Abstract
Identification and quantification of spatial genetic structure (SGS) within populations remains a central element of understanding population structure at the local scale. Understanding such structure can inform on aspects of the species’ biology, such as establishment patterns and gene dispersal distance, in addition to sampling design for genetic resource management and conservation. However, recent work has identified that variation in factors such as sampling methodology, population characteristics and marker system can all lead to significant variation in SGS estimates. Consequently, the extent to which estimates of SGS can be relied on to inform on the biology of a species or differentiate between experimental treatments is open to doubt. Following on from a recent report of unusually extensive SGS when assessed using amplified fragment length polymorphisms in the tree Fagus sylvatica, we explored whether this marker system led to similarly high estimates of SGS extent in other apparently similar populations of this species. In the three populations assessed, SGS extent was even stronger than this previously reported maximum, extending up to 360 m, an increase in up to 800% in comparison with the generally accepted maximum of 30–40 m based on the literature. Within this species, wide variation in SGS estimates exists, whether quantified as SGS intensity, extent or the Sp parameter. Consequently, we argue that greater standardization should be applied in sample design and SGS estimation and highlight five steps that can be taken to maximize the comparability between SGS estimates.
Similar content being viewed by others
Introduction
The identification and description of patterns in the distribution of biodiversity is key to understanding species–environment relationships across multiple spatial and temporal scales (Legendre, 1993; de Knegt et al., 2010). Such information can further our understanding of species from their biology to community ecology and broad-scale biogeography (Legendre, 1993). Within communities, resource distribution, interspecific interactions, dispersal limitation and disturbance can all lead to the non-random distribution of individuals (Legendre, 1993; de Knegt et al., 2010). Within populations, non-random distribution of individual genotypes can also occur. In plants, such spatial genetic structure (SGS) is commonly believed to result from restricted pollen and seed dispersal leading to deviation from random mating and offspring being more likely to establish close to the parent plants. Consequently, since it results from breeding and dispersal syndromes, SGS is expected to be comparable between groups of species sharing similar traits (Vekemans and Hardy, 2004).
Determination of SGS within natural populations is important because it can allow us to estimate the extent of gene dispersal within populations (Rousset 2000; Vekemans and Hardy, 2004) and, therefore, to indirectly quantify the dispersal distance of the species (Oddou-Muratorio et al., 2010) and predict the implications for local breeding and evolution (Smouse and Peakall, 1999). Furthermore, such fine-scale characterization of the genetic structure of populations can inform appropriate strategies for stand management and conservation, including in-situ and ex-situ conservation of genetic resources and collection strategies for genetic resource harvesting (Gapare and Aitken, 2005). Conversely, overlooking such structure in reserve design or resource collection might lead to over-representing some genotypes whilst under-representing population genetic diversity in collected material (Gapare and Aitken, 2005).
Given the significance of determining SGS for both theoretical aspects of population genetics and evolution and their application in species conservation and management, it is important to understand how SGS relates to the biology of the species and the degree of inter and intraspecific variation in this parameter. Previous meta-analysis has shown a significant relationship between SGS and plant life form, breeding system and population density (Vekemans and Hardy, 2004). In tree species with restricted seed and pollen dispersal, low gene flow is expected to result in significant genetic differentiation within continuous populations (Cavers et al., 2005; Hardy et al., 2006), whereas high gene flow within populations of wind-pollinated species is expected to result in them demonstrating relatively low levels of SGS (Leonardi and Menozzi, 1996; Streiff et al., 1998). Indeed, in wind-pollinated forest tree species such as the common ash, Fraxinus excelsior (Heuertz et al., 2003), the oaks Quercus petraea and Quercus robur (Bacilieri et al., 1994; Streiff et al., 1998), and the beeches Fagus sylvatica and Fagus crenata (Merzeau et al., 1994; Leonardi and Menozzi, 1996; Takahashi et al., 2000; Asuka et al., 2004; Vornam et al., 2004; Scalfi et al., 2005; Chybicki et al., 2009), significant SGS is rarely detectable beyond approximately 30–40 m. However, recent work has demonstrated that SGS can show marked variation within a single species because of variation in the density of individuals within the population or their life stage. For example, Vekemans and Hardy (2004) found higher SGS in low-density populations based on an analysis of 47 plant species differing in their life form and ecology. SGS is not expected to remain constant over time, instead declining in intensity and/or extent as trees age and the population thins (Chung et al., 2003; Hardesty et al., 2005; Vaughan et al., 2007), although some studies also report contrasting results (for example, Oddou-Muratorio et al., 2010). Tree density and age class may also interact. In the study by Jolivet et al. (2011) on the wild cherry, Prunus avium, SGS was found to be more intense in saplings than adult trees, but only in low-density populations. Furthermore, SGS has been shown to vary across the geographical range of a species with SGS being absent in populations that are part of continuous forest distribution in the core region of the geographical distribution of Sitka spruce, Picea sitchensis despite being relatively strong elsewhere (Gapare and Aitken, 2005). This geographical impact on SGS was interpreted as resulting from higher density of core populations reducing SGS, in agreement with the meta-analysis performed by Vekemans and Hardy (2004) referred to above.
The studies cited above demonstrate that variation in SGS within species can be substantial. Furthermore, such variation can be exacerbated by comparing across different molecular marker types, because the number of markers and degree of polymorphism of individual markers can also influence the results (Cavers et al., 2005; Jump and Peñuelas, 2007; Jolivet et al., 2011). Consequently, single estimates of SGS are unlikely to be reliable to accurately represent SGS for individual species. This potentially high variation becomes a problem, because many studies assume that SGS is a species-level trait. Consequently, the implications of SGS variability for the quantification of dispersal, distribution of diversity within populations or other aspects of the species biology risk being overlooked. Work that has explored variation in SGS across species has been highly valuable in allowing us to understand the trait relationships behind such variation (Vekemans and Hardy 2004; Hardy et al., 2006), however, within such studies, species generally remain represented by single values.
An example of wide variation in SGS within a single species comes from work on P. sitchensis, where the extent of SGS based on sequence tagged site molecular markers was significantly >0 for up to 500 m in some populations and yet remained effectively 0 in others (Gapare and Aitken, 2005). In F. sylvatica, initial work using allozyme and microsatellite markers revealed SGS to be approximately 30–40 m. (Merzeau et al., 1994; Leonardi and Menozzi, 1996; Vornam et al., 2004; Scalfi et al., 2005), while subsequent work identified that SGS extended to up to approximately 110 m when assessments were based on amplified fragment length polymorphism (AFLP) markers (Jump and Peñuelas, 2007).
Given the potential for wide variation in SGS within single species indicated by different studies and marker types, we first sought to determine if the application of AFLP in F. sylvatica leads to consistently high SGS estimates in this species. Subsequently, we aimed to quantify the variation in maximum extent of SGS reported for this species and recommend practical steps to increase the standardization of SGS estimation and maximize the comparability of this parameter between studies.
Materials and methods
Study species
F. sylvatica (European beech) is a monoecious, diploid, late-successional tree that dominates temperate forests over ca 17 million ha of Europe. It is highly outcrossing and largely self-incompatible with irregular synchronous flowering (masting) events. Reproduction does not begin until the species is 40–50 years old (Wagner et al., 2010). Seed is primarily gravity dispersed with average seed dispersal estimated to be <25 m (Gregorius and Kownatzki, 2005), although rare long distance seed dispersal can occur over distances as much as 3 km, effected primarily by the European jay (Garrulus glandarius) (Nilsson, 1985; Kunstler et al., 2007). Pollen dispersal generally occurs over distances up to 250 m, with the vast majority of pollen deposition estimated to occur within 1800 m (Poska and Pidek, 2010; Wagner et al., 2010).
Sites and sampling
This work was conducted in the Catalan Pyrenees in north-east Spain. The F. sylvatica forest is naturally occurring uneven-aged high forest where this species is monodominant with a relatively even distribution of individual trees. Populations were sampled in three valleys, Berguedà, Ripollès and Vall d’Aran. Further details of these populations are given in Table 1.
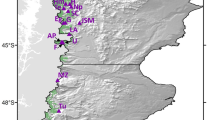
We sampled 150 young trees (< ca 80 years old) from each population in forest with no evidence of any recent disturbance. Trees were sampled from the largest expanse of continuous forest during spring 2009 and the location of each tree was mapped. Samples were collected at random along a series of transects (following SGS sampling recommendations of Zeng et al., 2010) from trees covering the full altitudinal distribution of the species in each location (Figure 1). Leaf material collected from each tree was dried immediately in fine-grain silica gel.
Location of individual F. sylvatica trees samples for assessment of SGS. Data for the Montseny population are taken from Jump and Peñuelas (2007). Gridlines are numbered with Universal Transverse Mercator (UTM) coordinates in kilometers.
DNA extraction and purification
Approximately 0.5 cm2 leaf tissue was ground in liquid nitrogen for 30 s at 30 revolutions per second using a mixer mill (Tissue Lyser, Qiagen Inc., Valencia, CA, USA) and two glass beads. Genomic DNA was extracted from ground tissue using a DNeasy Plant Mini Kit (Qiagen) and quantified using a NanoDrop ND-1000 spectrophotometer running software v3.0.1 (NanoDrop Technologies, Wilmington, DE, USA) following the manufacturer's instructions.
AFLP analysis
AFLP molecular markers were used for this study owing to their high utility for investigating SGS (Cavers et al., 2005; Jump and Peñuelas, 2007). AFLP analysis followed a modified version of the original protocol published by Vos et al. (1995) as detailed below.
Restriction digests and ligation of adapters
DNA (0.3 μg) was digested at 65 °C for 1 h with 5 units TrueI (Roche Applied Science, Basel, Switzerland) and 50 ng μl–1 bovine serum albumin in 35 μl of the manufacturer's buffer (composition confidential). Then 7.5 units of EcoRI (Roche Applied Science) were added and incubation continued at 37 °C for 2 h.
PCR adapters were ligated to the cut fragments by adding to the restriction digest 5 μl of the manufacturer's buffer (composition confidential), 25 pmol EcoRI adapter, 50 pmol MseI adapter and 1 unit T4 DNA ligase (Roche Applied Science). Ligation was allowed to proceed for 3 h at 37 °C and then overnight at 4 °C. The mix was then diluted 5 × with sterile H2O. Oligonucleotide sequences for PCR adapters and primers are those used in the original protocol.
PCR preamplification
In all, 5 μl of the diluted ligation mix was used as the template for the preamplification. PCR was performed with 30 ng Eco-A and 30 ng of Mse-C primers with 0.2 units Expand High Fidelity PCR System (Roche Applied Science) and 0.2 mM each dATP, dCTP, dGTP, dTTP with 2.5 mM MgCl2 in a total of 20 μl of the manufacturer's buffer (composition confidential). The PCR programme consisted of 28 cycles of (94 °C for 30 s, 56 °C for 60 s, 72 °C for 60 s). PCR products were diluted 10 × with sterile H2O.
Selective AFLP amplification
Six Eco-ANN/Mse-CNN primer combinations were tested for the selective amplification (where N represents any nucleotide). Eco-ANN primer extensions were: AAG and ACC. Mse-CNN primer extensions were: CTC, CAG and CAC. Combinations selected for the final genotyping on the grounds of giving the greatest number of polymorphic bands and most easily scored banding profile were, Eco-AAG+Mse-CTC and Eco-ACC+Mse-CAC. The Eco-ANN primer carried a VIC fluorochrome (Applied Biosystems—ABI, Foster City, CA, USA).
In total, 2.5 μl of the diluted PCR products was used as the template for the selective amplification. PCR was performed using 5 ng Eco-ACG primer and 25 ng Mse-CNN primer with 0.16 units Expand High Fidelity PCR System and 0.2 mM each dATP, dCTP, dGTP, dTTP with 2 mM MgCl2 in a total of 10 μl of the manufacturer's buffer. The PCR programme consisted 36 cycles in total: 13 cycles of (94 °C for 30 s, t °C for 30 s, 72 °C for 60 s, where t drops from 65 to 56 in 0.7 °C steps), followed by 23 cycles of (94 °C for 30 s, 56 °C for 30 s, 72 °C for 60 s).
For analysis of the selective AFLP-PCR products, 2 μl of PCR product was mixed with 12 μl Hi-Di Formamide (ABI) 0.4 μl Liz-600 size standard (ABI). Samples were denatured at 94 °C for 3 min and electrophoresis performed using an ABI3130xl genetic analyzer according to the manufacturer's instructions. Fragment sizes were determined with reference to the size standard using GeneMarker v 1.85 (Softgenetics LLC, State College, PA, USA). Fragment presence or absence was scored by hand and subsequently checked by a second investigator without reference to sample ID. A binary matrix of band presence/absence was then created. Each set of 96 reactions included 2 positive (known genotype duplicates) and 2 negative (H2O or PCR mix without DNA) controls carried from restriction digest through to selective AFLP-PCR. Individuals that repeatedly failed to amplify or showed poor amplification were eliminated from the data set. Any band occurring in a sample was excluded from the analysis if it also occurred in a negative control. Only those markers with a maximum allele frequency of 95% or less were included in the final SGS analyses.
Statistical analysis
Population genetic analysis
Population genetic analyses were conducted using AFLP-SURV v 1.0 (Vekemans et al., 2002) with allele frequencies estimated using the Bayesian method with non-uniform prior distribution and Fis of 0.088 (as calculated for a nearby population of this species by Jump and Peñuelas (2007)). Genetic diversity was estimated as expected heterozygosity and genetic divergence between the three populations was estimated by Fst.
Spatial genetic structure
Analyses of SGS were conducted based on the calculation of the kinship coefficient between individuals (Fij), which summarizes the genetic co-ancestry between individuals i and j and can be defined as Fij=(Qij–Qm)/(1–Qm) (where Qij is the probability of identity in state for random genes from i and j, and Qm is the average probability of identity in state for genes coming from random individuals from the sample). Kinship coefficients were calculated from AFLP data according to the dominant marker estimator described by Hardy (2003). For estimation of kinship coefficients, the inbreeding coefficient (Fis) was set to 0.088, however, Hardy (2003) notes that estimation of kinship using this method is relatively robust to errors in Fis estimation.
The mean estimate of Fij over pairs of individuals within a given distance interval (F(d)) was computed based on 20 equal intervals rising by 20 m from 0 to 400 m and plotted against distance to visualize spatial autocorrelation patterns. In all, 95% confidence intervals for F(d) were calculated based on 10 000 random permutations of individuals among geographical locations. To test the hypothesis that there was significant spatial structure, the observed regression slope of Fij on ln(rij), b, was compared with those obtained after 10 000 random permutations of individuals among locations and statistical significance determined using a Mantel test, (where ln(rij) represents the natural logarithm of the physical distance between individuals i and j). This procedure has the advantage that all of the information is contained in a single test statistic and the results are not dependent on arbitrarily set distance intervals (Vekemans and Hardy, 2004). SGS was also quantified by the Sp statistic, which represents the rate of decrease in pairwise kinship with distance (Vekemans and Hardy, 2004). Sp is calculated as –b/(1–F(1)) (where F(1) is the mean Fij between individuals in the first distance class). Following Hardy et al. (2006), we present the standard error (s.e.) of b (calculated by jackknifing over loci) as an estimate of the variability of Sp. This estimate does not take into account the s.e. of F(1), because its impact on the SE of Sp is very small in comparison with that of b (OJ Hardy, personal communication). All analyses of SGS were conducted using the program SPAGeDi V1.2d (Hardy and Vekemans, 2002).
In order to compare the data for the three populations sampled here with the previous results found for the nearby Montseny population by Jump and Peñuelas (2007), we randomly excluded loci and individuals from the data set of Jump and Peñuelas (2007) to give a population size of 150 individuals genotyped at 193 AFLP loci. The Montseny population was then subject to the same data analysis to that described above for the three Pyrenean populations.
Results
Population genetic analyses
A total of 317 AFLP polymorphic markers (allele frequency <1) were scored over 414 individuals from the Ripollès, Berguedà and Vall d’Aran populations. The number of markers scored per primer combination was 155 for Eco-AAG/Mse-CTC and 162 for Eco-ACC/Mse-CAC. In all, 193 markers polymorphic at the 95% criterion and with an error rate of 5.3% (Pompanon et al., 2005) were used for subsequent analyses. Expected heterozygosity ranged from 0.173 for the Berguedà population to 0.189 for Vall d’Aran (Table 1). Fst calculated between all three populations was 0.115.
Spatial genetic structure
Significant SGS was detected over all individuals sampled within each study population (P<0.001, Figure 2). The mean number of pairwise comparisons per distance class was 79 (24–101) for Ripollès, 83 (45–103) for Berguedà and 91 (59–127) for Vall d’Aran (means are followed by the range in parentheses). The mean sample size per distance class for the reanalyzed Montseny data was 75 (60–95).
Analysis of SGS in F. sylvatica forest based on AFLP data. Solid lines indicate the mean kinship coefficient per distance class (F(d)) and dashed lines the limits of its 95% confidence interval. Individuals are significantly more similar than would be expected by chance when F(d) lies above its 95% confidence limit. Patterns of SGS in all species differ significantly from a random distribution of individuals at P<0.001 based on the regression slope of individual pairwise kinship coefficients on the corresponding logarithmic geographical distance between individuals (see text). Sp represents the rate of decrease in pairwise kinship with distance. The Montseny analysis is based on data from Jump and Peñuelas (2007).
The lowest maximum intensity of SGS (F(max), 0.062 based on the reanalyzed Montseny data) was 36% of the value found for the population with the highest maximum intensity (0.173, Berguedà; Table 1; Figure 2). In the Ripollès and Montseny populations, this maximum was reached in the first distance class (F(1), 0–20 m) whereas in the Berguedà and Vall d’Aran populations maximum SGS intensity was recorded in the second distance class (F(2), 20–40 m; Figure 2). For all populations, the mean kinship coefficient per distance class, F(d), declines with increasing distance. However, the extent of SGS also varies between populations with the minimum estimate (SGSmin, the point at which F(d) first becomes statistically indistinguishable from 0 at P=0.05) varying from 200 m in the Vall d’Aran population to 80 m (40% of the Vall d’Aran value) at Montseny (Table 1). The maximum estimate of SGS extent (SGSmax, the greatest distance at which F(d)>0 at P=0.05 before F(d) crosses the x axis) in the population with the lowest value for this SGS parameter (Montseny, 180 m) is 50% of the highest value of this parameter (360 m, Berguedà). Values of Sp calculated from our full data sets varied from 0.0054 for Montseny to 0.0222 at Berguedà, with the minimum value being only 24% of the maximum reported value.
Discussion
The populations that we investigated show pronounced variation in the intensity and extent of their SGS. Of the populations newly investigated here, Berguedà shows the greatest intensity of SGS and extent of SGSmax as well as the highest value of Sp whereas the Vall d’Aran population shows the greatest extent of SGSmin. Tree density varies little between the populations studied, with the least-dense population (Ripollès) having a density some 80% that of the most-dense (Berguedà) based on data taken from forest inventory records (Table 1). Here, SGS is generally lowest in the lowest density population with a 25% increase in tree density being associated with an increase in SGS of 38% (when quantified by SGSmax) – 120% (when quantified by Sp) when minimum and maximum values are compared for each parameter. When we include the reanalyzed data for the Montseny population in these comparisons, an increase in tree density of 27% is associated with an increase in SGS of 100% (SGSmax) – 340% (Sp). Although the data sets for the Montseny population and the other populations reported here are partially different, the Eco-AAG + Mse-CTC primer combination is common to both data sets. Furthermore, the subsampling study conducted by Jump and Peñuelas (2007) indicates that, given the large number of markers used, the difference in identity of the individual AFLP loci is likely to have very little impact on the correlograms produced.
Our results clearly show that pronounced variation in SGS exists between the apparently similar F. sylvatica populations included in this study. However, the most striking implications of these results arise when these new quantifications of SGS in F. sylvatica are compared with previous estimates for this and other wind-pollinated tree species. The consensus from the literature is that SGS in such species is weak. For example, the extent of SGS is generally accepted to be limited to 30–40 m in F. sylvatica (Merzeau et al., 1994; Leonardi and Menozzi, 1996; Vornam et al., 2004; Oddou-Muratorio et al., 2010), and Q. petraea (Bacilieri et al., 1994; Streiff et al., 1998) and up to 60 m in Q. robur (Bacilieri et al., 1994; Streiff et al., 1998; Hampe et al., 2010) and Fraxinus excelsior (Heuertz et al., 2003). The values given in this study represent an increase in detected SGS extent for F. sylvatica of 567–800% depending on whether maximum values for SGSmin or SGSmax are compared. Variation in other SGS parameters is not so extreme, with F(max) increasing by 111% in comparison with values presented by Oddou-Muratorio et al. (2010), while the maximum value of Sp reported by Oddou-Muratorio et al. (2010) (0.0354) is 61% higher than the maximum value that we report here.
Given this wide variation in SGS reported from within a single species, clear issues arise related to the reporting, comparison and application of SGS estimates. The much lower values for SGS extent reported above for wind-pollinated trees (30–40 m for F. sylvatica and Q. petraea, 60 m for Q. robur and F. excelsior) were all derived using SSR markers, whereas the much more extensive SGS (up to 360 m) reported here was assessed using AFLP. Given this order of magnitude difference between estimates, the comparability of SGS estimates from different markers seems doubtful. Although it might be argued that the new estimates of SGS presented here are high because of unusually strong SGS rather than an effect of the marker system, a previous study in F. sylvatica demonstrated that even assessing the same individuals with different markers lead to SGS extent that was 267% higher when assessed using AFLP as opposed to SSR markers (Jump and Peñuelas, 2007). Studies comparing estimates of population differentiation based on AFLP and SSR markers often report higher divergence when calculated based on AFLP data (Mariette et al., 2002; Gaudeul et al., 2004; Hollingsworth and Ennos, 2004; Woodhead et al., 2005). Higher differentiation might be detected with AFLP markers if they are localized within the chloroplast or mitochondrial genome, and because there is a greater likelihood that some AFLPs will be linked to adaptive traits (Maguire et al., 2002; Mariette et al., 2002; Gaudeul et al., 2004). However, neither explanation sufficiently explains the large differences in SGS identified using these markers, which might be better explained by the greater genome coverage of the more numerous AFLP markers and the greater polymorphism of the individual SSR Loci (Hollingsworth and Ennos, 2004; Woodhead et al., 2005; Jump and Peñuelas, 2007; Skrede et al., 2009).
Sp represents an integrated estimate of both SGS intensity and extent and is believed to be the most readily comparable estimate of SGS between sites and species (Vekemans and Hardy, 2004). However, recent work demonstrates that Sp can also vary by an order of magnitude within the same species, for example, from a lower estimate of 0.0037 (Jump and Peñuelas, 2007) to 0.0354 (Oddou-Muratorio et al., 2010), both based on SSR data in F. sylvatica. The extent of reported variation of Sp within F. sylvatica is, therefore, 2.6 times greater than the variation across five wind-pollinated tree species (0.0020–0.0108) listed by Vekemans and Hardy (2004) and 12% of the total variation in Sp shown across all 47 species with diverse reproductive and dispersal ecology included in this meta-analysis. Such wide variation in Sp may be due, in part, to the observation that, if the relationship between F(d) and ln(r) departs strongly from linearity, estimates of Sp can also vary within a population depending on the maximum distance sampled (Vekemans and Hardy, 2004). The linearity of this relationship is rarely tested in studies reporting Sp. However, recent work demonstrates that not accounting for such deviation can lead to estimates of Sp varying by up to 280% within a single population (Jump and Peñuelas, 2007).
Despite using the same molecular markers and a broadly equivalent sampling regime SGS varied by 38–120% between the populations newly sampled for this study implying that, in many cases, differences reported between species could fall within the amount of variability expected between conspecific populations. Given that SGS within a species will be impacted by a wide range of factors that affect the distribution of individuals, their establishment success and longevity (Chung et al., 2003; Oddou-Muratorio et al., 2004; Gapare and Aitken, 2005; Hardesty et al., 2005; Vaughan et al., 2007; Hampe et al., 2010), it is unsurprising that intraspecific variation in SGS occurs. However, such wide variation within a single species highlights the dangers of characterizing SGS at the species level based on very few values and, in some cases, only a single estimate. Consequently, we caution against the uncritical comparison of SGS between studies, whether as SGS extent, intensity, or Sp and argue that in order to compare SGS between studies, a much greater degree of standardization in reporting is necessary. With this aim, we outline five points that should be taken in order to maximize the SGS comparability:
-
1)
Compare SGS estimates only within marker types and for similar marker numbers—thereby removing the variation associated with non-comparability of estimates based on different classes of markers and maximizing comparability within markers.
-
2)
Calculate Sp only over distances up to the maximum distance for which the relationship between F(d) and ln(r) is demonstrably linear—thereby removing methodologically based variation because of non-comparability in Sp calculation.
-
3)
When comparing between sites or treatments, the sampling of individuals should be standardized in order to maximize comparability, such as in the recent study of Q. robur by Hampe et al. (2010).
-
4)
When quantifying SGS or comparing different estimates, ensure that the number of markers and individuals used to derive estimates of SGS are sufficient to detect SGS where it exists (for example, using the subsampling approach of Cavers et al. (2005). Thereby avoiding the potential of falsely rejecting the occurrence of SGS in one or more populations because of a lack of statistical power, rather than a real absence of SGS. The use of a sub-optimal sampling design might still give informative comparisons within studies, assuming that other aspects of sampling design are held constant, although it is likely to limit cross-study comparability.
-
5)
When using SGS estimates to describe ecological characteristics at the species level (for example, the extent of gene flow, neighborhoods for collection of genetic material), give the mean and associated variation and the number of single population estimates from which the mean is derived.
Conclusions
Three superficially similar populations of F. sylvatica investigated for SGS for this study show SGS variation of up to 120%, despite similar tree density. These new estimates of SGS result in an increase in the generally accepted maximum extent of SGS by up to 800% in this species, with individuals being more genetically similar than would be expected by chance up to 360 m from each individual tree. In conjunction with estimates from earlier studies, it is clear that wide variation in SGS estimates exist whether quantified as SGS intensity, extent, or the Sp parameter. Furthermore, SGS estimates appear to show low comparability between AFLP and other marker types with much greater SGS extent observed using AFLP markers. Consequently, much greater caution should be applied when comparing SGS estimates between studies within or between species and greater standardization should be applied in sample design and SGS estimation and reporting. Such caution is particularly important when applying SGS estimates to inform on the biology of a species for conservation or management purposes, such as the estimation of gene flow within species or in sampling design for the collection or conservation of genetic resources.
Data archiving
Genotype data have been deposited at Dryad, doi:10.5061/dryad.6qg398d1.
References
Asuka Y, Tomaru N, Nisimura N, Tsumura Y, Yamamoto S (2004). Heterogeneous genetic structure in a Fagus crenata population in an old-growth beech forest revealed by microsatellite markers. Mol Ecol 13: 1241–1250.
Bacilieri R, Labbe T, Kremer A (1994). Intraspecific genetic structure in a mixed population of Quercus petraea (Matt) Leibl and Quercus robur L. Heredity 73: 130–141.
Cavers S, Degen B, Caron H, Lemes MR, Margis R, Salgueiro F et al. (2005). Optimal sampling strategy for estimation of spatial genetic structure in tree populations. Heredity 95: 281–289.
Chung MY, Epperson BK, Chung MG (2003). Genetic structure of age classes in Camellia japonica (Theaceae). Evolution 57: 62–73.
Chybicki IJ, Trojankiewicz M, Oleksa A, Dzialuk A, Burczyk J (2009). Isolation-by-distance within naturally established populations of European beech (Fagus sylvatica). Botany 87: 791–798.
de Knegt HJ, van Langevelde F, Coughenour MB, Skidmore AK, de Boer WF, Heitkönig IMA et al. (2010). Spatial autocorrelation and the scaling of species-environment relationships. Ecology 91: 2455–2465.
Gapare WJ, Aitken SN (2005). Strong spatial genetic structure in peripheral but not core populations of Sitka spruce [Picea sitchensis (Bong.) Carr.]. Mol Ecol 14: 2659–2667.
Gaudeul M, Till-Bottraud I, Barjon F, Manel S (2004). Genetic diversity and differentiation in Eryngium alpinum L. (Apiaceae): comparison of AFLP and microsatellite markers. Heredity 92: 508–518.
Gregorius H-R, Kownatzki D (2005). Spatiogenetic characteristics of beech stands with different degrees of autochthony. BMC Ecol 5:8 (doi:10.1186/1472-6785-5-8).
Hampe A, El Masri L, Petit RJ (2010). Origin of spatial genetic structure in an expanding oak population. Mol Ecol 19: 459–471.
Hardesty BD, Dick DW, Kremer A, Hubbell S, Bermingham E (2005). Spatial genetic structure of Simarouba amara Aubl. (Simaroubaceae), a dioecious, animal-dispersed Neotropical tree, on Barro Colorado Island, Panama. Heredity 95: 290–297.
Hardy OJ (2003). Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Mol Ecol 12: 1577–1588.
Hardy OJ, Maggia L, Bandou E, Breyne P, Caron H, Chevallier MH et al. (2006). Fine-scale genetic structure and gene dispersal inferences in 10 Neotropical tree species. Mol Ecol 15: 559–571.
Hardy OJ, Vekemans X (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Heuertz M, Vekemans X, Hausman JF, Palada M, Hardy OJ (2003). Estimating seed vs. pollen dispersal from spatial genetic structure in the common ash. Mol Ecol 12: 2483–2495.
Hollingsworth PM, Ennos RA (2004). Neighbour joining trees, dominant markers and population genetic structure. Heredity 92: 490–498.
Jolivet C, Holtken AM, Liesebach H, Steiner W, Degen B (2011). Spatial genetic structure in wild cherry (Prunus avium L.): I. variation among natural populations of different density. Tree Genet Genomes 7: 271–283.
Jump AS, Peñuelas J (2007). Extensive spatial genetic structure revealed by AFLP but not SSR molecular markers in the wind-pollinated tree, Fagus sylvatica. Mol Ecol 16: 925–936.
Kunstler G, Thuiller W, Curt T, Bouchaud M, Jouvie R, Deruette F et al. (2007). Fagus sylvatica L. recruitment across a fragmented Mediterranean Landscape, importance of long distance effective dispersal, abiotic conditions and biotic interactions. Diversity Distrib 13: 799–807.
Legendre P (1993). Spatial autocorrelation: trouble or new paradigm? Ecology 74: 1659–1673.
Leonardi S, Menozzi P (1996). Spatial structure of genetic variability in natural stands of Fagus sylvatica L (beech) in Italy. Heredity 77: 359–368.
Maguire TL, Peakall R, Saenger P (2002). Comparative analysis of genetic diversity in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae) detected by AFLPs and SSRs. Theor Appl Genet 104: 388–398.
Mariette S, Cottrell J, Csaikl UM, Goikoechea P, Konig A, Lowe AJ et al. (2002). Comparison of levels of genetic diversity detected with AFLP and microsatellite markers within and among mixed Q. petraea (MATT.) LIEBL. and Q. robur L. stands. Silvae Genet 51: 72–79.
Merzeau D, Comps B, Thiebaut B, Cuguen J, Letouzey J (1994). Genetic structure of natural stands Of Fagus sylvatica L (beech). Heredity 72: 269–277.
Nilsson SG (1985). Ecological and evolutionary interactions between reproduction of beech Fagus sylvatica and seed eating animals. Oikos 44: 157–164.
Oddou-Muratorio S, Bontemps A, Klein EK, Chybicki I, Vendramin GG, Suyama Y (2010). Comparison of direct and indirect genetic methods for estimating seed and pollen dispersal in Fagus sylvatica and Fagus crenata. Forest Ecol Manag 259: 2151–2159.
Oddou-Muratorio S, Demesure-Musch B, Pelissier R, Gouyon PH (2004). Impacts of gene flow and logging history on the local genetic structure of a scattered tree species, Sorbus torminalis L. Crantz. Mol Ecol 13: 3689–3702.
Pompanon F, Bonin A, Bellemain E, Taberlet P (2005). Genotyping errors: causes, consequences and solutions. Nat Rev Genet 6: 847–859.
Poska A, Pidek I (2010). Pollen dispersal and deposition characteristics of Abies alba, Fagus sylvatica and Pinus sylvestris, Roztocze region (SE Poland). Veg Hist Archaeobotany 19: 91–101.
Rousset F (2000). Genetic differentiation between individuals. J Evol Biol 13: 58–62.
Scalfi M, Piovani P, Piotti A, Leonardi S, Menozzi P. (2005). Effects of habitat fragmentation on genetic structure of beech populations in central Italy. In: The Role of Biotechnology for the Characterisation and Conservation of Crop, Forestry, Animal and Fishery Genetic Resources, FAO electronic forum on biotechnology in food and agriculture, Turin, Italy, http://www.fao.org/biotech/torino05.htm.
Skrede I, Borgen L, Brochmann C (2009). Genetic structuring in three closely related circumpolar plant species: AFLP versus microsatellite markers and high-arctic versus arctic-alpine distributions. Heredity 102: 293–302.
Smouse PE, Peakall R (1999). Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82: 561–573.
Streiff R, Labbe T, Bacilieri R, Steinkellner H, Glossl J, Kremer A (1998). Within-population genetic structure in Quercus robur L. and Quercus petraea (Matt.) Liebl. assessed with isozymes and microsatellites. Mol Ecol 7: 317–328.
Takahashi M, Mukouda M, Koono K (2000). Differences in genetic structure between two Japanese beech (Fagus crenata Blume) stands. Heredity 84: 103–115.
Vaughan SP, Cottrell JE, Moodley DJ, Connolly T, Russell K (2007). Distribution and fine-scale spatial-genetic structure in British wild cherry (Prunus avium L.). Heredity 98: 274–283.
Vekemans X, Beauwens T, Lemaire M, Roldan-Ruiz I (2002). Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11: 139–151.
Vekemans X, Hardy OJ (2004). New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13: 921–935.
Vornam B, Decarli N, Gailing O (2004). Spatial distribution of genetic variation in a natural beech stand (Fagus sylvatica L. based on microsatellite markers. Conserv Genet 5: 561–570.
Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M et al. (1995). AFLP - a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414.
Wagner S, Collet C, Madsen P, Nakashizuka T, Nyland RD, Sagheb-Talebi K (2010). Beech regeneration research: from ecological to silvicultural aspects. For Ecol Manag 259: 2172–2182.
Woodhead M, Russell J, Squirrell J, Hollingsworth PM, Mackenzie K, Gibby M et al. (2005). Comparative analysis of population genetic structure in Athryium distentifolium (Pteridophyta) using AFLPs and SSRs from anonymous and transcribed genomic regions. Mol Ecol 14: 1681–1695.
Zeng LY, Xu LL, Tang SQ, Tersing T, Geng YP, Zhong Y (2010). Effect of sampling strategy on estimation of fine-scale spatial genetic structure in Androsace tapete (Primulaceae), an alpine plant endemic to Qinghai-Tibetan Plateau. J Syst Evol 48: 257–264.
Acknowledgements
This work was funded by grants from the Catalan Government (SGR2009-458), the Spanish Ministry of Education and Science (CGL2006-04025/BOS, CGL2010-17172/BOS and Consolider - Ingenio MONTES CSD 2008-00040) and via Natural Environment Research Council (NERC) grant NE/G002118/1 for ERA-net BiodivERsA project, Beech Forests for the Future.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Jump, A., Rico, L., Coll, M. et al. Wide variation in spatial genetic structure between natural populations of the European beech (Fagus sylvatica) and its implications for SGS comparability. Heredity 108, 633–639 (2012). https://doi.org/10.1038/hdy.2012.1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2012.1
Keywords
This article is cited by
-
Factors determining fine-scale spatial genetic structure within coexisting populations of common beech (Fagus sylvatica L.), pedunculate oak (Quercus robur L.), and sessile oak (Q. petraea (Matt.) Liebl.)
Annals of Forest Science (2024)
-
A novel synthesis of two decades of microsatellite studies on European beech reveals decreasing genetic diversity from glacial refugia
Tree Genetics & Genomes (2023)
-
High gene flow through pollen partially compensates spatial limited gene flow by seeds for a Neotropical tree in forest conservation and restoration areas
Conservation Genetics (2021)
-
Fine-scale analysis reveals a potential influence of forest management on the spatial genetic structure of Eremanthus erythropappus
Journal of Forestry Research (2021)
-
Patterns of genetic variation in leading-edge populations of Quercus robur: genetic patchiness due to family clusters
Tree Genetics & Genomes (2020)