for which no variation in mutation rate has been documented).
Second, we tested for neutrality of the observed polymorphisms by computing Tajima’s D (Tajima, 1989), which is based on the difference between π and θw, and Fu and Li’s D (Fu and Li, 1993), which is based on differences between the total number of mutations in the external branches of the genealogy (with S. latifolia as an outgroup) and the overall number of mutations. These two tests were performed using DnaSP version 5 (Librado and Rozas, 2009). We then used a maximum-likelihood-ratio test of the standard neutral model, using multilocus data on polymorphism within species and divergence between species. This model (MLHKA) is based on the HKA test, which evaluates the fit of polymorphism and divergence to expectations under the neutral theory, even if the mutation rates differ between two species (Hudson et al., 1987), but allows for an explicit test of selection at individual loci in a multilocus framework. Under the neutral theory, within-species diversity should correlate with between-species divergence (Kimura, 1983); an unexpectedly high divergence can therefore suggest positive selection, whereas an excess level of within-species polymorphism can detect balancing selection (Hudson et al., 1987). The MLHKA approach compares the relative extents of polymorphism and divergence across loci, and assesses the overall fit of the data to a neutral model that assumes the same ratios of polymorphism and divergence at all loci. We used this approach to compare the polymorphism to divergence ratio between S. nutans and the outgroup species S. latifolia with that between S. otites and the same outgroup, combining likelihood across all gene sequences of S. nutans and S. otites for a given genome. The version used was developed by Wright and Charlesworth (2004) and is available from http://labs.eeb.utoronto.ca/wright/Stephen_I._Wright/Programs.html. The program was run under a strictly neutral model for a total of one million chains, followed by a ‘selection’ model in which the S. nutans loci were designated candidates to test for the action of selection, again for a total of one million chains. Significance was assessed using the likelihood-ratio test where minus twice the difference in log-likelihood between the nested models is approximately chi-squared distributed with a number of degrees of freedom equal to the number of genes tested.
Neighbour-Joining (NJ) trees were built using the software MEGA version 4.1 (Kumar et al., 2004) with Kimura’s two parameters model (Kimura, 1980) and a uniform gamma value, including transitions and transversions.
Results
Editing assessment
To accurately evaluate the non-synonymous polymorphism in our data set, we used the online resource PREP-Mt (Mower, 2005) (with a cutoff value of 0.2) to detect potential edited sites on non-synonymous variants. Only one site was predicted to be edited: site 747 of cox1 (but that still remains non-synonymous after editing: G747C748G749/G747T748G749 translated (A249/V249) becomes after editing G747T748G749/G747T748T749 translated (A249/V249)]. The amino-acid sequences of both genes were thus deduced and revealed several variable sites, generating, after editing, four different cob and seven different cox1 amino-acid sequences (Supplementary Table 3). Two peptide sequences from the sequences of the cob gene were shared by both species, which was not the case for the peptide sequences of cox1.
Neutrality tests
With only three exceptions, all in S. otites, the frequency spectra suggested no strong departures from neutrality in either species (Table 1). However, for S. otites, significantly positive Tajima's D was found for the mitochondrial cob gene, and the nuclear X4 gene, and significantly positive Fu and Li's D value for the nuclear ATUB gene. Overall, across the different loci studied, Tajima's D tended to be more negative in S. nutans than in S. otites, suggesting possible recent population growth in the former, and/ or a recent bottleneck in the latter.
Phylogenetic relationships between the two species
We built NJ trees of haplotypes using S. latifolia as an outgroup. As the two species were closed, we used the same outgroup for them. The NJ trees revealed that ABCtrp, X4, cob and the chloroplastic sequences, clustered according to the species (Supplementary Figure 1). For ATUB, ELF and cox, the NJ trees exhibited an incomplete lineage sorting of haplotypes. For ABCtrp, X4 and ELF sequences, the haplotypes of S. otites were a subset of those seen in S. nutans. Therefore, we evaluated the level of shared polymorphism between the two species.
One shared mutation between S. nutans and S. otites was found in matK, in LF and in cob gene (Supplementary Table 4). We found no fixed sites between the two species for the cox1 sequences, but detected two shared polymorphisms. The concatenated chloroplast sequences showed one shared mutation. These observations suggested either that the two species have recently diverged, or that introgression had occurred between them.
Similar nuclear diversity in both species
For the nuclear genes, the numbers of haplotypes were identical in S. nutans and S. otites for every locus analyzed, and ranged from 4 to 38 (Table 1); a one-tailed paired Wilcoxon signed-rank test revealed no significant difference. There was also no difference in the number of segregating sites (S) (V=3; P-value=0.18). θw was also very similar between the two species, except for the X4 gene, with more variable sites in S. nutans than S. otites (9.15±1.08 vs 2.39±1.15, respectively), mostly due to the presence in S. nutans of two singleton haplotypes that contributed 22 out of a total of 23 polymorphic sites. MLHKA tests did not detect any diversity difference between the two species for the nuclear genes (−2.deltaL=6.2482, df=4, P-value=0.1813; Table 3). Taken together, the results from the nuclear genes suggest that any difference in cytoplasmic diversity between the two species should not be ascribed to a difference in their demographic history.
Test for mitochondrial mutation rate differences
The S. nutans and S. otites chloroplast sequences showed similar silent site divergence from S. latifolia (Ks=97.8 × 10−3 and 92.4 × 10−3, respectively). In contrast, both S. nutans mitochondrial genes were less diverged from S. latifolia than those of S. otites (at synonymous sites Ks=19.8 × 10−3 and 16.8 × 10−3 for S. nutans cob and cox1, respectively, vs S. otites values of Ks=36.1 × 10−3 and 31.3 × 10−3 for cob and cox1, respectively), suggesting neutral substitution rate in S. nutans half that in S. otites, and therefore a lower mutation rate. Thus, higher diversity in the S. nutans mitochondrial genome (see next section) is unlikely to be caused by a higher mutation rate.
Comparison of cytoplasmic diversity between the gynodioecious and dioecious species
The level of diversity for the cytoplasmic genes was strikingly different between the two species. The number of haplotypes was higher in S. nutans than in S. otites for both mitochondrial loci (Table 1). The cob gene had 11 distinct haplotypes in S. nutans, vs only 4 in S. otites (Supplementary Table 3; Table 2). For cox1, S. nutans had 16 haplotypes, twice the number in S. otites (8). The number of polymorphic sites was also twice as high for S. nutans as S. otites for both genes (9 vs 4 and 18 vs 9, for cob and cox1, respectively). In line with the mitochondrial results, the concatenated chloroplast sequences also had more haplotypes in S. nutans than S. otites (11 vs 6) (Table 1). Across all the cytoplasmic loci, one-sided paired Wilcoxon signed-rank tests revealed a significant difference in the number of haplotypes (V=21; P-value=0.018), but not for the number of segregating sites (V=17; P-value=0.104). However the latter result is due mainly to a single chloroplast gene (LF), and excluding this gene resulted in a significant difference (V=15; P-value=0.028) between the two species.
Interestingly, the elevated diversity observed in S. nutans as compared to S. otites was much more pronounced for the mitochondrial genes than the chloroplast genes. Indeed, for the mitochondrial genes studied, cob and cox1, both the nucleotide diversity measures, θw and π, were higher in S. nutans than in S. otites, as were the polymorphism/divergence ratios (Table 2). The MLHKA program estimated a 3.88-fold elevation of diversity in S. nutans compared with S. otites, which was close to significance, for these two mitochondrial genes (−2.deltaL=58.5, df=2, P-value=0.053; Table 3). Chloroplast diversity was also higher in S. nutans than in S. otites, but there was only a 1.34-fold estimated difference, and only three of the four chloroplast fragments showed higher π in the gynodioecious species, and only two had a larger θw. Nevertheless, the MLHKA test using all four sequences still indicated a significant difference (−2.deltaL=10.70, df=4, P-value=0.030; Table 3).
The lesser elevation in diversity in S. nutans for the chloroplast than the mitochondrial genes is consistent with incomplete LD between variants in the cytoplasmic genomes, which could result through occasional paternal leakage leading to heteroplasmy. Although mitochondrial inheritance is probably largely uniparental, there is evidence of heteroplasmy in S. vulgaris (McCauley et al., 2005; McCauley and Ellis, 2008; Pearl et al., 2009) and recombination in mitochondrial genes of several gynodioecious Silene species (Städler and Delph, 2002; Houliston and Olson, 2006; Touzet and Delph, 2009). Four-gamete tests (Hudson and Kaplan, 1985) indeed revealed clear evidence for recombination within as well as between mitochondrial and chloroplast genomes for both species. The minimum number of recombination events Rm detected between the mitochondrial gene cob and the concatenated chloroplast sequences was 1 for both S. nutans and S. otites. No recombination was detected between cox1 and the chloroplast sequences in either species. Recombination was also apparent within mitochondrial genes, with at least two and one recombination events for S. nutans and S. otites within cob, respectively, and even more for cox1, with at least five and two recombination events for S. nutans and S. otites, respectively.
In line with this observation, significant breakdown of LD was observed between chloroplastic and mitochondrial genomes in S. nutans (mean LD within genomes=0.947 vs mean LD between genomes=0.853, P<0.01). No such difference was observed in S. otites (0.979 vs 0.964, respectively, P>0.05).
Discussion
What can we conclude about the evolutionary processes maintaining gynodioecy in S nutans? This gynodioecious species exhibits higher diversity in its cytoplasmic genes, compared with the dioecious S. otites. Interestingly, this diversity difference is the opposite of the mitochondrial mutation rate difference, as the rate is lower in S. nutans. Altogether, these results are consistent with the ‘balancing selection’ scenario, in which natural selection maintains cytoplasmic haplotypes over long periods of time specifically in the gynodioecious species.
Previous studies on Silene species suggested balancing selection as the most probable dynamics maintaining nuclear-cytoplasmic gynodioecy. In particular, Touzet and Delph (2009) showed that gynodioecious species exhibited more mitochondrial haplotypes and more divergent ones when compared with hermaphroditic or dioecious species. However, the question remained whether the result could not be explained by a variation in the mitochondrial mutation rate, which can be high among Silene species, as pointed out later by several studies (Barr et al., 2007; Mower et al., 2007; Sloan et al., 2008; Sloan et al., 2009). This is particularly critical when one considers that the species that exhibited the highest diversity (S. acaulis and S. nutans) belong to the same subgenus clade, while the non-gynodioecious species belong to another clade. For the current study, we chose a pair of phylogenetically closely related species, gynodioecious S. nutans and dioecious S. otites, to limit this phenomenon. Using a sample representative of both species, we assessed the nucleotide diversity of multiple genes in the three genomes, to control any demographic effect with the nuclear data and any variation of mitochondrial mutation rate with the chloroplastic data. Convincingly, thanks to the chosen methodology, we showed that mutation rate is not the proximal cause of the higher cytoplasmic diversity found in S. nutans and therefore that balancing selection maintains gynodioecy in populations. Our results apparently exhibit some discrepancy with a former study conducted by Sloan et al. (2009) that found, by using a phylogenetic approach, that the mitochondrial mutation rate was higher in S. nutans compared with S. otites. However, this higher rate in S. nutans was mainly due to an increased rate specific to atp1, illustrating, as pointed out by the authors, the large variation in the estimated mutation rate among the genes studied (nad9, cox3, atp1 and atp9 in this case). Because none of these genes were included in the current study, these two sets of results are not necessarily contradictory.
More generally, our results complement and partly confirm conclusions drawn by studies that used other methodological approaches to investigate the evolutionary dynamics driving the evolution of gynodioecy and found variation in sex ratio among populations that fits expectations under balancing selection (for example, Dufay et al. (2009) in Beta vulgaris) and empirical evidence for frequency-dependent individual reproductive success, that is a necessary condition for such dynamics to occur (for example, Graff (1999) in Sidalcea malviflora; Williams et al. (2000) in Geranium richardsonii; McCauley and Brock (1998) and Miyake and Olson (2009) in Silene vulgaris and De Cauwer et al. (2010a, 2010b) in Beta vulgaris).
The non-gynodioecious sister species to which the nucleotide diversity of S. nutans was compared in this study is dioecious (with males and females). This reproductive system has evolved many times independently in flowering plants (reviewed by Renner and Ricklefs (1995)). Gynodioecy may sometimes be a step in the evolutionary route from hermaphroditism to dioecy (reviewed by Barrett (2002)) and several theoretical studies have shown that nucleo-cytoplasmic gynodioecy (as in S. nutans) can evolve towards dioecy, through the replacement of hermaphrodites by males (Maurice et al., 1994; Schultz, 1994). Although this evolutionary transition has received little empirical support (Spigler and Ashman, 2012), it could have occurred in the genus Silene. Gynodioecy is the ancestral mating system in the genus, and at least two independent transitions from gynodioecy towards dioecy have probably occurred: one leading to the S. latifolia group, and one to the S. otites one (Desfeux et al., 1996; Mrackova et al., 2008; Marais et al., 2011). Dioecy in S. otites, is thought to have evolved from gynodioecy only recently, because (i) intermediate stages between the two mating systems have been reported, with occasional hermaphroditic individuals being found (Desfeux et al., 1996) and (ii) the S. otites sex-determining homomorphic chromosomes seem to be at an evolutionarily much younger stage than those of dioecious S. latifolia (Mrackova et al., 2008). Käfer et al. (2012) tested recently whether dioecious species suffered from a less efficient purifying selection in comparison with non-dioecious ones in Silene due to an expected reduction of their effective population size. Contrarily to Silene latifolia, which exhibited the expected effect, they did not find any trace of it in S. otites, suggesting also a recent transition to dioecy in the species. This view is consistent with several of our results, such as the fact that S. otites haplotypes are often a subset of the S. nutans ones and the shared polymorphism for most of the genes studied between the two species.
If the hypothesis of evolution of dioecy in S. otites from nuclear-cytoplasmic gynodioecy has not been formally established in the literature at this point, this does not affect our conclusion that balancing selection is probably involved in S. nutans. One should note, however, that when dioecy evolves in such models, balancing selection on the mitochondrial genome should not continue in the dioecious species, which usually becomes fixed for the genotype. Consistently with our findings, such transition from gynodioecy to dioecy should thus lead to loss of diversity in the cytoplasmic genome, even in a newly evolved dioecious species. For a better understanding of the transition from gynodioecy to dioecy, it would be interesting to investigate S. acaulis genetic diversity, as a recent study by Marais et al. (2011) suggests that S. acaulis is indeed the closest relative to dioecious S. otites.
Data archiving
Data deposited in the Dryad repository: doi:10.5061/dryad.gd93s and in Genbank: accession numbers: KC211324 to KC211517.
Accession codes
References
Adams KL, Qiu YL, Stoutemyer M, Palmer JD . (2002). Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci USA 99: 9905–9912.
Atanassov I, Delichère C, Filatov DA, Charlesworth D, Negrutiu I, Monéger F . (2001). A putative monofunctional fructose-2,6-bisphosphatase gene has functional copies located on the X and Y sex chromosomes in white campion (Silene latifolia). Mol Biol Evol 18: 2162–2168.
Barr CM, Keller SR, Ingvarsson PK, Sloan DB, Taylor DR . (2007). Variation in mutation rate and polymorphism among mitochondrial genes of Silene vulgaris. Mol Biol Evol 24: 1783–1791.
Barrett SCH . (2002). The evolution of plant sexual diversity. Nat Rev Genet 3: 274–284.
Barrett SCH . (2010). Understanding plant reproductive diversity. Phil Trans R Soc B 365: 99–109.
Brennicke A, Marchfelder A, Binder S . (1999). RNA editing. FEMS Microbiol Rev 23: 297–316.
Charlesworth D . (1981). A further study of the problem of the maintenance of females in gynodioecious species. Heredity 46: 27–39.
Charlesworth D . (2002). What maintains male-sterility factors in plant populations. Heredity 89: 408–409.
Charlesworth D, Wright SI . (2001). Breeding systems and genome evolution. Curr Opin Genet Dev 11: 685–690.
Chase CD . (2007). Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet 23: 81–90.
Cosmides LM, Tooby J . (1981). Cytoplasmic inheritance and intragenomic conflict. J Theor Biol 89: 83–129.
Couvet D, Ronce O, Gliddon C . (1998). Maintenance of nucleo-cytoplasmic polymorphism in a metapopulation: the case of gynodioecy. Am Nat 152: 59–70.
Darwin C . (1877) The Different Forms of Flowers on Plants of the Same Species. John Murray: London.
De Cauwer I, Arnaud JF, Schmidt E, Dufay M . (2010a). Fine-scale sex structure and pollen limitation of female reproductive success in the gynodioecious wind-pollinated Beta vulgaris spp. maritima. J Evol Biol 23: 2636–2647.
De Cauwer I, Dufay M, Cuguen J, Arnaud JF . (2010b). Effects of fine-scale genetic structuring on male mating success in a gynodioecious species. Mol Ecol 19: 1540–1558.
Delph LF, Touzet P, Bailey MF . (2007). Merging theory and mechanism in studies of gynodioecy. Trends Ecol Evol 22: 17–24.
Desfeux C, Maurice S, Henry JP, Lejeune B, Gouyon PH . (1996). Evolution of reproductive systems in the genus Silene. Proc Roy Soc Lond B 263: 409–414.
Dufay M, Touzet P, Maurice S, Cuguen J . (2007). Modelling the maintenance of male-fertile cytoplasm in a gynodioecious population. Heredity 99: 349–356.
Dufay M, Cuguen J, Arnaud JF, Touzet P . (2009). Sex ratio variation in gynodioecious populations of wild beet: can it be explained by frequency-dependent selection? Evolution 63: 1483–1497.
Dufay M, Lahiani E, Brachi B . (2010). Gender variation and inbreeding depression in gynodioecious- gynomonoecious Silene nutans. Int J Plant Sci 171: 53–62.
Frank SA . (1989). The evolutionary dynamics of cytoplasmic male sterility. Am Nat 133: 345–376.
Fu YX, Li WH . (1993). Statistical tests of neutrality of mutations. Genetics 133: 693–709.
Garraud C, Brachi B, Dufaÿ M, Touzet P, Shykoff J . (2011). Genetic determination of male sterility in gynodioecious Silene nutans. Heredity 106: 757–764.
Glémin S, Bazin E, Charlesworth D . (2006). Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc R Soc Lond B 273: 3011–3019.
Gouyon PH, Vichot F, Van Damme JMM . (1991). Nuclear-cytoplasmic male sterility: single point equilibria versus limit cycles. Am Nat 137: 498–514.
Graff A . (1999). Population sex structure and reproductive fitness in gynodioecious Sidalcea malviflora malviflora (Malvaceae). Evolution 53: 1714–1722.
Gray MW, Covello PS . (1993). RNA editing in plant mitochondria and chloroplasts. FASEB J 7: 64–71.
Gray MW, Burger G, Lang BF . (1999). Mitochondrial Evolution. Science 283: 1476–1481.
Hall TA . (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98.
Hegi G . (1979) Illustrierte Flora von Mittel-Europa Band III Teil Parey: Berlin.
Hilu KW, Borsch T, Müller K, Soltis DE, Soltis PS, Savolainen V et al. (2003). Angiosperm phylogeny based on matK sequence information. Am J Bot 90: 1758–1776.
Houliston GJ, Olson MS . (2006). Nonneutral evolution of organelle genes in Silene vulgaris. Genetics 174: 1983–1994.
Hudson RR, Kaplan NL . (1985). Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164.
Hudson RR, Kreitman M, Aguade M . (1987). A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159.
Hudson RR, Kaplan NL . (1988). The coalescent process in models with selection and recombination. Genetics 120: 831–840.
Ingvarsson PK, Taylor DR . (2002). Genealogical evidence for epidemics of selfish genes. Proc Natl Acad Sci USA 99: 11265–11269.
Jürgens A, Witt T, Gottsberger G . (2002). Pollen grain numbers, ovule numbers and pollen-ovule ratios in Caryophylloideae: relation to breeding system, pollination, life form, style number, and sexual system. Sex Plant Reprod 14: 279–289.
Käfer J, Talianová M, Bigot T, Michu E, Guéguen L, Widmer A et al. (2012). Patterns of molecular evolution in dioecious and non-dioecious Silene. J Evol Biol 26: 335–346.
Kimura M . (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120.
Kimura M . (1983) The neutral theory of molecular evolution. Cambridge University Press: Cambridge.
Kumar S, Tamura K, Nei M . (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163.
Lewis D . (1941). Male sterility in natural populations of hermaphrodite plants. New Phytol 40: 56–63.
Lewontin RC . (1964). The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49: 49–67.
Librado P, Rozas J . (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452.
Maier RM, Zeltz P, Kössel HH, Bonnard G, Gualberto JM, Grienenberger JM . (1996). RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol 32: 343–365.
Marais G, Forrest A, Kamau E, Käfer J, Daubin V, Charlesworth D . (2011). Multiple nuclear gene phylogenetic analysis of the evolution of dioecy and sex chromosomes in the genus Silene. PLoS ONE 6: e 21915.
Maurice S, Belhassen E, Couvet D, Gouyon PH . (1994). Evolution of dioecy: can nuclear-cytoplasmic interactions select for maleness? Heredity 73: 346–354.
McCauley DE, Brock MT . (1998). Frequency-dependent fitness in Silene vulgaris, a gynodioecious plant. Evolution 52: 30–36.
McCauley DE, Bailey MF, Sherman NA, Darnell MZ . (2005). Evidence for paternal transmission and heteroplasmy in the mitochondrial genome of Silene vulgaris, a gynodioecious plant. Heredity 95: 50–58.
McCauley DE, Ellis JR . (2008). Recombination and linkage disequilibrium among mitochondrial genes in structured populations of the gynodioecious plant Silene vulgaris. Evolution 62: 823–832.
Miyake K, Olson MS . (2009). Experimental evidence for frequency dependent self-fertilization in the gynodioecious plant, Silene vulgaris. Evolution 63: 1644–1652.
Mohr G, Perlman PS, Lambowitz AM . (1993). Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucl Acid Res 21: 4991–4997.
Mower JP . (2005). PREP-Mt: predictive RNA editor for plant mitochondrial genes. BMC Bioinform 6: 96.
Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD . (2007). Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol 7: 135.
Mrackova M, Nicolas M, Hobza R, Negrutiu I, Moneger F, Widmer A et al. (2008). Independent origin of sex chromosomes in two species of the genus Silene. Genetics 179: 1129–1133.
Nei M . (1987) Molecular Evolutionary Genetics. Columbia University Press: New York.
Neuhaus H, Link G . (1987). The chloroplast tRNALys:(UUU) gene from mustard (Sinapsis alba) contains a class II intron potentially coding for a maturase polypeptide. Curr Genet 11: 251–257.
Pearl SA, Welch ME, McCauley DE . (2009). Mitochondrial heteroplasmy and paternal leakage in natural populations of Silene vulgaris, a gynodioecious plant. Mol Biol Evol 26: 537–545.
Renner SS, Ricklefs RE . (1995). Dioecy and its correlates in the flowering plants. Am J Bot 82: 596–606.
Saumitou-Laprade P, Cuguen J, Vernet P . (1994). Cytoplasmic male sterility in plants: molecular evidence and the nucleocytoplasmic conflict. Trends Ecol Evol 9: 431–435.
Schultz S . (1994). Nucleo-cytoplasmic male sterility and alternative routes to dioecy. Evolution 48: 1933–1945.
Sloan DB, Barr CM, Olson MS, Keller SR, Taylor DR . (2008). Evolutionary rate variation at multiple levels of biological organization in plant mitochondrial DNA. Mol Biol Evol 25: 243–246.
Sloan DB, Oxelman B, Rautenberg A, Taylor DR . (2009). Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol Biol 9: 260.
Spigler RB, Ashman TL . (2012). Gynodioecy to dioecy: are we there yet? Ann Bot 109: 531–543.
Städler T, Delph LF . (2002). Ancient mitochondrial haplotypes and evidence for intragenic recombination in a gynodioecious plant. Proc Natl Acad Sci USA 99: 11730–11735.
Tajima F . (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595.
Touzet P, Delph LF . (2009). The effect of breeding system on polymorphism in mitochondrial genes of Silene. Genetics 181: 631–644.
Valdeyron G, Dommée B, Valdeyron A . (1973). Gynodioecy: another computer simulation model. Am Nat 107: 454–459.
Van Rossum F, De Bilde J, Lefèbvre C . (1996). Barriers to hybridization in calcicolous and silicicolous populations of Silene nutans from Belgium. Belg J Bot 129: 13–18.
Van Rossum F, Meerts P, Gratia E, Tanghe M . (1999). Ecological amplitude in Silene nutans in relation to allozyme variation at the western margin of its distribution. J Veg Sci 10: 253–260.
Vogel WF, Gish GD, Alves F, Pawson T . (1997). The discoidin domain receptor tyrosine kinases are activated by collagen. Mol Cell 1: 13–23.
Watterson GA . (1975). On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7: 256–276.
Williams CF, Kuchenreuther MA, Drew A . (2000). Floral dimorphism, pollination, and self-fertilization in gynodioecious Geranium richardsonii (Geraniaceae). Am J Bot 87: 661–669.
Wright SI, Charlesworth B . (2004). The HKA test revisited: a maximum likelihood ratio test of the standard neutral model. Genetics 168: 1071–1076.
Acknowledgements
We wish to thank R Bergero R and S Qiu for sharing information on nuclear gene primers and S. latifolia sequences, and Meise Botanical Garden for providing leaf samples of S. otites from the herbarium. This work was supported by Agence Nationale de la Recherche (ANR-06-JCJC-0074) and a grant from Région Nord Pas de Calais and the European Community (Arcir PLANT-TEQ6) to PT and MD, a PhD fellowship from Centre National de la Recherche Scientifique and Région Nord Pas de Calais to EL and a Postdoctoral Fellowship from National Fund for Research Luxembourg to SLC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Rights and permissions
About this article
Cite this article
Lahiani, E., Dufaÿ, M., Castric, V. et al. Disentangling the effects of mating systems and mutation rates on cytoplamic diversity in gynodioecious Silene nutans and dioecious Silene otites. Heredity 111, 157–164 (2013). https://doi.org/10.1038/hdy.2013.32
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2013.32
Keywords
This article is cited by
-
Homologous recombination changes the context of Cytochrome b transcription in the mitochondrial genome of Silene vulgaris KRA
BMC Genomics (2018)
-
Phylogeographic pattern of range expansion provides evidence for cryptic species lineages in Silene nutans in Western Europe
Heredity (2016)
-
The structure of allozyme variation in Silene nutans (Caryophyllaceae) in Denmark and in north-western Europe
Plant Systematics and Evolution (2016)