Abstract
Patagonian forests are the southernmost temperate forests in the world, and Nothofagus pumilio is one of their most ecologically important tree species (i.e., a foundation species). It presents great adaptability and a wide distribution range, making it a suitable model for predicting the performance of trees facing global climate change. N. pumilio forests are increasingly threatened by extreme climatic events and anthropogenic activities. This study aims to identify priority conservation areas and Genetic Zones (GZs) for N. pumilio, promoting the implementation of specific practices to ensure its management and long-term preservation. Thirty-five populations (965 trees) sampled across its distribution (more than 2200 km latitudinally) were genotyped with SSRs, and geographical patterns of genetic variation were identified using Bayesian approaches. The phylogeographic patterns of the species and geomorphological history of the region were also considered. Six priority conservation areas were identified, which hold high allelic richness and/or exclusive allelic variants. Eighteen GZs were delineated based on the genetic structure of this species, and maps showing their distributional range were drawn up. Overall, this study defines management units based on genetic data for N. pumilio for the first time, which will facilitate the establishment of sustainable practices and highlight priorities for investment of conservation funding.
Similar content being viewed by others
Introduction
Patagonian forests represent the most southern temperate forests in the world, and were isolated from other continental forests approximately 40–2.5 million years before present1. The biological value of these forests lies partly in their high level of endemism and frequency of taxonomically isolated genera, despite low species richness in comparison with other South American forests1. Together with some conifer species, southern beeches of the Nothofagus genus constitute the main components of Andean Forests, which are recognized as providing a wide range of ecological, economic, and social benefits and services. Therefore, the management of these native forests is required in order to maintain the ecological dynamics of this unique ecosystem. Several studies have documented the role of climatic variability during the Quaternary in shaping the current distribution of Patagonian forests2,3,4. In addition, extreme climatic events have recently been suggested as one of the causal agents of current forest decline in Northern Patagonia5, e.g. severe droughts occurring after a wet period triggered N. pumilio decline6. In this regard, an increase in mean maximum summer temperature, overall warming, and a marked decrease in annual precipitation were detected in this region throughout the twentieth century7,8, threatening the persistence of these forests. Besides, anthropogenic and natural causes such as logging and more frequent fires9,10 also endanger these forests.
Nothofagus pumilio, commonly called lenga, is one of the most important species of Patagonian forests, since it has the largest distribution area of all native trees in the region, covering about 1.636 million hectares and occupying a range of about 20° latitude (from 36°S to 55°S) in Argentina. This species has a similar distributional range to the west of the Andes mountain range, in Chile11. Moreover, it occurs in heterogeneous environments along altitudinal, latitudinal, and longitudinal gradients that imply diverse and contrasting thermal, photoperiodic, and precipitation conditions, respectively. N. pumilio shows great adaptation or plasticity to extreme and contrasting environments, since it grows close to the timberline (cold-tolerant species) and thrives in a wide range of precipitation levels (from 3000 to 300 mm average annual precipitation)12. In addition, several phenotypic variations have been described for this species across environmental clines; e.g., bud phenology variation and seedling growth associated with altitude13,14, and wood density related to soil moisture gradients15. Since N. pumilio exhibits great adaptability, it serves as a suitable model for prediction of the performance and fitness of trees to cope with global climate change in the southern hemisphere. Consequently, a research is currently being undertaken to determine whether local adaptation of this Southern beech evolved in a convergent way with respect to European tree species that are also distributed along broad environmental gradients (bi-national project, grant number 410759149, German Research Foundation-DFG). Natural hybridization between N. pumilio and N. antarctica has been observed in sympatric populations16,17 and it has been suggested that the latter acts predominantly as pollen receptor18. The introgression of the chloroplast genome from N. pumilio to N. antarctica would have occurred as a consequence of repeated backcrosses between hybrids and parents, and the former is clearly distinguished at the nuclear level19.
The importance of defining operational genetic management units and priority conservation areas in N. pumilio forests
Government directives regulating the criteria and indicators for defining management units, such as Conservation Units (CU), became important worldwide during the last decades of the twentieth century. Appropriate management of native resources depends on understanding the nature and distribution of genetic diversity among and within species, and highlighting how this genetic variation is structured across their distribution range. Since most native forests are widely distributed and composed of hundreds of natural populations, management of their genetic resources cannot be based on single-population units. Thus, the establishment of operational genetic management units (OGMUs) is necessary for implementation of specific practices arising from conservation or restoration programs. In Argentina, OGMUs were delineated in three native species based on genetic criteria: Austrocedrus chilensis20,21, Nothofagus nervosa and Nothofagus obliqua22,23. According to the National Law N° 26331 on Minimum Requirements for Environmental Protection of Native Forests, most of the Argentinean N. pumilio forests (up to 97%) were included in areas of high (38%) or moderate (59.1%) conservation value24. Therefore, forest management is highly encouraged for this species, particularly the identification of priority conservation areas, in order to ensure compliance with this law and their long-term preservation. The identification of Genetic Zones (GZs) is one alternative of OGMU delineation. GZs are genetically homogeneous regions within which seeds can be transferred with low risk of causing change in the original genetic structure25,26. A requirement for the identification of GZs is the characterization of the population structure of a species, resulting from genetic signatures of evolutionary history, which can be assessed by neutral molecular markers. A second, well-known alternative for OGMU delineation is the definition of Provenance Regions (PRs), which aims to ensure the safe transference of propagation material without risk of maladaptation since it is based on variation in adaptive characters27. In addition, assessment of species’ genetic diversity with molecular markers could facilitate the identification of priority conservation areas28,29. Several authors have suggested that areas containing high allelic richness and/or exclusive allelic variants are meaningful for conservation purposes30,31,32, particularly because they play a key role as genetic diversity reservoirs or sources of locally-adapted genotypes.
The aim of this work is to identify priority conservation areas in N. pumilio forests. Genetically homogeneous regions (i.e., GZs) were first delineated by assessing the neutral variation of the target species across its natural distribution range in Argentina. Based on this, geographical patterns of N. pumilio genetic variation were recognized. In addition, areas holding high allelic richness and/or exclusive allelic variants were identified. Previous information from N. pumilio phylogeographic patterns33 was considered and new information from SSR markers was added.
Results and discussion
Intra-species genetic diversity
The characterization of intraspecific genetic diversity patterns is crucial for development of sustainable forest management prescriptions34, due to its usefulness in determining the persistence and adaptive potential of a species. Therefore, indicators of genetic diversity should be considered when developing criteria and guidelines for forest management35,36,37. A set of genetic parameters was used here in order to characterize the N. pumilio populations (Table 1, Fig. 1).
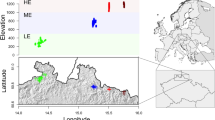
Current distribution range of Nothofagus pumilio in Argentina and location of the sampled populations used in this study. Population IDs correspond to those presented in Table 1. The map was created using ArcGIS 9.2 (ESRI, https://www.esri.com).
Significant differences in unbiased heterozygosity were detected among populations using Wilcoxon’s test (p < 0.05). In particular, HE Nei for Tr population (0.757) was significantly higher than the HE Nei obtained for PG (0.581), Pi (0.573), BL (0.537), and HP (0.398) populations. In addition, both BL and HP exhibited the lowest heterozygosity values, suggesting that they hold lower genetic diversity than the other populations analyzed.
Inbreeding and bottlenecks are demographic events that shape population genetic variation, specifically, reducing allelic variants. Signs of inbreeding were detected in 12 populations (Table 1), but only Np showed a significant inbreeding coefficient after standard errors of the estimates were calculated by jackknifing across loci (FIS” = 0.1461, Z > 1.96). Since this population is isolated from the main forest masses (western side of the species’ distribution) and located in an area that was covered by an ice field at the Last Glacial Maximum (LGM), we hypothesize that few individuals contributed to the recolonization process. In addition, a population bottleneck was identified in Sierra Colorada (SC), since it showed significant heterozygosis excess under both Infinite Allele Model -IAM and Two Phase Model -TPM mutation models (Wilcoxon’s test, p < 0.05).
The differentiation coefficient for N. pumilio was estimated from the FST index with and without implementing the ‘exclusion of null alleles’ (ENA) method (0.077 [0.050–0.113] and 0.078 [0.053–0.115], respectively). The indexes did not differ substantially, indicating that null alleles did not strongly bias the FST value. At the same time, the standardized Hedrick’s G′STH (calculated based on the GST coefficient = 0.075) was 0.201. These differentiation coefficients (FST and GST) were similar to those previously reported in other Nothofagus species such as N. nervosa (0.061) and N. obliqua (0.049)22.
An isolation-by-distance (IBD) pattern was not supported by the Mantel test and the Spatial Genetic Structure (SGS) test (see Supplementary Fig. S1), in accordance with a previous study that involved 20 N. pumilio populations19. Based on these results, an estimation of population structure was carried out, implementing spatial clustering methods as recommended by Perez et al.38. In contrast, the genetic and geographic distances do correlate with each other when using maternally inherited cpDNA markers33. In this regard, it was suggested that the pollen dispersal capacity of N. pumilio greatly exceeds that of seed dispersal (75–100 times greater)39, as in other wind-pollinated trees40,41, which could partly explain the differences found between cpDNA and SSR results from IBD analyses.
Cluster definition and genetic zone delineation
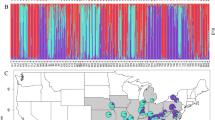
Our results showed that a main division between northern and southern chloroplast lineages took place between Cholila (42° 30′S) and Huemules (42° 50′S) in N. pumilio forests, since all individuals analyzed from these two sites exhibited either northern or southern haplotypes, respectively. The evolutionary significance of this lineage segregation prompted us to consider these findings as the first division criterion for exploration of the genetic structure of Argentinean N. pumilio forests located to the north and south of these boundaries (Fig. 2).
From the genetic structure analysis (BAPS) of the ‘north’ subset, four main clusters were identified which grouped the populations with an adjustment probability of 0.941 (Supplementary Fig. S2 Ia). In addition, the admixture patterns exhibited in each cluster were taken into account in defining the number of genetic zones that fit best with the structure of N. pumilio northern populations (Supplementary Fig. S2 Ib and Ic). Based on this, an extra division was considered within the largest cluster that extends between 37° 51′S and 41° 58′S. In agreement with these results, five genetic zones were defined which grouped the populations as follows: GZ1 Epu; GZ2 Cav, BM, and Tr; GZ3 Q, Pu, Ot, V, and Pi; GZ4 HP; and GZ5 M and Ch (Fig. 3). At the time the genetic diversity for these GZs was studied, it was found that GZ2 and GZ3 presented high AR, LCA, and Pa (Table 2). In this regard, several studies suggested that the glacial setting in northern Patagonia led to a higher occurrence of potential refuges at these latitudes than in the southern area42,43, as well as more favorable slope-down habitats44. The Lácar Lake region (included in GZ3) was reported as a putative relictual zone33 since it exhibits high genetic diversity in plants (e.g., Calceolaria polyrhiza45) and animals (e.g., lizards46).
Genetic Zones (GZs) for Nothofagus pumilio based on genetic criteria. LGM line corresponds to the Last Glacial Maximum advance (based on Glasser et al.49). Maps were created using ArcGIS 9.2 (ESRI, https://www.esri.com).
Since the ‘south’ subset comprises a large area with a diverse evolutionary history (i. e., the presence or absence of glacial refugia and post-glacial recolonization patterns), other division criteria were also considered in defining the optimum number of clusters. A particular topographic configuration occurs at 44°S where there are changes in the orientation of the watersheds (north–south oriented mountain chains between 42°S and 44°S shift afterward to a west–east orientation). This feature contributed to different ice mass development during the glacial period47, which led to a particular arrangement of ecological niches and refugia48. Because of these peculiarities, the populations located between 42° 30′S and 44°S (seven populations) were analyzed separately (Fig. 2). The genetic structure analysis of these populations allows us to identify the three main clusters containing them (K = 3 with a probability of 0.865) (Supplementary Fig. S2 IIa). In addition, the admixture test suggested a greater degree of division by identifying a particular situation in Huemules (Hm) (located in the ‘Cordón Rivadavia’ area) which exhibits an almost balanced admixture of K1 and K2 clusters (Supplementary Fig. S2 IIb and IIc). In this way, four GZs were delineated for the area between 42° 30′S and 44°S: GZ6 Te and Np; GZ7 Hm; GZ8 H and JSM; and GZ9 SC, G, and Eg (Fig. 3). GZ6 (including Np and Te populations) showed high genetic diversity (Table 2), which could be explained, at least in part, by the occurrence of hybridization between N. pumilio and N. antarctica sympatric populations, as reported by Soliani et al.19. In addition, high LCA and Pa were found in GZ9 (Table 2).
The glacial history south of 44°S showed different processes than the north, leading to the development of a greater ice mass at LGM in this area2, 4, 49. In addition, palynological records suggest a different paleoclimatic history for the area south of 50°S, which shaped the forest expansion in a different way (and could also have imprinted the genetic patterns) in southern Patagonia and Tierra del Fuego during the postglacial period3. Furthermore, a particular recolonization process was suggested for Tierra del Fuego Island, mainly led by trees from local periglacial refugia50. Currently, the N. pumilio Argentinean forest south of 44°S is highly fragmented along a narrow range on the Andes hillsides. Nonetheless, an exception to this configuration occurs on Tierra del Fuego Island, where N. pumilio forest probably retained or recovered its continuity. Moreover, this forest mass is present at low altitudes (less than 600 m a. s. l.) and is highly influenced by seasonal storm track patterns. In this study the five populations located in Tierra del Fuego were analyzed separately (Fig. 2), considering all the information previously mentioned.
N. pumilio populations located south of 44°S, excluding those from Tierra del Fuego, were grouped in four clusters by BAPS (Supplementary Fig. S2 IIIa). In addition, admixture among gene pools was found in the ‘Alto Río Senguer’ basin (around 44° 50′S) (Supplementary S2. S2 IIIb and IIIc). In particular, the Union (U) population displayed genetic mixing between Arroyo Perdido (AP) and Fontana (F) gene pools. Based on this, six genetic zones were delineated between 44°S and 52°S: GZ10 LA; GZ11 F; GZ12 AP; GZ13 U; GZ14 MZ, Tu, N, and Sa; and GZ15 CC and MI (Fig. 3). Within this large area, a high LCA (2.14) was found only in GZ14 (Table 2), although allelic richness was at least similar in all GZs and even smaller than those obtained for northern GZs.
Populations located on Tierra del Fuego Island were structured in three main clusters (Supplementary Fig. S2 IVa) which also exhibited three distinctive genetic patterns in the STRUCTURE analysis (Supplementary Fig. S2 IVb and IVc). Therefore, three genetic zones were delineated: GZ16 TdFN, TdFC, and TdFE; GZ17 PG (under provincial jurisdiction), and GZ18 BL (situated within a national park) (Fig. 3). N. pumilio forest located in the northern region of the island, belonging to GZ16, exhibited high LCA and an absence of Pa (Table 2). Additionally, this area (GZ16) was categorized by the provincial government as having medium conservation value, and is currently under management by the forestry industry.
The optimum number of genetic management units (in this case GZs) in N. pumilio was confirmed by assessing the hierarchical analysis of variance (AMOVA), where the proportion of variance due to both the ‘among- and within-’ group components were used as elements of comparison. Our proposal of eighteen GZs had a greater among-group than within-group component of variance, which is evidence of correct grouping (Table 3). The phylogenetic relationships among all inferred clusters are presented in Supplementary Figure S3. In addition, some genetic parameters used to characterize these ‘ad hoc’ GZs (Table 2) are considered in the next section in order to define specific operating practices.
Priority conservation areas and operational management considerations
The delineated GZs could facilitate the implementation of specific practices related to conservation programs by determining priority areas based on the genetic diversity of each OGMU. The genetic structure of N. pumilio was assessed here by neutral molecular markers (SSRs) and was taken into account when delineating N. pumilio GZs in Argentina. This approach has been successfully used to define management units for several species (e.g.,25,51,52), since microsatellite diversity was correlated with genomic diversity at population level, although not at individual level53. In addition, considering that microsatellite diversity may not reflect adaptive diversity54, when defining Conservation Units (CUs) and Provenance Regions (PRs) it would be appropriate to determine the genetic variation data based on adaptive traits30,55. Even though Chilean N. pumilio populations were not sampled, the current status of these forests could reflect similar variation to the Argentinean side, since they are probably subject to comparable threats. However, it may be difficult to achieve common management criteria on both sides of the mountain range (Andes Cordillera), since each country has specific regulations.
Standardized allelic richness (AR) is commonly chosen as a genetic criterion in order to define priority areas for conservation31,32. This parameter is independent of sample size31,56 and suitable for determining historical processes such as bottleneck or admixture56, and inferring putative relictual zones31. Accordingly, GZ2, GZ3, and GZ6 require special attention due to their high genetic diversity (high standardized AR, Table 2). Therefore, these areas become meaningful for in situ and ex situ conservation purposes, especially since they play a key role as genetic diversity reservoirs. In this sense, a high number of genetic variants with great potential for adapting to climate change could be present in these reservoir areas57,58. They are therefore essential as a source of planting material, to ensure the long-term success of active restoration programs59,60 and to enrich low-intensity breeding programs.
Numerous studies suggest that priority should also be given to areas exhibiting locally common or exclusive alleles (i.e. Pa), which could be evidence of genotypes adapted to specific environments30,60. Furthermore, a high LCA value could be indicative of genetic isolation30 and thus help to identify the putative location of species–specific refugia61,62. Based on the high LCA and/or Pa detected in this study (Table 2), GZ2, GZ3, GZ9, GZ14, and GZ16 should be highlighted as holding exclusive allelic variants. In this regard, in situ conservation management is highly encouraged for these genetic zones. Furthermore, tree-planting activities targeting these areas should preferably use reproductive material collected from local sources.
In addition, since a relatively recent demographic bottleneck was inferred in the N. pumilio forest located close to Rosario Lake (SC population), particular consideration should be given to this area, which is included within GZ9. This demographic event leads to a reduction in genetic diversity, which could threaten the adaptive potential of this population and, in an extreme case, lead to local extinction63. For this reason, strict in situ conservation management is highly recommended for this area, in order to prevent the loss of these genetic variants.
Finally, GZ7 and GZ13 displayed specific genetic composition, based on their genetic structure and admixture patterns. In particular, these zones combine an almost balanced mixture of two surrounding gene pools, and thus, special consideration is suggested for their operational forest management. Local sources must be chosen if reproductive materials are required for active restoration in these GZs, in order to preserve their characteristic genetic structures. In the case of severe loss of the GZ7 and GZ13 local forests (e.g., caused by an intensive fire), a balanced seed pool from the surrounding GZs is suggested for future tree-planting activities.
Final remarks
This study presents the first scientific evidence for the definition of a N. pumilio management plan based on genetic knowledge and considering its entire distribution range (more than 2200 km). Six priority areas (GZ2, GZ3, GZ6, GZ9, GZ14 and GZ16) were identified that could lead conservation action, specifically targeting in situ conservation. Furthermore, this study enabled the delineation of 18 N. pumilio GZs in Argentina, thus providing preliminary operational genetic management units that best fit the genetic structure of the target species. Maps showing the distributional range of each GZ in Argentina were drawn up. Reproductive material can thus be transferred within these ad hoc GZs with reduced risk of altering the original genetic structure. This possibility is important in light of the numerous active ecological restoration projects that have been developing in the N. pumilio forests on both sides of the Andes Range in the last decade. In addition, the recommendations suggested here are highly important for regulatory institutions that have to enforce compliance with national Law 26331, on the protection of native forests. Moreover, the 18 GZs constitute a first step towards the definition of both PRs and CUs, thus promoting preservation of the southernmost forest ecosystem of the world.
Materials and methods
Plant material and molecular analysis
Of the 35 N. pumilio populations (965 adult trees) included in this study (Table 1, Fig. 1), 20 had been previously analyzed to determine the species’ phylogeographic patterns19,33, while the remaining 15 populations were used exclusively in the present work. The sampled sites covered the natural distribution area of the target species in Argentina from 36°S to 55°S in the latitudinal range (more than 2200 km) and from 71° 56′ to 66° 37′ in the longitudinal range. A minimum of 20 trees were sampled in each population, with the exception of Cholila and El Chalten sites where 18 and 19 samples were obtained (respectively) due to limited access. In every site, a minimum distance of 50 m between sampled trees was maintained in order to preclude the collection of closely-related individuals. In addition, only N. pumilio phenotypes were considered at the time of population sampling (thus, avoiding possible or ambiguous intermediate phenotypes between this species and related ones).
Total DNA was extracted from bud tissue following the procedure described by Dumolin et al.64. Samples were stored at − 20 °C until PCR amplifications were carried out. Seven polymorphic loci (Npum1, Npum3, Npum9, Npum10, Npum13, Npum17a, and Npum18) were analyzed using a specific set of microsatellite markers (nuSSRs) developed for this species by Soliani et al.65. SSR amplification conditions and PCR thermal profiles were set following the recommendations given by the authors65. Amplification reactions were performed with 20 ng of template DNA in a final volume of 11 µl, using 1X Green Buffer and GoTaq DNA Polymerase (Promega, Wisconsin, USA) for all primers, with the exception of Npum10 that was amplified with Platinum Taq (Invitrogen, California, USA). The M13 protocol66 was applied and PCR products were visualized on 1% agarose gel stained with Sybr Safe (Invitrogen). Individual genotyping was done using a 3130xl Genetic Analyzer sequencer (Applied Biosystems, California, USA) and the allele assignment with GeneMarker V2.7.4 (SoftGenetics, demo version).
Genetic characterization of the populations
A set of genetic parameters was first obtained in order to characterize the populations under study (Table 1). These parameters included the number of different alleles (Na), the effective number of alleles (Ne), and the expected heterozygosity that considers the null allele effect (HE null), which were obtained with GenAlEx 6.5 software67. Unbiased heterozygosity described by Nei68 was also estimated with Info-Gen software69. The Wilcoxon test was implemented as an a posteriori multiple-comparison analysis, to identify the significant differences among populations. The presence of null alleles at both locus and population levels was tested with MICRO-CHECKER70 and their frequencies were estimated using FreeNA71. The presence of null alleles was considered in the estimation of inbreeding coefficients (FIS), which was calculated with INEST 2.272. Two approaches were tested for obtaining FIS: 1. an Individual Inbreeding Model (IIM) using a Bayesian approach (FIS′), and 2. a Maximum Likelihood approach (FIS”). In both cases, three different models were tested (i. e., considering the occurrence of null alleles, genotyping errors, and inbreeding events) at population level, and the model with the lowest Deviance Information Criterion (DIC) was considered as best fitting the data. Once the inbreeding model had been selected, the mean inbreeding coefficients (FIS′) were calculated. In addition, a jackknife procedure was performed to estimate the standard error (SE) of the FIS” values, which were then used to assess the significance of this coefficient using the Z-test based on the normal distribution theory.
In order to identify populations experiencing a relatively recent reduction in effective population size, a demographic bottleneck test was carried out using BOTTLENECK software73. This approach computes the distribution of the expected heterozygosity from the observed number of alleles at population level, assuming the mutation-drift equilibrium model. A significant heterozygosity excess or deficiency is confirmed by performing a Wilcoxon sign-rank test for every mutation model (i.e., Infinite Allele Model -IAM, Stepwise Mutation Model -SMM, and Two Phase Model -TPM). In this study, a past population bottleneck was inferred when heterozygosity excess was significant (p < 0.05) in at least two of the above-mentioned mutational models.
Genetic differentiation among populations was determined by estimating both FST 74 and G′ST75 indexes. FST was then recalculated by implementing the ‘exclusion of null alleles’ (ENA) method in order to avoid a possible bias introduced by the presence of null alleles71. Confidence intervals (95% level) were obtained through a bootstrap re-sampling procedure. In addition, to evaluate the occurrence of an isolation-by-distance (IBD) pattern, a correlation between the genetic and geographic distances between populations was assessed using two approaches: a. The Mantel test, performed with Info-Gen and using the correlation between FST estimates and the logarithm of the geographic distances between population pairs; b. The Spatial Genetic Structure (SGC) test by GenAlEx 6.4, with which spatial autocorrelation coefficients (r76,) were obtained for ten distance classes (each one covering 200 km). In both approaches, 1000 permutations were considered when calculating statistical significance. In the SGC test, the upper and lower error limits for the 95% confidence interval were determined by carrying out 10,000 bootstrap samples.
Population genetic structure and cluster definition
Due to the large latitudinal span of the area under study (more than 2200 km long) and the diverse evolutionary history of the region, multiple division criteria were considered (Fig. 2). Firstly, previous information from chloroplast DNA (cpDNA) markers(33, see Fig. 2a) was used, and the haplotypes of new samples from the Cholila population (~ 42°S) were determined due to its proximity to a transition zone between the northern and southern lineages. In a second stage, a broad-scale clustering analysis was performed (considering the 35 populations) using a Bayesian approach by implementing BAPS 6.0 software77. This analysis showed weak grouping, since the adjusted probability of the inferred number of clusters was 0.58632. This reinforced the decision of considering alternative division criteria before performing the final clustering.
A phylogeographic break around 42°S latitude was previously reported for some species of Nothofagus subgenus Nothofagus, which was inferred from genetic studies using cpDNA markers19,33,39,78. It was suggested that northern and southern lineages were geographically segregated around 42°S and were probably isolated for a prolonged period, which indicates a different evolutionary history. In addition, different glaciation patterns were described for the areas located between 38°–42°S and areas southward of 45°S49. A particular distribution of N. pumilio forests is currently characteristic from 41°S northward, since it occurs as a continuous forest mass in the subalpine zone, forming the upper tree limits, whereas at the southern latitudes it also occurs at lower elevations11. Based on all this information, the studied populations were divided into two subsets for further analyses: 1. ‘North’ including the 12 populations with northern cpDNA haplotypes, located between Epulauquen (36° 49′S) and Cholila, and 2. ‘South’ including the remaining 23 populations that exhibited southern cpDNA haplotypes and are located between Huemules and Bahía Lapataia (54° 50′S). The genetic structure of N. pumilio populations from each north and south group was then determined by a spatial clustering method using BAPS software. This approach has the advantage of considering the geographic locations of pre-assigned groups of individuals when defining the best number of clusters (K)77. In addition to this analysis, an admixture model within each inferred K was tested with STRUCTURE 2.2 software79 by running 500,000 Markov chain Monte Carlo (MCMC) steps, a burn-in of 100,000 iterations and five replicates for each inferred K in the entire analysis. Complementarily, UPGMA (Unweighted Pair Group Method using Arithmetic averages) dendrograms based on Nei’s distance (averaged over loci) were constructed to explore the relationships among the identified clusters.
A grid-based spatial analysis was carried out with a resolution of 2 min to visualize the geographical distribution and admixture pattern of inferred clusters. Separate raster maps were produced, considering a circular neighborhood cell of 20 min diameter around every sampled location, following van Zonneveld et al.32 and Thomas et al.80. In this case, individual admixture coefficients for each inferred cluster (obtained by STRUCTURE) larger than 0.7 were considered. Each cell was assigned with the mean membership value of the individuals, enclosed by a circular neighborhood of 20 min diameter constructed around its center to graphically represent the inferred clusters. Raster package 2.14 was used to generate these maps in R software environment (R Core Team) and they were edited in ArcGIS 9.3 (ESRI, Redlands, California, USA).
Delineation and characterization of the genetic zones
Delineation of GZs was performed based on the previously-mentioned analyses and summarized in Fig. 2. Information on historical processes such as the putative location of glacial refugia and post-glacial recolonization patterns was taken into account in identifying the optimum number of clusters. In addition, other criteria such as topography, mountain height and the presence of water bodies (e.g., lakes) were considered.
A hierarchical analysis of molecular variance (AMOVA) was performed using Arlequin 3.581, in order to evaluate the number of clusters that best grouped the studied populations. The optimum number of genetic zones was confirmed based on the significant differences among and within the obtained clusters, as proposed by Pastorino and Gallo20; i.e., genetic variance should be maximum between populations of different groups and minimum within the groups.
To obtain valuable information for the definition of management plans, some genetic parameters for each defined GZ were calculated with GenAlEx and Info-Gen: private alleles (Pa), the expected heterozygosity considering the null allele effect (HE null), and locally common alleles (LCA). The LCA average for each GZ was calculated considering alleles with frequencies higher than 5% and, at the same time, an occurrence probability of less than 25% of the defined GZs. In addition, genetic diversity was estimated by calculating allelic richness (AR) after rarefaction to a common sample size82 using FSTAT software83.
References
Silander, J. A. Temperate Forests. In: Encyclopedia of Biodiversity (Second Edition) (ed. Simon A Levin), 112–227 (Academic Press, 2001).
Glasser, N. F., Harrison, S., Winchester, V. & Aniya, M. Late pleistocene and holocene palaeoclimate and glacier fluctuations in patagonia. Glob. Planet. Change 43, 79–101 (2004).
Markgraf, V. Paleoenvironments and paleoclimates in Tierra del Fuego and southernmost Patagonia, South America. Palaeogeogr. Palaeoclimatol. Palaeoecol. 102, 53–67 (1993).
Markgraf, V., McGlone, M. & Hope, G. Neogene paleoenvironmental and paleoclimatic change in southern temperate ecosystems—a southern perspective. Trends Ecol. Evol. 10(4), 143–147 (1995).
Amoroso, M. M., Rodríguez-Catón, M., Villalba, R. & Daniels, L. D. Forest Decline in Northern Patagonia: The Role of Climatic Variability. In: Dendroecology, Ecological Studies (Analysis and Synthesis): volume 231 (ed. Amoroso, M. M.; Daniels, L. D.; Baker, P. J.; Camarero, J. J.), 325–342 (Springer, 2007).
Rodríguez-Catón, M., Villalba, R., Morales, M. & Srur, A. Influence of droughts on Nothofagus pumilio forest decline across northern Patagonia, Argentina. Ecosphere 7(7), e01390. https://doi.org/10.1002/ecs2.1390 (2016).
Barros, V. R. et al. Climate change in Argentina: trends, projections, impacts and adaptation. Wiley Interdiscip. Rev. Clim. Change 6, 151–169 (2005).
Rusticucci, M. & Barrucand, M. Observed trends and changes in temperature extremes over Argentina. J. Clim. 17, 4099–4107 (2004).
Mundo, I. A. et al. Fire history in southern Patagonia: human and climate influences on fire activity in Nothofagus pumilio forests. Ecosphere 8(9), e01932. https://doi.org/10.1002/ecs2.1932 (2017).
Mohr-Bell, F. D. Superficies afectadas por incendios en la región bosque Andino Patagónico (BAP) durante los veranos de 2013–2014 y 2014–2015. Patagon. For. 21, 34–41 (2015).
Veblen, T. T., Hill, R. S. & Read, J. Ecology of Southern Chilean and Argentinean Nothofagus Forests. In: The Ecology and Biogeography of Nothofagus Forests, pp. 293–353 (Yale University, USA, 1996).
Donoso Zegers, C. Las Especies Arbóreas de los Bosques Templados de Chile y Argentina: Autoecología. 678p (María Cuneo Ediciones, 2006)
Soliani, C. & Aparicio, A. G. Evidence of genetic determination in the growth habit of Nothofagus pumilio (Poepp. & Endl.) Krasser at the extremes of an elevation gradient. Scand. J. For. Res. 35 (5–6), 211–220 (2020).
Rusch, V. E. Altitudinal variation in the phenology of Nothofagus pumilio in Argentina. Rev. Chil. Hist. Nat. 66, 131–141 (1993).
Fajardo, A. & Piper, F. I. Intraspecific trait variation and covariation in a widespread tree species (Nothofagus pumilio) in southern Chile. N. Phytol. 189, 259–271 (2011).
Burns, S. L., Cellini, J. M., Lencinas, M. V., Martínez Pastur, G. J. & Rivera, S. M. Description of possible natural hybrids between Nothofagus pumilio and N. antarctica at South Patagonia (Argentina). Bosque 31(1), 9–16 (2010).
Quiroga, P., Vidal Russel, R. & Premoli, A. C. Evidencia morfológica e isoenzimática de hibridación natural entre Nothofagus antarctica y N. pumilio en el noroeste patagónico. Bosque 26(2), 25–32 (2005).
Acosta, M. C. & Premoli, A. C. Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Mol. Phylogenet. Evol. 54, 235–242 (2010).
Soliani, C. et al. Halfway encounters: meeting points of colonization routes among the southern beeches Nothofagus pumilio and N. antarctica. Mol. Phylogenet. Evol. 85, 197–207 (2015).
Pastorino, M. J. & Gallo, L. A. Preliminary operational genetic management units of a highly fragmented forest tree species of southern South America. For. Ecol. Manag. 257, 2350–2358 (2009).
Pastorino, M. J., Aparicio, A. & Azpilicueta, M. M. Regiones de Procedencia del Ciprés de la Cordillera y Bases Conceptuales para el Manejo de sus Recursos Genéticos en Argentina.108 p (Ediciones INTA, 2015).
Azpilicueta, M. M. et al. Management of Nothofagus genetic resources: definition of genetic zones based on a combination of nuclear and chloroplast marker data. For. Ecol. Manag. 302, 414–424 (2013).
Azpilicueta et al. Zonas Genéticas de Raulí y Roble Pellín en Argentina: Herramientas para la Conservación y el Manejo de la Diversidad Genética (ed. M.M. Azpilicueta, P. Marchelli) 50 p (Ediciones INTA, 2016).
OTBN. Ordenamiento Territorial de Bosque Nativo/Mapa Legal CREA. https://www.crea.org.ar/mapalegal/otbn
Bucci, G. & Vendramin, G. G. Delineation of genetic zones in the European Norway spruce natural range: preliminary evidence. Mol. Ecol. 9, 923–934 (2000).
McKay, J. K., Christian, C. E., Harrison, S. & Rice, K. J. “How Local Is Local?”—a review of practical and conceptual issues in the genetics of restoration. Restor. Ecol. 13, 432–440 (2005).
Williams, M. I. & Dumroese, R. K. Preparing for climate change: forestry and assisted migration. J. For. 111(4), 287–297 (2013).
Geburek, T. Isozymes and DNA markers in gene conservation of forest trees. Biodivers. Conserv. 6, 1639–1654 (1997).
Ballesteros-Mejia, L., Lima, J. S. & Collevatti, R. G. Spatially-explicit analyses reveal the distribution of genetic diversity and plant conservation status in Cerrado biome. Biodivers. Conserv. 29, 1537–1554 (2018).
Frankel, O. H., Brown, A. H. D. & Bordon, J. The Genetic Diversity of Wild Plants. In: The Conservation of Plant Biodiversity. (Cambridge University Press, Cambridge, 1995).
Petit, R. J., El Mousadik, A. & Pons, O. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12, 844–855 (1998).
van Zonneveld, M. et al. Mapping genetic diversity of cherimoya (Annona cherimola Mill.): application of spatial analysis for conservation and use of plant genetic resources. PLoS ONE 7, e29845. https://doi.org/10.1371/journal.pone.0029845 (2010).
Soliani, C., Gallo, L. & Marchelli, P. Phylogeography of two hybridizing southern beeches (Nothofagus spp.) with different adaptive abilities. Tree Genet. Genomes 8, 659–673 (2012).
Laikre, L. et al. Neglect of genetic diversity in implementation of the convention on biological diversity. Conserv. Biol. 24(1), 86–88 (2009).
Fady, B. et al. Forests and global change: what can genetics contribute to the major forest management and policy challenges of the twenty-first century?. Reg. Environ. Change 16, 927–939 (2016).
Graudal, L. et al. Global to local genetic diversity indicators of evolutionary potential in tree species within and outside forests. For. Ecol. Manag. 333, 35–51 (2014).
Sgrò, C. M., Lowe, A. J. & Hoffmann, A. A. Building evolutionary resilience for conserving biodiversity under climate change. Evol. Appl. 4, 326–337 (2011).
Perez, et al. Assessing population structure in the face of isolation by distance: Are we neglecting the problem?. Divers. Distrib. 24(12), 1883–1889 (2018).
Mathiasen, P. & Premoli, A. C. Out in the cold: genetic variation of Nothofagus pumilio (Nothofagaceae) provides evidence for latitudinally distinct evolutionary histories in austral South America. Mol. Ecol. 19, 371–385 (2010).
Jump, A. & Peñuelas, J. Extensive spatial genetic structure revealed by AFLP but not SSR molecular markers in the wind-pollinated tree, Fagus sylvatica. Mol. Ecol. 16, 925–936 (2007).
Oddou-Muratorio, S. et al. Comparison of direct and indirect genetic methods for estimating seed and pollen dispersal in Fagus sylvatica and Fagus crenata. For. Ecol. Manag. 259, 2151–2159 (2010).
Marchelli, P. & Gallo, L. Multiple ice-age refugia in a southern beech of South America as evidenced by chloroplast DNA markers. Conserv. Genet. 7, 591–603 (2006).
Pastorino, M. J. & Gallo, L. A. Quaternary evolutionary history of Austrocedrus chilensis, a cypress native to the Andean-Patagonian forest. J. Biogeogr. 29, 1167–1178 (2002).
Villagrán, C. Un modelo de la historia de la vegetación de la cordillera de la costa de Chile central-sur: la hipótesis glacial de Darwin. Rev. Chil. Hist. Nat. 74, 793–803 (2001).
Cosacov, A., Sersic, A., Sosa, V., Johnson, L. & Cocucci, A. Multiple periglacial refugia in the Patagonian steppe and post-glacial colonization of the Andes: the phylogeography of Calceolaria polyrhiza. J. Biogeogr. 37, 1463–1477 (2010).
Breitman, M. F., Avila, L. J., Sites, J. W. & Morando, M. Lizards from the end of the world: phylogenetic relationships of the Liolaemus lineomaculatus section (Squamata: Iguania: Liolaemini). Mol. Phylogenet. Evol. 59, 364–376 (2011).
Flint, R. F. & Fidalgo, F. Glacial drift in the eastern argentine Andes between latitude 41° 10’ S. and latitude 43° 10’ S. GSA Bull. 80, 1043–1052 (1969).
Holderegger, R. & Thiel-Egenter, C. A discussion of different types of glacial refugia used in mountain biogeography and phylogeography. J. Biogeogr. 36, 476–480 (2009).
Glasser, N. F., Jansson, K., Harrison, S. & Kleman, J. The glacial geomorphology and Pleistocene history of South America between 38°S and 56°S. Quaternary Sci. Rev. 27(3), 365–390 (2008).
Premoli, A. C., Mathiasen, P. & Kitzberger, T. Southern-most Nothofagus trees enduring ice ages: genetic evidence and ecological niche retrodiction reveal high latitude (54°S) glacial refugia. Palaeogeogr. Palaeoclimatol. Palaeoecol. 298, 247–256 (2010).
Derory, J. et al. What can nuclear microsatellites tell us about maritime pine genetic resources conservation and provenance certification strategies?. Ann. For. Sci. 59, 699–708 (2002).
Honjo, M. et al. Management units of the endangered herb Primula sieboldii based on microsatellite variation among and within populations throughout Japan. Conserv. Genet. 10, 257–267 (2009).
Väli, U., Einarsson, A., Waits, L. & Ellegren, H. To what extent do microsatellite markers reflect genome-wide genetic diversity in natural populations?. Mol Ecol. 17(17), 3808–3817 (2018).
Reed, D. H. & Frankham, R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evol. 55(6), 1095–1103 (2001).
Crandall, K. A., Bininda-Emonds, O. R. P., Mace, G. M. & Wayne, R. K. Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 15, 290–295 (2000).
Widmer, A. & Lexer, C. Glacial refugia: sanctuaries for allelic richness, but not for gene diversity. Trends Ecol. Evol. 16, 267–269 (2001).
Reed, D. H. & Frankham, R. Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237 (2003).
Jump, A. S., Marchant, R. & Peñuelas, J. Environmental change and the option value of genetic diversity. Trends Plant Sci. 14, 51–58 (2009).
Prober, S. et al. Climate-adjusted provenancing: a strategy for climate-resilient ecological restoration. Front. Ecol. Evol. 3, 65 (2015).
Thomas, E. et al. Genetic considerations in ecosystem restoration using native tree species. For. Ecol. Manag. 333, 66–75 (2014).
Marchelli, P., Thomas, E., Azpilicueta, M. M., van Zonneveld, M. & Gallo, L. Integrating genetics and suitability modelling to bolster climate change adaptation planning in Patagonian Nothofagus forests. Tree Genet. Genomes 13, 119 (2017).
Thomas, E. et al. Genetic diversity of Enterolobium cyclocarpum in Colombian seasonally dry tropical forest: implications for conservation and restoration. Biodivers. Conserv. 26(4), 825–842 (2016).
Jump, A. S. & Peñuelas, J. Running to stand still: adaptation and the response of plants to rapid climate change. Ecol. Lett. 8, 1010–1020 (2005).
Dumolin, S., Demesure, B. & Petit, R. J. Inheritance of chloroplast and mitochondrial genomes in pedunculated oak investigated with an efficient PCR method. Theor. Appl. Genet. 91, 1253–1256 (1995).
Soliani, C., Sebastiani, F., Marchelli, P., Gallo, L. & Giovanni, G. Development of novel genomic microsatellite markers in the southern beech Nothofagus pumilio (Poepp. et Endl.) Krasser. Mol. Ecol. Resour. 10, 404–408 (2010).
Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18, 233–234 (2000).
Peakall, R. & Smouse, P. E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. J. Bioinform. 28, 2537–2539 (2012).
Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89, 583–590 (1978).
Balzarini, M. & Di Rienzo, J. Info-Gen: Software para Análisis Estadístico de Datos Genéticos. Facultad de Ciencia Agropecuarias. Universidad Nacional de Córdoba. Argentina. https://www.info-gen.com.ar/. (2003).
Oosterhout, C. V., Hutchinson, W. F., Wills, D. P. M. & Shipley, P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538 (2004).
Chapuis, M. P. & Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 24, 621–631 (2006).
Chybicki, I. J. & Burczyk, J. Simultaneous estimation of null alleles and inbreeding coefficients. J. Heredity 100, 106–113 (2009).
Cornuet, J. M. & Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144, 2001–2014 (1996).
Wright, S. The genetical structure of populations. Ann. Eugen. 15, 323–354 (1949).
Hedrick, P. W. A standardized genetic differentiation measure. Evolution 59, 1633–1638 (2005).
Smouse, P. E. & Peakall, R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. J. Heredity 82, 561–573 (1999).
Corander, J., Waldmann, P. & Sillanpää, M. J. Bayesian analysis of genetic differentiation between populations. Genetics 163, 367–374 (2003).
Pastorino, M. J., Marchelli, P., Milleron, M., Soliani, C. & Gallo, L. A. The effect of different glaciation patterns over the current genetic structure of the southern beech Nothofagus antarctica. Genetica 136, 79–88 (2009).
Pritchard, J., Stephens, M. & Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. https://www.genetics.org/content/155/2/945.long. (2000).
Thomas, E. et al. Present spatial diversity patterns of Theobroma cacao L. in the neotropics reflect genetic differentiation in pleistocene refugia followed by human-influenced dispersal. PLoS ONE 7, e47676. https://doi.org/10.1371/journal.pone.0047676 (2012).
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
El Mousadik, A. & Petit, R. J. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor. Appl. Genet. 92, 832–839 (1996).
Goudet, J. FSTAT: a Program to Estimate and Test Gene Diversities and Fixation Indices (version 2.9.3.2) https://www.unil.ch/izea/softwares/fstat.html. (2001).
Acknowledgements
We would like to thank Victor Mondino, Jorge Bozzi, Pablo Peri, Francisco Mattenet and Guillermo Martínez-Pastur for their assistance with material collection, and Evert Thomas for kindly providing R scripts to run neighboring analysis.
Funding
This work was funded by Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT - FONCyT), Argentina [Grant Numbers PICT 2015-1193]. The first author has a fellowship from ANPCyT.
Author information
Authors and Affiliations
Contributions
M.G.M.: resources (plant material collection), experiment development, data curation, analysis, interpretation, writing of original draft, and preparation of figures and supplementary material. M. J. P.: conceptualization, resources (plant material collection), interpretation, and funding acquisition. M. V. L.: resources (computing software), preparation of figures, and supplementary material. P. M.: conceptualization and interpretation. C. S.: conceptualization, resources (plant material collection, molecular marker development), validation, interpretation, supervision, and project administration. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mattera, M.G., Pastorino, M.J., Lantschner, M.V. et al. Genetic diversity and population structure in Nothofagus pumilio, a foundation species of Patagonian forests: defining priority conservation areas and management. Sci Rep 10, 19231 (2020). https://doi.org/10.1038/s41598-020-76096-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-76096-0
This article is cited by
-
Predicting the distribution of plant species from southern South America: are the hotspots of genetic diversity threatened by climate change?
Biodiversity and Conservation (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.