Abstract
Prevention of malaria transmission throughout much of Africa is dependent on bednets that are impregnated with pyrethroid insecticides. Anopheles arabiensis is the major malaria vector in Chad and efforts to control this vector are threatened by the emergence of pyrethroid resistance. WHO bioassays revealed that An. arabiensis from Ndjamena is resistant to pyrethroids and dichlorodiphenyltrichloroethane (DDT) but fully susceptible to carbamates and organophosphates. No 1014F or 1014S kdr alleles were detected in this population. To determine the mechanisms that are responsible for resistance, genetic crosses were established between the Ndja strain and an insecticide susceptible population from Mozambique. Resistance was inherited as an autosomal trait and quantitative trait locus (QTL) mapping identified a single major locus on chromosome 2R, which explained 24.4% of the variance in resistance. This QTL is enriched in P450 genes including 25 cytochrome P450s in total. One of these, Cyp6p4 is 22-fold upregulated in the Ndja strain compared with the susceptible. Piperonyl butoxide (PBO) synergist and biochemical assays further support a role for P450s in conferring pyrethroid resistance in this population.
Similar content being viewed by others
Introduction
Malaria is endemic in the Chad republic (Central Africa) affecting >95% of the overall population and is the main cause of morbidity and mortality (Kerah-Hinzoumbe et al., 2009).
Anopheles arabiensis is the main malaria vector in this region and a recent study in the Southwest of Chad estimated the annual entomological inoculation rate at 311, of which An. arabiensis accounted for 84.5% (Kerah-Hinzoumbe et al., 2009).
The National Malaria Control Programme in Chad is currently promoting the use of long-lasting insecticide nets in order to reduce the burden of the disease. However, in 2008 pyrethroid resistance was reported in four sites in the Southwest of the country (Ranson et al., 2009). Full susceptibility to bendiocarb and fenitrothion was observed in all populations, with resistance to DDT in only one population (from a cotton growing site). A similar resistance profile was reported previously in An. arabiensis from Bongor and Guelendeng, (Kerah-Hinzoumbe et al., 2008). In this study, genotyping for the known target-site mutations found in An. gambiae s.l, L1014F and L1014S, indicated the absence of these ‘kdr’ mutations in this population.
Pyrethroid resistance has been reported in An. arabiensis populations in West Africa (Diabate et al., 2004), East Africa (Matambo et al., 2007), Southern Africa (Hargreaves et al., 2003; Casimiro et al., 2006) and Central Africa (Wondji et al., 2005; Müller et al., 2008a). In Sudan and Ethiopia, this resistance has been associated with the 1014F kdr allele (Abdalla et al., 2008; Yewhalaw et al., 2010). Other studies, however, reported reduced susceptibility and resistance to pyrethroids in An. arabiensis populations from Cameroon and South Africa in the absence of the kdr allele (Chouaïbou et al., 2008; Mouatcho et al., 2009).
Elevated activity of three enzyme families, the cytochrome P450s, esterases and glutathione S-transferases, have been associated with metabolic resistance to insecticides (Hemingway and Ranson, 2000) and, more recently, several cytochrome P450 enzymes, with proven ability to bind and/or metabolise pyrethroids have been found to be upregulated in pyrethroid-resistant populations of An. gambiae (Müller et al., 2008b; Stevenson et al., 2011). As much less is known about pyrethroid resistance mechanisms in An. arabiensis, we used a combination of genetic mapping, bioassays and biochemical and molecular assays to characterise the causes of this resistance in a strain colonised from Ndjamena in Chad.
Materials and methods
Field collection
Gravid or blood fed An. gambiae s.l. adult females resting indoors were collected in houses between 0600 and 0000 hours in Ndjamena (12° 6′ 47′′ North, 15° 2′ 57′′ East), capital city of Chad, in September 2009 using aspirators and torches. They were transferred to cages and left to lay eggs, which were transported to the Liverpool School of Tropical Medicine (LSTM), UK. The eggs were hatched and larvae fed with Tetramin baby fish food (Tetra, Melle, Germany) and a colony, named Ndja, was established. In addition, a similar sampling of females of An. gambiae s.l. was carried out from Chokwe (24° 33′ 37′′ S, 33° 1′ 20′′ E) in southern Mozambique in July 2009. Eggs were also transported to LSTM for rearing and establishment of a colony named Moz.
All field-collected females used to establish the laboratory colony were morphologically identified as belonging to the An. gambiae species complex according to the key of (Gillies and Coetzee, 1987) and identified as An. arabiensis by PCR (Scott et al., 1993).
Insecticide susceptibility assays
Insecticide susceptibility assays were carried out using 2- to 5-day-old F1 adults from both colonies following the WHO protocol (WHO, 1998). Approximately 20–25 mosquitoes per tube were exposed to insecticide-impregnated filter paper for 1 h and then transferred to a clean holding tube supplied with 10% sugar solution. Mortality was determined after 24 h. After phenotyping, surviving, resistant mosquitoes were stored at −80 °C for subsequent DNA and RNA extraction, and dead mosquitoes were stored dried on silica gel. The following insecticides were tested: the pyrethroids permethrin (0.75%), and deltamethrin (0.05%); the carbamate bendiocarb (0.1%); the organophosphate malathion (5%); and the organochlorines DDT (4%) and dieldrin (4%).
For permethrin and DDT, the effect of a 1-h exposure with the synergist piperonyl butoxide (4%), immediately before 1-h insecticide exposure was also assessed.
Biochemical assays
Biochemical assays were carried out using 2- to 3-day-old adults (25 females and 25 males) from both strains. Glutathione transferase, acetylcholinesterase, cytochrome P450 and general esterase activity (with substrates p-nitrophenyl acetate; α- and β-napthyl acetate) were assayed using methods described by (Penilla et al., 1998). A two-sample t-test was used to compare the results between the two strains following an adjustment for total protein content.
Kdr and ace-1 genotyping using pyrosequencing method
The 1014L, 1014F and 1014S kdr alleles were genotyped in a set of permethrin, deltamethrin and DDT-resistant mosquitoes from the Ndja strain (n=82) using the pyrosequencing method (Wondji et al., 2007). In addition, all the mosquitoes alive after bioassays with the organophosphate malathion (n=25) were screened for the presence of the acetylcholinesterase target-site mutation G119S (ace-1) associated with carbamate and organophosphate resistance using the pyrosequencing method.
Pyrosequencing reactions were performed as described by (Wondji et al., 2007) according to the manufacturer’s instructions using the PSQ 96 SNP Reagent Kit (Biotage AB, Uppsala, Sweden). The genotype was determined using the SNP Software (Biotage AB). Ambiguous kdr genotypes after pyrosequencing were confirmed by sequence analysis.
Software provided by Pyrosequencing AB (Biotage, Uppsala, Sweden) was used to design three sequence-specific primers for kdr (L1014F and L1014S mutations are detected in the same assay) and ace-1 mutations. The primer sequences and sequences to analyse for the genotyping of both mutations are found in the Supplementary material in Supplementary Table S1.
Mosquito crosses
Reciprocal crosses were established between virgin Ndja and Moz females with males from the alternative strain. Mosquitoes were allowed to mate for 4 days after which they were blood fed, and females were left to oviposit singly. Eggs from each crossing were collected and reared separately to adults. A subset of the F1 offspring was left to intercross to generate F2 progeny to produce isofemale lines. Further subsets were backcrossed to the resistant or susceptible parental strains.
The F2 progeny (3- to 5-day-old adults) from the isofemale lines were phenotyped for resistance to permethrin using the standard WHO bioassay method (1-h exposure to 0.75% permethrin). In addition, the remaining F1, F2 and backcross progeny were also phenotyped to investigate the mode of inheritance of the trait. Surviving and dead mosquitoes were preserved on silica gel for later DNA extraction and genotype determination.
Genotyping of molecular markers
F2 progeny of two families were used for genetic mapping; family 2 originated from a Ndja female × Moz male cross and family 3 from the reciprocal Moz female × Ndja male cross. Only female progeny were used for genotyping, and these were sampled equally from alive and dead progeny (24 susceptible and 24 permethrin resistant for each family). DNA was extracted using the LIVAK method (Collins et al., 1987) and microsatellite genotyping performed using the Beckman CEQ8000 (Beckman Coulter, Inc., Brea, CA, USA). Informative microsatellite markers were selected after genotyping the parental female F0 and three randomly selected F1 individuals. Microsatellite loci were initially screened using a tailed primer system. Primer pairs for informative markers were re-synthesised to directly incorporate a fluorescent dye. The majority of markers used have been described previously (Zheng et al, 1996). Microsatellites not previously described are listed in the Supplementary material in Supplementary Table S2.
In addition to the microsatellite markers, four single-nucleotide polymorphism markers were identified de novo by sequence analysis of PCR products of the parental female F0 and three F1 individuals. These single-nucleotide polymorphisms included a synonymous substitution in exon 27 of the sodium channel gene. The single-nucleotide polymorphism markers were genotyped by pyrosequencing (Supplementary Table S1).
Quantitative trait locus (QTL) mapping
The JoinMap 2.0 software package (Stam and van Ooijen, 1996) was used to build genetic linkage maps for each individual family and for the combined genotyping data from both families. Genotype data for each marker were tested for conformity to Mendelian ratios (Hardy–Weinberg equilibrium) with a χ2 goodness-of-fit analysis using the JoinMap JMSLA procedure and a χ2 test for genotype–phenotype association was performed using Haploview 4.2 (Barrett et al., 2005). Loci were separated into linkage groups with JMGRP and JMSPL procedures with minimum and maximum logarithm of the odds (LOD) thresholds of 1.0 and 4.0, respectively and LOD increments of 0.1. The JoinMap JMREC was used to estimate pairwise cM distances between all pairs of informative loci in each linkage group and JMMAP to find the maximum likelihood map using the Kosambi distances. Genetic maps were plotted with MapChart 2.1 software (Voorrips, 2002).
The JoinMap linkage positions along with the genotype–phenotype data were entered into Windows QTL Cartographer 2.5 (Wang et al., 2005). Interval mapping (IM; Lander and Botstein, 1989), composite IM (CIM; Zeng, 1993) and multiple IM (MIM; Zeng et al., 1999) procedures were performed for each family separately and for the combined data. The MapChart 2.1 software (Voorrips, 2002) was used to plot the LOD graphs next to the respective linkage maps. MIM analyzes multiple marker intervals simultaneously to fit multiple putative QTLs. An initial MIM model was estimated by forward and backward marker selection with a probability of a partial r2 set to 0.01. The model was refined by optimising QTL positions, searching for new and testing for existing QTLs.
Gene expression analysis
For quantitative reverse transcriptase-PCR RNA was extracted from three pools of 10 three-day-old unmated, female mosquitoes from Ndja resistant to permethrin (survivors after 1-h exposure to 0.75% permethrin) and from the susceptible Moz strain (un-exposed) (snap-frozen and stored at −80 °C). The PicoPure RNA Isolation Kit (Arcturus, Applied Biosystems, Mountain View, CA, USA) was used and single-stranded complementary DNA was synthesised using Superscript III (Invitrogen, Carlsbad, CA, USA) with oligo-dT20 and RNase H (New England Biolabs, Ipswich, MA, USA), according to the manufacturer’s instructions.
Real-time quantitative reverse transcriptase-PCR was performed on a set of nine candidate genes using the Stratagene Mx3005P qPCR system (Agilent Technologies, Inc., Santa Clara, CA, USA) and analysed using Agilent’s qPCR software, MxPro. A serial dilution of complementary DNA was used to establish standard curves for each gene in order to assess PCR efficiency and quantitative differences between samples. The real-time quantitative PCR reaction mixture was prepared using the Agilent Ultra-Fast SYBR Green qPCR Master Mix (Agilent Technologies, Inc.) according to manufacturer’s instructions using 1 μl template complementary DNA (diluted to 10 μM). The thermal cycling protocol was as follows: initial denaturation for 3 min at 95 °C followed by 40 cycles of 10 s at 95 °C, 10 s at 60 °C and 30 s at 72 °C. The fluorescence signal was measured at the end of each extension step at 72 °C. For each sample, three biological replicates were run alongside three technical replicates of the standard curves. Fold change was calculated through the delta-delta Ct method (Pfaffl, 2001) using the geometric means of two control genes RSP7 (AGAP010592) and Elongation Factor gene (AGAP005128). Primer details are listed in Supplementary Table S3.
Results
Insecticide susceptibility assays
The An. arabiensis population from Ndjamena was resistant to permethrin with a mortality rate of 54.3% for females and 75.6% for males (Table 1). Females exhibit reduced susceptibility to deltamethrin (90.6% mortality) although the males were susceptible to this insecticide. The An. arabiensis population from Chokwe, Mozambique, was fully susceptible to both pyrethroids.
Both strains showed some survival after DDT exposure (69.1% in Ndja and 89.5% in Moz for females but the difference between strains was not significant (P=0.33)). There were also some indications of reduced susceptibility to malathion (95% mortality) and dieldrin (90.1% mortality) in the Ndja strain but only in the females. Both strains are susceptible to bendiocarb.
The synergist PBO fully restored permethrin susceptibility in the Ndja strain but did not significantly (P=0.35) affect DDT mortality (84% mortality after PBO exposure).
Target-site mutations
Sixty-two mosquitoes from the Ndja strain that had survived DDT or pyrethroid exposure and 30 susceptible individuals (20 from Ndja, 10 from Moz strain) were genotyped for mutations at codon 1014 of the sodium channel. Pyrosequencing unambiguously assigned 90 of these individuals as wild type (that is, 1014 codon TTA). The remaining two gave ambiguous results but sequencing confirmed that these individuals were also homozygote for the wild type.
The four mosquitoes from the Ndja strain that survived malathion exposure and 21 non-exposed mosquitoes were genotyped for the ace-1 mutation. All 25 individuals were homozygous for the wild-type allele and the 119S ace-1 mutation was not detected. Pyrograms for the kdr and ace-1 assay are shown in Supplementary material Figure S1.
Biochemical assays
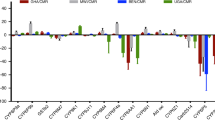
A significant difference in esterase activity was observed between females of the Ndja and Moz strain with the substrates p-nitrophenyl acetate (1.86-fold higher in Ndja, P<0.0038) and α-napthyl acetate (2.71-fold higher, P<0.0011) but no difference was observed between the strains when using β-napthyl acetate substrate (Figure 1, Table 2).
Range of enzyme activity for An. arabiensis females from Ndja (left) and Moz (right) for (a) P450s; (b) glutathione S-transferase (GST); and esterases with the substrate (c) p-nitrophenyl acetate (pNPA); (d) α-napthyl acetate (units as in legend to Table 2).
AChE inhibition rates by propoxur are close to identical between the two strains, ranging from 88 to 100% for the females. This, plus the ace-1 genotyping results and the bioassay data all support absence of any carbamate resistance.
A significant increase in the level of glutathione S-transferase activity as measured with CDNB (1-chloro-2,4-dinitrobenzene) was observed in the Ndja strain compared with the susceptible Moz strain with mean CDNB levels 3.09-fold higher in Ndja females (P<0.001; Figure 1, Table 2). Similarly, cytochrome P450 levels were elevated in the Ndja females compared with the Moz strain (2.57-fold, P<0.003; Figure 1, Table 2).
Inheritance mode of permethrin resistance
There was no significant difference (two samples t-test; P=0.47) between the F1 progeny resulting from the reciprocal crosses between Ndja and Moz (Table 3) indicating that the resistance to permethrin was inherited autosomally in the Ndja strain of An. arabiensis. Both F1 hybrid populations showed intermediate resistance between the parental strains. To test whether permethrin resistance in An. arabiensis was controlled by a single gene (monogenically inherited), expected mortalities from F2 backcrosses and intercrosses were calculated and compared with observed mortalities (Table 3). The expected mortality ratio was estimated for the backcross to resistant as equal to ½ RS+½ RR mortalities while for the backcross to susceptible, the mortality ratio was estimated as being ½ RS+½ SS. One sample t-test carried out either under the assumption of complete dominance or complete recessivity, showed that most observed mortalities were significantly different from the expected mortalities rejecting the hypothesis of monogenic mode of inheritance.
Under the assumption of a dominant resistance gene, the backcross progeny to Ndja (resistant) and to Moz (susceptible) are expected to show mortalities of 61% and 80.5%, respectively, and assuming complete recessiveness, expected mortalities would be 80.5% and 100%, respectively. Actual observed mortality means for the backcross to the resistant Ndja were 27.0% (B1+B5) (dominance: P=0.0102, recessiveness P=0.0042) and 56.4% (B3+B7) (dominance P=0.18, recessiveness P=0.0082) 24-h post exposure. For the backcross to the susceptible parent, observed mortality means were 56.0% (B2+B6) (dominance: P=0.07 and recessiveness P=0.0032) and 18.4% (B4+B8) (dominance: P=0.021 and recessiveness P=0.0008) 24-h post exposure. In both cases, both crosses differ significantly from the recessive model whereas only one cross differs from the dominant one (B3+B7 and B4+B8; Table 3).
The observed mortalities for the F2 intercrosses for family 3 and family 2 of 72.1% and 77.4% (Table 3) do not significantly differ from each other (two sample t-test, P=0.413). They do not differ significantly from the dominant model (one sample t-test, P=0.51 for family 3 and P=0.06 for family 2) but the recessive model can be rejected (one sample t-test, P=0.0006 for family 3 and P=0.0034 for family 2; Table 3).
Overall, these results indicate that permethrin resistance in the Ndja-resistant strain is not monogenically inherited but is expressed as an incompletely dominant trait.
Linkage mapping
Linkage maps were established for the two individual isofemale families. Three linkage groups were resolved each corresponding to one of the three chromosomes (not shown). Overall map sizes and resolutions are similar for both families with 7.1 cM per marker for family 2 and 5.3 cM per marker for family 3.
Data from the two families were used to build combined genetic linkage maps (Figure 2), which consisted of genotypes of 98 individuals at 31 loci. For the combined data, the average resolution for each chromosome is 12.4, 5.8 and 4.5 cM per marker, respectively, for chromosome X, 2 and 3. The total length of the current map is 198.1 cM with an average resolution of 6.4 cM per marker. The marker order on the combined map for chromosome 2 correlates with the optimal physical order according to Vectorbase. The small number of exceptions on the X chromosome and chromosome 3 are indicated on Figure 2.
Genetic maps of An. arabiensis using the combined data from the two families. A scale of genetic distance in centi Morgan (cM) is shown on the left. Positions of the centromeres are between markers 786 and ex27 on chromosome 2 and 3R_arab and 811 on chromosome 3. The stars indicate where the marker order differs from the physical order of markers in An. gambiae s. s. (according to vectorbase).
A LOD significance threshold of 2.5 (α=0.05) was calculated based on the size of the linkage maps for the combined data using the criteria described in (van Ooijen, 1999).
QTL mapping
A χ2 goodness-of fit test was used to identify markers statistically (P⩽0.05) associated with pyrethroid resistance in each family, the null hypothesis being that likelihood of survival or death after permethrin exposure is equal for each genotype class.
For family 3, significant associations were found between ten markers and permethrin resistance, eight markers on chromosome 2R, one on 2L and one on the X chromosome. The eight markers on chromosome 2R are arranged sequentially but only five of these are fully informative (Supplementary Table S4).
For family 2, only marker 590 is significantly (P⩽0.05) associated with permethrin resistance and four flanking, semi-informative markers show a similar trend (Supplementary Table S4).
All four markers are found sequentially on chromosome 2R and overlap with the set found significant in family 3.
These results give an important indication that a locus responsible for resistance is located within this region. The complete non-association of marker 786 in both families indicates this marker lies outside the boundaries of association. Plotting resistance, measured as mortality, against genotypes suggests that the alleles in these loci are additive in their effect on resistance (Figure 3).
IM and CIM were used to predict the location of this QTL. The two families were analysed individually and combined.
No QTL were identified in family 2. However, significant LOD scores were observed on chromosome 2R in family 3 and this QTL was further strengthened in the combined family analysis (Figure 4a). In the combined data, a single QTL was identified on chromosome 2R with a maximum LOD of 4.6 (CIM) and 3.8 (IM). This QTL was named rp1 for resistance to permethrin 1. The most closely associated markers with this QTL are indicated in Figure 4b where the chromosome 2 LOD scores of CIM are plotted alongside the genetic map. The boundaries of the rp1 QTL are ∼10 cM with the flanking markers 757 and 590 spanning chromosomal divisions 10–13 and a genome region of ∼14 Mb.
(a) Plot of LOD scores associated with pyrethroid resistance for chromosomes X, 2 and 3 in the combined maps. Solid lines represent LOD estimated by CIM and dashed lines represent LOD estimated by interval mapping. The straight line along the top of each graph represents the threshold value for LOD calculated according to (van Ooijen, 1999). (b) Plot of LOD scores (CIM) associated with permethrin resistance along the linkage map of chromosome 2 including marker names to orient QTL position. The dashed line across the graph represents the threshold value for LOD.
Multiple interval mapping (MIM) confirms the presence of a single QTL on chromosome 2R, rp1, in both families and estimates its genetic variance (δg2) as 14.4% and 18.9% of the phenotypic variance (δp2) of permethrin resistance for family 2 and family 3, respectively. For the combined data, this QTL explains 24.4% of the phenotypic variance (resulting from 0.5% for additive and 23.9% for dominance effect). No additional QTLs were detected using MIM.
Analysis of the expression pattern of candidate genes spanning the rp1 QTL
The QTL detected on the 2R chromosome spans several clusters of P450s (Supplementary Table S5). Real-time quantitative reverse transcriptase-PCR was performed on a subset of these genes (Cyp6p1, Cyp6p2, Cyp6p3, Cyp6p4, Cyp6p5, Cyp6aa1, Cyp6ad1 and Cyp6z4). These genes were selected as members of the same subclass have previously been associated with insecticide resistance in other Anopheles species (Müller et al., 2008b; Wondji et al., 2009; Duangkaew et al., 2011). In addition, the expression profile of Cyp6m2 was also investigated. This gene is located on chromosome 3R and hence not linked to resistance in this An. arabiensis population but the gene was included as Cyp6m2 is strongly associated with pyrethroid resistance in the sister species, An. gambiae (Müller et al., 2008b). Despite extensive attempts at optimisation, reproducible standard curves could not be obtained for Cyp6p5, Cyp6ad1 and Cyp6z4 and these three genes were removed from subsequent analysis.
A significant upregulation was observed for the Cyp6p4 gene (P<0.01) with 22.2 fold-change in the Ndja-resistant strain compared with the Moz susceptible strain (Figure 5). No significant difference in transcription levels was observed between these strains for other genes including Cyp6m2 gene on 3R chromosome (Figure 5).
Quantitative reverse transcriptase-PCR expression pattern of six candidate genes in permethrin-resistant Ndja females and the susceptible Moz strain. The relative fold change of the 2−ΔΔCt of each gene between Ndja and Moz are represented on the Y axis after normalisation against the mean of RSP7 and elongation factor (EF) genes. Error bars are based on upper and lower 95% confidence interval.
Discussion
In this study, we provide an assessment of the susceptibility status of an An. arabiensis strain from Chad against different insecticides and investigate the underlying mechanisms involved in conferring the observed permethrin resistance. QTL mapping of permethrin resistance was performed on F2 progeny resulting from crossing the Chad population with a susceptible strain from Mozambique to provide the first insight into the genetic architecture of resistance in An. arabiensis.
The Ndja strain is resistant to type I pyrethroids (permethrin) and a low prevalence of resistance to the type II pyrethroid, deltamethrin was observed in females of this strain.
An. arabiensis is the major malaria vector in Chad and, although the source of the selection pressure that has led to the pyrethroid resistance in this vector is unknown, the potential impact of this pyrethroid resistance on the efficacy of malaria control in Chad should be monitored to ensure their continuous efficacy. The resistance profile described here for the Ndja strain is comparable to that described for samples of An. arabiensis in Ndjamena in 2008 (Ranson et al., 2009). Given that the level of gene flow between An. arabiensis populations in Central Africa is relatively high (Simard et al., 2000; Wondji et al., 2005), this resistance may spread rapidly throughout the region.
In contrast, the Moz strain was found completely susceptible to both type I and II pyrethroids. This result is in line with a previous study on An. arabiensis in Mozambique (Casimiro et al., 2006) where populations of this species from Chokwe, the same location where the Moz strain was collected, was found to be fully susceptible to all pyrethroids.
The pyrethroid resistance in the Ndja strain is not caused by target-site mutations as no kdr mutation was detected in this strain and no QTL was detected in the vicinity of this locus (situated on chromosome 2L). Metabolic resistance has already been implicated as the main mechanism of pyrethroid resistance in an An. arabiensis populations in Maga from North Cameroon around 200 miles from Ndjamena (Wondji et al., 2005) and the synergist and biochemical data from this study support the suggestion that this mechanism is common to populations of this species in Central Africa.
In order to identify loci associated with permethrin resistance linkage maps of all three Anopheles chromosomes were constructed using combined data from two families generated from reciprocal crosses between the permethrin-resistant Ndja and the susceptible Moz strain. The maps generated cover the An. arabiensis genome with similar marker resolution on the autosomes (5.8 and 4.5 cM per marker for chromosomes 2 and 3, respectively) and lower resolution for the X chromosome (12.4 cM per marker). This compares with previously established genetic maps for An. funestus (Wondji et al., 2007) that also reported lower marker resolution for the X chromosome. This is most likely due to the limited availability of informative markers for the X chromosome as well as restricted recombination in sex chromosomes in general. The average resolution of 6.4 cM per marker of this first An. arabiensis map is much lower than the resolution of the published An. gambiae s.s. microsatellite map (Zheng et al. 1996) but was sufficient for initial QTL mapping.
Single-family analysis identified a QTL associated with permethrin resistance in family 3 only. However, as the strength of this QTL was increased in the combined data set, it is likely that the same locus is associated with resistance in both families but there was insufficient resolution to detect this in family 2 in this study. Many of the markers in the vicinity of the QTL were only semi-informative for family 2 and this will substantially reduce the power of the analysis. The difference in marker density on chromosome 2 for the two families (6.1 cM per marker in family 2 compared with 2.9 cM per marker in family 3) could also explain the absence of a QTL in family 2.
This study identified a single QTL for permethrin resistance on chromosome 2R explaining 24% of the genetic variance. A previous genetic mapping study of permethrin resistance in An. gambiae s.s. identified two major QTL in populations possessing target-site and metabolic resistance (Ranson et al., 2004). It is likely that other minor QTLs for permethrin resistance are present in the Ndja strain but this study lacked the power to detect these. The existence of potential undetected QTLs of smaller genetic variance than rp1 is supported by the study of the inheritance mode that shows that resistance in this strain is under the control of more than one loci.
The use of F2 generations, limitations in sample sizes and marker density all limit the resolution that can be obtained in the QTL analysis. In An. funestus a single major QTL for pyrethroid resistance was detected in the F2 generation but when advanced intercross lines at generations F6 and F8 were analysed, two additional minor QTLs where detected (Wondji et al., 2007, 2009). Therefore, future studies to improve the resolution of the An. arabiensis rp1 QTL and detect potential new QTLs should use such an advanced intercross line experimental approach.
At least 47 cytochrome P450 genes are encoded on chromosome 2R (Ranson et al., 2002) with the majority of these located in three clusters (belonging to gene families cyp4, cyp325 and cyp6; Ranson et al., 2002). The boundaries of the identified QTL are ∼10 cM with the flanking markers 757 and 590 spanning a genome region of ∼14 Mb including 910 annotated genes (vectorbase biomart 03/2012). This region is significantly enriched in P450s with an enrichment of 4.2-fold (DAVID; Gene Functional Classification software v.6.7; Huang et al., 2009). The QTL boundaries encompass a total of 25 P450 genes. This, plus the elevated levels of cytochrome P450s observed in the Ndja strain from the biochemical assays, synergist data, plus the defined role of P450s in pyrethroid resistance in other species, leads us to suggest that the rp1 QTL may represent the overexpression of one or more P450 genes. This is supported by the significant upregulation of the Cyp6p4 P450 gene in the Ndja-resistant strain compared with the susceptible Moz strain. Cyp6p4 is also upregulated in a DDT/pyrethroid-resistant population of An. arabiensis from Chad (Ranson, unpublished data) and the ortholog of Cyp6p4 has previously been associated with pyrethroid resistance in the malaria vector An. funestus (Wondji et al., 2009), suggesting that this gene could probably be responsible for the permethrin resistance explains by the rp1 QTL. Functional characterisation of Cyp6p4 is now necessary to determine whether it can metabolise pyrethroid insecticides.
The elucidation of the inheritance of this permethrin resistance in An. arabiensis from Chad will aid the tracking of the spread of resistance alleles in this population, which is important to monitor the success of resistance management strategies.
A full susceptibility to the carbamate bendiocarb was observed in the Ndja strain as well as a nearly full susceptibility to the organophosphate malathion. The absence of the ace-1 mutation in this strain (even in the few specimens alive after malathion exposure) further confirms this susceptibility profile. Therefore, carbamate or organophosphate insecticides represent a suitable alternative for control of An. arabiensis in Chad through indoor residual spraying (IRS) if this resistance to pyrethroids proves widespread in the country.
Data Archiving
Data deposited in the Dryad repository: doi:10.5061/dryad.jf880.
References
Abdalla H, Matambo TS, Koekemoer LL, Mnzava AP, Hunt RH, Coetzee M (2008). Insecticide susceptibility and vector status of natural populations of Anopheles arabiensis from Sudan. Trans R Soc Trop Med Hyg 102: 263–271.
Barrett JC, Fry B, Maller J, Daly MJ (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265.
Casimiro S, Coleman M, Hemingway J, Sharp B (2006). Insecticide resistance in Anopheles arabiensis and Anopheles gambiae from Mozambique. J Med Entomol 43: 276–282.
Chouaïbou M, Etang J, Brévault T, Nwane P, Hinzoumbé CK, Mimpfoundi R et al (2008). Dynamics of insecticide resistance in the malaria vector Anopheles gambiae s.l. from an area of extensive cotton cultivation in Northern Cameroon. Trop Med Int Health 13: 476–486.
Collins FH, Mendez MA, Rasmussen MO, Mehaffey PC, Besansky NJ, Finnerty V (1987). A ribosomal RNA gene probe Ddifferentiates mmber secies of the Anopheles gambiae complex. Am J Trop Med Hyg 37: 37–41.
Diabate A, Baldet T, Chandre F, Dabire KR, Simard F, Ouedraogo JB et al (2004). First report of a kdr mutation in Anopheles arabiensis from Burkina Faso, West Africa. J Am Mosq Control Assoc 20: 195–196.
Duangkaew P, Pethuan S, Kaewpa D, Boonsuepsakul S, Sarapusit S, Rongnoparut P (2011). Characterization of mosquito CYP6P7 and CYP6AA3: differences in substrate preference and kinetic properties. Arch Insect Biochem Physiol 76: 236–248.
Gillies M, Coetzee M (1987) A supplement to the anophelinae of Africa south of the Sahara (Afrotropical region) Vol. 55. Publ South African Inst Med Res.
Hargreaves K, Hunt RH, Brooke BD, Mthembu J, Weeto MM, Awolola TS et al (2003). Anopheles arabiensis and An. quadriannulatus resistance to DDT in South Africa. Med Vet Entomol 17: 417–422.
Hemingway J, Ranson H (2000). Insecticide resistance in insect vectors of human disease. Annu Rev Entomol 45: 371–391.
Huang DW, Sherman BT, Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57.
Kerah-Hinzoumbe C, Peka M, Antonio-Nkondjio C, Donan-Gouni I, Awono-Ambene P, Samè-Ekobo A et al (2009). Malaria vectors and transmission dynamics in Goulmoun, a rural city in south-western Chad. BMC Infect Dis 9: 71.
Kerah-Hinzoumbe C, Peka M, Nwane P, Donan-Gouni I, Etang J, Samè-Ekobo A et al (2008). Insecticide resistance in Anopheles gambiae from south-western Chad, Central Africa. Malar J 7: 192.
Lander ES, Botstein D (1989). Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199.
Matambo TS, Abdalla H, Brooke BD, Koekemoer LL, Mnzava A, Hunt RH et al (2007). Insecticide resistance in the malarial mosquito Anopheles arabiensis and association with the kdr mutation. Med Vet Entomol 21: 97–102.
Mouatcho JC, Munhenga G, Hargreaves K, Brooke BD, Coetzee M, Koekemoer LL (2009). Pyrethroid resistance in a major African malaria vector Anopheles arabiensis from Mamfene, northern KwaZulu-Natal, South Africa. S Afr J Sci 105: 127–131.
Müller P, ChouaÏBou M, Pignatelli P, Etang J, Walker ED, Donnelly MJ et al (2008a). Pyrethroid tolerance is associated with elevated expression of antioxidants and agricultural practice in Anopheles arabiensis sampled from an area of cotton fields in Northern Cameroon. Mol Ecol 17: 1145–1155.
Müller P, Warr E, Stevenson BJ, Pignatelli PM, Morgan JC, Steven A et al (2008b). Field-caught permethrin-resistant Anopheles gambiae overexpress CYP6P3, a P450 that metabolises pyrethroids. Plos Genet 4: e1000286.
Penilla PR, Rodríguez AD, Hemingway J, Torres JL, Arredondo-Jiménez JI, Rodríguez MH (1998). Resistance management strategies in malaria vector mosquito control. Baseline data for a large-scale field trial against Anopheles albimanus in Mexico. Med Vet Entomol 12: 217–233.
Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45.
Ranson H, Abdallah H, Badolo A, Guelbeogo WM, Kerah-Hinzoumbé C, Yangalbé-Kalnoné E et al (2009). Insecticide resistance in Anopheles gambiae: data from the first year of a multi-country study highlight the extent of the problem. Malar J 8: 299.
Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV et al (2002). Evolution of supergene families associated with insecticide resistance. Science 298: 179–181.
Ranson H, Paton MG, Jensen B, McCarroll L, Vaughan A, Hogan JR et al (2004). Genetic mapping of genes conferring permethrin resistance in the malaria vector, Anopheles gambiae. Insect Mol Biol 13: 379–386.
Scott JA, Brogdon WG, Collins FH (1993). Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg 49: 520–529.
Simard F, Lehmann T, Lemasson J-J, Diatta M, Fontenille D (2000). Persistence of Anopheles arabiensis during the severe dry season conditions in Senegal: an indirect approach using microsatellite loci. Insect Mol Biol 9: 467–479.
Stam R, van Ooijen JW (1996) JoinMap (TM) Version 2.0: Software for the Calculation of Genetic Linkage Maps. CPRO-DLO: Wageningen.
Stevenson BJ, Bibby J, Pignatelli P, Muangnoicharoen S, O’Neill PM, Lian LY et al (2011). Cytochrome P450 6M2 from the malaria vector Anopheles gambiae metabolizes pyrethroids: sequential metabolism of deltamethrin revealed. Insect Biochem Mol Biol 41: 492–502.
van Ooijen JW (1999). LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83: 613–624.
Voorrips RE (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93: 77–78.
Wang S, Basten C, Zeng Z (2005) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University: Raleigh, NC.
WHO (1998) Test Procedures for Insecticide Resistance Monitoring in Malaria Vectors, Bio-Efficacy and Persistence of Insecticides on Treated Surfaces: Report of the WHO Informal Consultation. World Health Organization: Geneva.
Wondji C, Simard F, Lehmann T, Fondjo E, Samè-Ekobo A, Fontenille D (2005). Impact of insecticide-treated bed nets implementation on the genetic structure of Anopheles arabiensis in an area of irrigated rice fields in the Sahelian region of Cameroon. Mol Ecol 14: 3683–3693.
Wondji CS, Irving H, Morgan J, Lobo NF, Collins FH, Hunt RH et al (2009). Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome Res 19: 452–459.
Wondji CS, Morgan J, Coetzee M, Hunt RH, Steen K, Black WC et al (2007). Mapping a quantitative trait locus (QTL) conferring pyrethroid resistance in the African malaria vector Anopheles funestus. BMC Genomics 8: 34.
Yewhalaw D, Bortel WV, Denis L, Coosemans M, Duchateau L, Speybroeck N (2010). First evidence of high knockdown resistance frequency in Anopheles arabiensis (Diptera: Culicidae) from Ethiopia. Am J Trop Med Hyg 83: 122–125.
Zeng Z-B (1993). Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Nat Acad Sci USA 90: 10972–10976.
Zeng Z-B, Kao C-H, Basten CJ (1999). Estimating the genetic architecture of quantitative traits. Genet Res 74: 279–289.
Zheng L, Benedict MQ, Cornel AJ, Collins FH, Kafatos FC (1996). An integrated genetic map of the African human malaria vector mosquito, Anopheles gambiae. Genetics 143: 941–952.
Acknowledgements
This work was supported by a Wellcome Trust Research Career Development Fellowship to CSW and the UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (WHO/TDR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Witzig, C., Parry, M., Morgan, J. et al. Genetic mapping identifies a major locus spanning P450 clusters associated with pyrethroid resistance in kdr-free Anopheles arabiensis from Chad. Heredity 110, 389–397 (2013). https://doi.org/10.1038/hdy.2012.112
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2012.112
Keywords
This article is cited by
-
Using sibship reconstructions to understand the relationship between larval habitat productivity and oviposition behaviour in Kenyan Anopheles arabiensis
Malaria Journal (2019)
-
High insecticide resistance in the major malaria vector Anopheles coluzzii in Chad Republic
Infectious Diseases of Poverty (2019)
-
Nationwide insecticide resistance status and biting behaviour of malaria vector species in the Democratic Republic of Congo
Malaria Journal (2018)
-
Investigating insecticide resistance and knock-down resistance (kdr) mutation in Dielmo, Senegal, an area under long lasting insecticidal-treated nets universal coverage for 10 years
Malaria Journal (2018)
-
Investigating knockdown resistance (kdr) mechanism against pyrethroids/DDT in the malaria vector Anopheles funestus across Africa
BMC Genetics (2017)