Abstract
A stable packaging cell line (Vero/BC-F) constitutively expressing fusion (F) protein of the human parainfluenza virus type 2 (hPIV2) was established for production of the F-defective and single round-infectious hPIV2 vector in a strategy for recombinant vaccine development. The F gene expression has not evoked cytostatic or cytotoxic effects on the Vero/BC-F cells and the F protein was physiologically active to induce syncytial formation with giant polykaryocytes when transfected with a plasmid expressing hPIV2 hemagglutinin-neuraminidase (HN). Transduction of the F-defective replicon RNA into the Vero/BC-F cells led to the release of the infectious particles that packaged the replicon RNA (named as hPIV2ΔF) without detectable mutations, limiting the infectivity to a single round. The maximal titer of the hPIV2ΔF was 6.0 × 108 median tissue culture infections dose per ml. The influenza A virus M2 gene was inserted into hPIV2ΔF, and the M2 protein was found to be highly expressed in a human lung cancer cell line after transduction. Furthermore, in vivo airway infection experiments revealed that the hPIV2ΔF was capable of delivering transgenes to hamster tracheal cells. Thus, non-transmissible or single round-infectious hPIV2 vector will be potentially applicable to human gene therapy or recombinant vaccine development.
Similar content being viewed by others
Introduction
Human parainfluenza virus type 2 (hPIV2), a member of the Paramyxovirdae, has a non-segmented negative-strand RNA genome of approximately 15 kb in length, encoding seven structural proteins, NP, P/V, M, F, HN and L in this order.1,2 hPIV2 is a respiratory pathogen that causes croup and other upper and lower respiratory tract diseases in young children at the incidence peak between 1 and 2 years of age,1,3 and a majority of children have been infected with hPIV2 by 5 years of age.1 hPIVs commonly reinfect children and adults; however, the illness in healthy children and adults is usually limited to the upper respiratory tract.1
hPIV2 has two surface glycoproteins, the hemagglutinin-neuraminidase (HN) and fusion (F) protein. hPIV2 attaches to the cell surface receptors via the HN protein, and F protein triggers the envelope-cell fusion. F protein-mediated fusion allows the viral nucleocapsid to enter a host cell. F protein also induces membrane fusion between host cells (syncytial formation).3 Syncytial formation is one of the characteristics of many enveloped viruses including the paramyxovirus, and has been reported as a potentially important mechanism for virus-induced cytotoxic effects.3 F protein of the human parainfluenza virus is generated as an inactive precursor (F0) and must be cleaved by endopeptidase to yield an active F protein, which is composed of two disulfide-linked molecules (F1 and F2) and is thought to be required for syncytial formation. The proteolytic cleavage of F protein varies among the different human parainfluenza virus types. In hPIV2 (Toshiba strain)4 F0 is cleaved into F1 and F2 without the addition of trypsin in the culture medium.4
The viral vectors derived from non-segmented negative-stranded RNA viruses such as hPIV2 are supposed to be superior to other transient expression systems because of their high transduction efficiency.5 In comparison with other viruses, the use of such RNA viruses as a vector is safer because they do not have a DNA phase throughout their life cycles and they replicate exclusively in the cytoplasm, thus avoiding unintended genetic modifications of host cell DNA. These characteristics make the negative-stranded RNA viruses useful as potential vectors for transient high expression.
A method to recover the infectious virus from cloned DNA was established in non-segmented negative-stranded RNA virus (reverse genetics method). The first success was in a rabies virus in which a plasmid DNA encoding a full length of antigenomic viral RNA was used.6 The development of reverse genetics has made it possible to explore the utility of negative-stranded RNA viruses to deliver foreign genes and has opened the way to gene transfer vectors for therapeutic purposes or vaccine development.7, 8, 9, 10 However, the wild-type hPIV2 vector harbors all the viral structural genes to produce multiple round-infectious hPIV2 replicon particles. Therefore, non-transmissible or a single round-infectious replicon form of the hPIV2 vector is desirable for clinical safety as biopharmaceuticals. It is considered that lack of either an F or HN envelope gene not only abolishes the progeny generation but also prevents deleterious fusion events to neighboring cells, thus conferring a safety property. Although having a high expression of F gene of the parainfluenza virus 5 was reported to be toxic to the infected cells,11 establishment of a stable packaging cell line expressing F gene is attractive for the simple and efficient manufacturing process of non-transmissible hPIV2 replicons.
Vero cell line was established from a normal kidney of an adult African green monkey (Cercopithecus) in 1962 by Y Yasumura and Y Kawakita at the Chiba University in Japan.12 Vero cells do not produce interferons,13 and are susceptible to infection and proliferation of SV-40, measles virus, arbovirus, reovirus, rubella virus, simian adenovirus, poliovirus, influenza virus, parainfluenza virus, respiratory syncytial virus, vaccinia virus and other viruses.14 A well-characterized Vero cell line has been used for the production of a number of vaccines against human diseases caused by viruses, including poliomyelitis,15,16 rabies16 and others. Therefore, we used the Vero cells to stably express the F gene of hPIV2.
Transduction of the replicon RNA lacking an F gene (hPIV2ΔF) into the stable Vero packaging cell line expressing F protein led to the release of single round-infectious particles that packaged the replicon RNA with high production efficiency. Here we show that the exogenous gene is expressed efficiently in human lung cancer cells in vitro and hamster respiratory tract cells in vivo via this novel production system of the recombinant hPIV2ΔF vector.
Results
Stable cell line expressing F protein of hPIV2
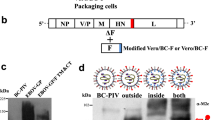
To generate a stable Vero cell line constitutively expressing F protein of hPIV2, a pCXN2,17 which consisted of cytomegalovirus immediate-early enhancer, chicken β-actin promoter and Neomycin-resistance gene, was used to construct pCXN2-hPIV2F (Figure 1a), and the expression vector was transfected into Vero cells. Twenty of the G418-resistant clones were isolated 2 to 4 weeks after transfection to evaluate mRNA for the F protein by one-step reverse transcription (RT)-PCR. Consequently, 8 out of 20 G418-resistant clones were selected as F-stable clone candidates, based on the F gene expression. Afterwards, the clone with best growth and the highest virion productivity out of the eight F-stable clone candidates was selected and further subjected to limiting dilution. The resultant cells derived from a single cell were designated as Vero/BC-F. The RT-PCR analysis indicated that the amount of F gene mRNA in Vero/BC-F cells was nearly equal to that in Vero cells infected with wild-type hPIV2 at a multiplicity of infection (MOI) of 0.2 for 24 h (Figure 1b).
Generation and characterization of the stable Vero cell line constitutively expressing hPIV2 F protein. (a) Schematic representation of the recombinant plasmid used in this experiment. A coding sequence of the F protein of hPIV2 was amplified by PCR and inserted into pCXN2. (b) Ascertainment of mRNA encoding the F protein in Vero/BC-F cells by one-step RT-PCR. A sample without RT was used as a negative control (labeled as (PCR)). (c) Cell fusion induced by coexpression of hPIV2 HN protein in Vero/BC-F cells. Control Vero cells were also transfected with pSRα-F and/or pSRα-HN, or pSRα. Arrows indicate syncytia. (d) Quantification of the syncytia formation induced by coexpression of hPIV2 F and HN proteins. Numbers of syncytia in Vero/BC-F cells transfected with pSRα-HN (black columns) and control Vero transfected with pSRα-F and pSRα-HN (white columns) were counted 20, 24 and 28 h after transfection. All the data are shown as mean±s.d.
We next investigated syncytial formation with the HN protein of hPIV2. The F protein was not reported to induce cell-to-cell fusion by itself, but to induce when coexpressed with the HN protein.3 Indeed, transient expression of the F gene in control Vero cells induced syncytial formation only with coexpression of the HN gene (Figure 1c, bottom panels), disclosing that the F and HN proteins produced after transfection are physiologically active in inducing syncytial formation in Vero cells. Therefore, Vero/BC-F cells were transfected with pSRα-HN.18,19 The transfected cells similarly made syncytia (giant polykaryocytes) (Figure 1c (upper panels) and d). These results indicate that Vero/BC-F cells produce physiologically active F protein.
Characteristics of the Vero/BC-F cells
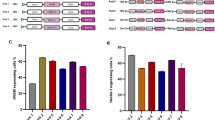
As shown in Figure 2a, the Vero/BC-F cells grew almost equally to control Vero cells in the growth medium, indicating that F protein expression did not affect the cell growth in these cells.
Characteristics of the Vero/BC-F cells. (a) Growth of the Vero/BC-F cells. Cells (5.0 × 104) were prepared in 6-well plates and cultured in the growth medium. The numbers of the cells were counted on various days. All the data are shown as mean±s.d. (b) Cell surface expression of the F protein in Vero/BC-F cells. The overlay histograms show expression of hPIV2 F proteins on the cell surface. Vero (gray area), Vero/BC-F (green line) and Vero cells infected with wild-type hPIV2 (red line) were incubated with the specific antibody for hPIV2 F protein. Vero cells infected with wild-type hPIV2, treated with control IgG, were shown as a black line. All the cells were labeled with an Alexa Fluor 488-conjugated anti-mouse IgG antibody. (c) The effect of the overexpressed F protein on the Vero/BC-F cells. The Vero/BC-F cells were transfected with the plasmid shown above each panel, and incubated at 37 °C for 48 h. Cytotoxicity was evaluated morphologically and by PI staining. (d) Flow cytometry analysis on cell damage by overexpression of hPIV2 F protein in the Vero/BC-F cells. The Vero/BC-F cells transfected with pSRα-F or pSRα were incubated for 48 h, and the cells were stained with PI without fixation (red line, pSRα-F; black line, pSRα). (e) Comparison of expression levels of hPIV2 F mRNA in short- and long-term cultured Vero/BC-F cells by RT-qPCR. The same amounts of total RNAs from short-term (fresh Vero/BC-F) and long-term (old Vero-BC-F) cultured cells, and from control Vero cells were reverse transcribed, followed by qPCR. Fresh Vero/BC-F cells were cultured within a few months, and old Vero/BC-F cells were cultured for more than 2.5 years. Relative quantities of hPIV2 F transcripts are shown as mean±s.d., and the statistical significance (*P<0.05) was determined by the Bonferroni test. (f) Comparison of virus recovery from short- and long-term cultured Vero/BC-F cells. The same new and old cells (1 × 106) as in Figure 2e were infected with the EGFP-hPIV2ΔF at an MOI of 0.1, and the supernatant was collected on 3 and 6 days after infection, followed by titration by the TCID50 method. All the data are shown as mean±s.d.
The flow cytometry analysis revealed the expression of F protein on the surface of Vero/BC-F cells with an anti-F (144-1A) monoclonal antibody (mAb)20 (Figure 2b). The amount of F protein in Vero/BC-F cells (red line) was nearly equal to that in Vero cells infected with wild-type hPIV2 at an MOI of 0.2 for 24 h (green line).
Good growth of Vero/BC-F cells was supposed to be attributed to the proper level of F protein expression. To test this, we investigated the effects of overexpression of F protein in Vero/BC-F cells. The cells were transfected with pSRα (pcDL-SRα296)18 or pSRα-F,19 and dying cells were stained with propidium iodide (PI). The cells transfected with pSRα-F showed significantly increased numbers of rounding cells and PI-positive cells (Figure 2c), indicating that overexpression of F protein elicited considerable cytotoxicity in Vero/BC-F cells. Furthermore, flow cytometry analysis corroborated the cytotoxicity of the overexpression of F protein (Figure 2d). Therefore, Vero cells could have a threshold within which they are tolerable to the cytotoxicity of F protein.
Subsequently, we examined whether the Vero/BC-F cells could continue to proliferate normally and maintain expressing physiologically active F protein for a long period, using the cells passaged continuously for at least 2.5 years. The cells could continue to grow normally. However, the amount of the F gene mRNA in the cells cultured for 2.5 years was decreased to approximately one-fifth of that in the fresh Vero/BC-F cells, by the RT-quantitive PCR (qPCR) analysis (Figure 2e). Accordingly, the efficiency of viral recovery from the long-term passaged Vero/BC-F cells was reduced to about one-sixth at day 3 and a half at day 6 after infection of EGFP-hPIV2ΔF at an MOI of 0.1, compared with that from the fresh cells (Figure 2f).
Recovery of the F-defective hPIV2 replicon particles with intact genomic structure
F-defective hPIV2 was constructed from pPIV221 with modifications, which harbors an hPIV2 genome of 15 654 nucleotides in length after resequencing. The F-defective hPIV2 cDNA with EGFP gene as a marker was constructed in accordance with the rule of six (Figure 3a).22,23 Vero/BC-F cells were transfected with the F-defective EGFP-hPIV2 cDNA driven by T7 RNA polymerase promoter together with the plasmids that express NP, P and L proteins (polymerase units), and T7 RNA polymerase, respectively, in 6-well plates. After 1 week, half of the culture supernatant was transferred to the culture medium of the fresh Vero/BC-F cells. After incubation for 3 days, some GFP-expressing cells were observed, indicating successful recovery of the F-defective hPIV2 replicon particles. The culture supernatant was collected and passed through the 0.45 μm filter to remove cells and cell debris, and then transferred to the fresh Vero/BC-F cells. After a few days, GFP fluorescence spread over the entire cell culture and the infectious particles were collected from the culture supernatant.
Recovery of non-transmissible or single round-infectious hPIV2 replicon particles from the Vero/BC-F cells. (a) Construction of hPIV2 cDNA defective in the gene encoding F protein. hPIV2 cDNA lacking the gene encoding the F protein was constructed by removing the entire F gene and adding the EGFP gene as a marker just upstream of the gene encoding nucleocapsid protein (NP). The PCR products shown in Table 1 are indicated by two-way arrows. (b) Certification by RNA analyses for generation of hPIV2 defective in the F gene. RNAs from virus particles of wild-type hPIV2/Vero and EGFP-hPIV2ΔF/Vero/BC-F were subjected to one-step RT-PCR using the various primer sets shown in Table 1. (c) Expression of F or HN protein on cell surface of Vero cells infected with wild-type hPIV2 or EGFP-hPIV2ΔF. Vero cells were infected with wild-type hPIV2 (hPIV2 wt) or the EGFP-hPIV2ΔF (hPIV2ΔF) at an MOI of 5, and cell surface proteins were biotinylated 24 and 72 (EGFP-hPIV2ΔF only) hours after infection, and were immunoprecipitated with the anti-F and anti-HN mAbs, respectively. The precipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by nitrocellulose membrane transfer. The biotinylated proteins on the membrane were detected by enhanced chemiluminescence. (d) Spread of GFP expression in the Vero/BC-F cells. Confluent monolayers of the control Vero cells and the Vero/BC-F cells in 6-well plates were inoculated with the EGFP-hPIV2ΔF at an MOI of 0.1, and cultured for the time indicated: upper row, Vero/BC-F cells; lower row, control Vero cells. (e) Time-dependent increase of EGFP expression in Vero/BC-F cells after infection with hPIV2ΔF. Mean fluorescence intensities (MFIs) of EGFP are shown in Vero/BC-F cells and control Vero cells, both of which were infected with hPIV2ΔF. Data are shown as means±s.d., and the statistical significance (*P<0.05) was determined by the Bonferroni test. (f) The growth of EGFP-hPIV2ΔF in comparison with that of wild-type hPIV2. Control Vero cells were infected with the EGFP-hPIV2ΔF or wild-type hPIV2, and the Vero/BC-F cells were infected with the EGFP-hPIV2ΔF, at an MOI of 0.1. The supernatant was collected on various days after infection, and the titration was performed by the TCID50 method. The growth of EGFP-hPIV2ΔF on control Vero cells, that of EGFP-hPIV2ΔF on Vero/BC-F cells and that of wild-type hPIV2 on control Vero cells are shown in open, closed and hatched columns, respectively. All the data are shown as mean ±s.d.
Subsequently, we examined whether the infectious particles contained the F-defective hPIV2 genome. One-step RT-PCR was carried out with the primers shown in Table 1. The primers were designed to span the entire F gene to amplify a 2.2 kbp fragment for the wild-type hPIV2 genome or a 0.37 kbp fragment for the F-defective hPIV2 genome. Also, the primers were designed to cover the M and F gene array to amplify a 0.36 kbp fragment for the wild-type hPIV2 genome or no fragment for the F-defective hPIV2 genome. The products of a 2.2 and a 0.36 kbp, and those of a 0.37 kbp and no amplification were observed on the wild-type hPIV2 genome and the F-defective hPIV2 genome, respectively (Figure 3b). Moreover, the amplified fragment within the L gene and that covering the NP and P genes confirmed that the particles have the genome of hPIV2 lacking the F gene (Figure 3b). In addition, the direct sequencing of the whole viral genome of the particles revealed that the entire F gene was deleted in the genome of the particles and the rest of the hPIV2 genome was correctly maintained even after 4, 7 and 10 rounds of viral passage in triplicate (data not shown). It was thus proven that the infectious particles have the genome of hPIV2 devoid of the whole F gene. The infectious particles were designated as EGFP-hPIV2ΔF.
Next, the expression of the HN or F protein was investigated in control Vero cells infected with the wild-type hPIV2 or the EGFP-hPIV2ΔF at an MOI of 5 by cell surface biotinylation and immunoprecipitation methods.24,25 The viral glycoproteins were immunoprecipitated from the cell lysate with either an anti-HN (108-S1)20 or the anti-F (144-1A) mAb. As shown in Figure 3c, the EGFP-hPIV2ΔF gave no detectable bands with the anti-F mAb at 24 and 72 h postinfection, but a weak band with the anti-HN mAb at 24 h, followed by a dense band at 72 h postinfection. In contrast, wild-type hPIV2 showed dense bands both with the anti-HN and anti-F mAbs at 24 h postinfection. These results indicate that because of the F gene deficiency, the hPIV2ΔF could not express the F protein from the genome, and required the F protein expressed in trans for infectious virus production. Moreover, it was considered that the assembly of the EGFP-hPIV2ΔF could be delayed relative to that of the wild-type hPIV2 at 24 h after infection (Figure 3c), as the expression level of the HN protein of the EGFP-hPIV2ΔF was significantly lower than that of the wild-type hPIV2 at 24 h after infection at an MOI of 5.
To examine whether the EGFP-hPIV2ΔF RNA could be encapsidated in single round-infectious particles in an F protein-dependent manner, the spread of GFP-positive cells was investigated in the Vero/BC-F cells and control Vero cells, which were infected with the EGFP-hPIV2ΔF at an MOI of 0.1. Two days after the infection, there was a small difference in the number of GFP-positive cells between both cells. Four and six days later, however, strong fluorescence significantly spread over the entire area in the Vero/BC-F cells, whereas it did not in the control Vero cells (Figure 3d). Furthermore, flow cytometry analysis disclosed markedly increased expression of GFP in Vero/BC-F cells, but not in Vero cells (Figure 3e). Therefore, the increase of GFP-positive cells depended on the F protein expression, indicating that the EGFP-hPIV2ΔF particles are non-transmissible replicon particles after single round infection.
Subsequently, the growth of the EGFP-hPIV2ΔF and wild-type hPIV2 was assessed. The EGFP-hPIV2ΔF showed no propagation when cultured in control Vero cells. On the other hand, the EGFP-hPIV2ΔF in Vero/BC-F cells and wild-type hPIV2 cultured in the control Vero cells propagated vigorously 2–4 days after infection to generate a maximal infectious titer of 6.0 × 108 and 9.0 × 108 median tissue culture infective dose (TCID50) per ml, respectively (Figure 3f). The viral titer of wild-type hPIV2 cultured in Vero/BC-F cells increased approximately 10-fold, compared with that in the parental Vero cells 3 days after infection, but that in Vero cells finally reached almost equal to that in Vero/BC-F cells by the sixth day after infection (data not shown).
Although homologous recombination has not been reported in the genome of non-segmented negative-stranded RNA viruses,26 the appearance of a recombinated hPIV2 genome was examined by GFP expression as an indicator. The number of GFP-positive cells did not increase in the control Vero cells infected at an MOI of 0.1 with the EGFP-hPIV2ΔF, which were passaged on the Vero/BC-F cells more than 10 times, whereas it increased in the Vero/BC-F cells (data not shown). Moreover, no extra bands suggesting acquisition of the F gene-containing fragments were detected by the one-step RT-PCR analysis with the primers iii shown in Figure 3a and Table 1 in the culture supernatant containing EGFP-hPIV2ΔF, which were passaged on Vero/BC-F cells more than 10 times. The same results were obtained by three independent experiments. Taken together, it was ascertained that the hPIV2ΔF is not likely to generate replication-competent hPIV2 by homologous recombination in the Vero/BC-F cells.
Expression of influenza M2 protein by M2-hPIV2ΔF in human cells
In an attempt to develop a potential universal influenza vaccine, we constructed a phPIV2ΔF harboring M2 gene of the influenza A virus (referred to as pM2-hPIV2ΔF). The M2-hPIV2ΔF viruses were successfully recovered by the reverse genetics method using pM2-hPIV2ΔF. To examine M2 protein expression from the M2-hPIV2ΔF, human lung cancer cell line A549 cells were infected with either the M2-hPIV2ΔF or the EGFP-hPIV2ΔF at an MOI of 5 for 2 days. A western blot analysis revealed the M2 protein expression only in M2-hPIV2ΔF-infected cells with strong intensity, whereas hPIV2 NP protein was detected both in M2-hPIV2ΔF- and EGFP-hPIV2ΔF-infected cells (Figure 4). The M2-hPIV2ΔF viruses were recovered in nearly the same high titer as in the EGFP-hPIV2ΔF viruses by using Vero/BC-F cells (data not shown).
Expression of influenza A virus M2 protein by hPIV2ΔF vector in human lung cancer A549 cells 2 days after infection. The cell lysates were denatured and separated on 5–20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto a nitrocellulose membrane. The membrane was probed with the mAbs specific to M2 (upper) of the influenza A virus and NP of the hPIV2 (lower), respectively. Vero/pcDNA-M2, Vero cells transfected with pcDNA-M2 as a positive control; A549, non-infected A549 cells as a negative control; A549/EGFP-hPIV2ΔF, A549 cells infected with EGFP-hPIV2ΔF; A549/M2-hPIV2ΔF, A549 cells infected with M2-hPIV2ΔF.
Transgene expression in the respiratory tract of the hamsters infected with EGFP-hPIV2ΔF
To investigate the infectivity of the hPIV2ΔF vector and property of the transgene expression in vivo, 4-week-old female Syrian hamsters were inoculated intranasally with 7.5 × 108 cell infectious units of the EGFP-hPIV2ΔF. Three days after infection, the tracheas and lungs were harvested and GFP expression was observed under fluorescence microscopy to evaluate the transduction efficiency. Tracheal cells of the EGFP-hPIV2ΔF-administered hamsters showed prominent GFP expression, but none were observed in PBS-administered hamsters (Figures 5a and b). The speckled pattern of the GFP expression in the tracheas is noteworthy because it suggests that the cell-to-cell spread seldom takes place in vivo in this system. GFP expression in the lungs was below the detection level (data not shown). These results indicate that the hPIV2ΔF vector is suitable to deliver and express an exogenous gene through the respiratory tract.
GFP expression in the tracheas of the hamsters after respiratory tract infection of EGFP-hPIV2ΔF. Syrian hamsters were administrated intranasally with EGFP-hPIV2ΔF (7.5 × 108 plaque-forming unit). Three days after infection, GFP expression in the tracheas was observed to evaluate the transduction efficiency. Tracheas were extirpated and the inside of incised tracheas were observed with a fluorescence stereoscopic microscope (Olympus, Tokyo, Japan; SZX7) (a) and an inverted fluorescence microscope (Olympus CKX41) (b, EGFP-hPIV2ΔF only).
Discussion
The application of RNA viral vectors for gene delivery has been hampered owing to difficulty in establishing stable packaging cell lines to generate sufficient amounts of single round-infectious viral vectors of high quality in the manufacturing process. Among human parainfluenza viruses, hPIV3 has been used as a vector for vaccine development, based on a replication-competent system,7 or on a live-attenuated chimeric virus.27 Live-attenuated vectors such as the vesicular stomatitis virus have been used extensively as an experimental vaccine, and these vectors easily grow to high titers and stimulate potent cellular and humoral immunity.28, 29, 30, 31, 32 However, obtaining regulatory approval of multiround infectious or replication-competent viral vectors is not easy because of concerns regarding potential pathogenicity. Therefore, RNA viral vectors for gene therapy or recombinant vaccines are expected to be engineered into single round-infectious virions to evade dissemination.
We have successfully generated a cell line (Vero/BC-F) that constitutively expresses the hPIV2 F protein to avoid a transient transfection step and a complex inducible gene expression system during the production of the viral vector.
The Vero/BC-F cells derived from a single cell by limiting dilution have been successfully adapted to culture with a serum-free medium, and a master cell bank of the clone has been established, based on the ICH Guideline (data not shown). We could obtain the EGFP-hPIV2ΔF by the reverse genetics method by using the Vero/BC-F cells in high yields. These results would be applicable to manufacture the recombinant vaccines on the regulation. Furthermore, EGFP-hPIV2ΔF vectors were capable of delivering transgenes to tracheal cells in hamsters.
Single round-infectious cytoplasmic RNA vector expressing the foreign gene is a desirable tool to use as a vector for recombinant vaccine development, because of its high efficiency of transduction, safety property and its effect of inducing dendritic cell maturation (adjuvant effect of the vector itself).5 As a foreign gene, we tried to express the M2 protein of the influenza A virus. M2 protein consists of 97 amino acids, works as a viral ion channel with a proton transport function and has been proposed as one of the candidates for the universal influenza vaccine.33, 34, 35 As shown in this study, hPIV2ΔF could efficiently express M2 protein in human lung cancer cells. This result as well as previous reports36, 37, 38 suggests that a respiratory viral vector-mediated delivery of the antigen is one of the good strategies for vaccine development against various respiratory diseases.
As for the other paramyxoviral vectors carrying a defective genome, the Sendai virus vector that was devoid of an F gene was previously generated in a trypsin-dependent manner with the packaging cell line inducibly expressing an F protein by using a Cre-loxP system.39 In contrast to this Sendai virus vector system, our hPIV2ΔF system uses the stably expressed F protein, and cleavage of F protein is trypsin-independent, thus being easy to handle.
One would consider the pre-existing immunity to hPIV2 in humans is an obstacle for the utility of the vector for recombinant vaccines. Although antibodies in serum and cellular immunity have a major role in reducing pathogenic reactions after respiratory tract infection, innate immunity and local mucosal immunity have key roles in the initial phase of protection in each infection. However, most mucosal responses are often short lived. Although a majority of children have been infected with hPIV2 by 5 years of age,1 hPIV reinfection is known to occur throughout human life,1,3 because of an incomplete immunity to hPIVs.3 Interestingly, the hPIV2 genome was reported to be detected in only 1–3% of cases among children with respiratory diseases,1,40 suggesting rarity of hPIV2 infection itself in comparison with other respiratory viruses, or weak pathogenicity of hPIV2. We have designed that hPIV2ΔF vector has a cloning site for exogenous gene insertion in the most upstream region within the vector genome to guarantee the preferential expression of the transgene compared with other viral backbone genes. Nevertheless, further studies are required to see the impact of pre-existing immunity to the vector on the efficiency as a recombinant vaccine.
As for the application of the vector to gene therapy, particularly for the treatment of hereditary diseases due to genetic defects, or cancer therapy, a long-lasting expression of therapeutic genes in high levels in wide varieties of cell types or organs is required. The hPIV2ΔF has an ability to deliver genes efficiently; however, transgene expression will be limited by dilution in actively dividing cells, or might be abrogated by induction of anti-hPIV2 immune responses in addition to pre-existing immunity to hPIV2. Therefore, non- or less-dividing cells would be appropriate targets in gene therapy by the hPIV2ΔF vector. Furthermore, we need to study the therapeutic effects and immune reactions in repeated infection of hPIV2ΔF in vivo.
hPIV2ΔF itself may also be used for another purpose. Currently, genome-modified human hPIV2, as a potential vaccine candidate delivered as nose drops, is in phase I of clinical trial for adults, children and infants. The virus has a 3′ genomic promoter mutation and L polymerase mutations that result in virus attenuation.41 In African green monkeys, immunization with the virus conferred a high level of protection in the upper and lower respiratory tracts of the challenged animals with wild-type hPIV2.41 Here, we advocate hPIV2ΔF as another potential vaccine candidate against the wild-type hPIV2 because of its property limiting to single round infectivity with no cell-to-cell spread.
In summary, we have generated an F-defective hPIV2 vector together with a high-efficiency packaging cell line for recombinant vaccine development.
Materials and methods
Cells, virus and plasmid construction
The Vero cell line was obtained from RIKEN BioResource center (Tsukuba, Japan) and cultured in a minimal essential medium (MEM) (Sigma, St Louis, MO, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific Inc., Waltham, MA, USA). The A549 human lung cancer cell line and MDCK (Madin–Darby canine kidney) cell line were cultured in MEM with 10% FBS. The hPIV2 used was a Toshiba strain.4 The influenza virus used an A/Puerto Rico/1934 (H1N1) (PR8 strain). The hPIV2 F gene was amplified from pPIV221 by PCR with tagged primers and cloned into a multicloning site of pCXN217 containing a neomycin-resistance gene to generate pCXN2-hPIV2F (Figure 1a).
An entire reading frame of the F gene was deleted from pPIV2 by using the two-step overlap-primer PCR method to generate F-defective hPIV2 cDNA driven by T7 promoter, which was designated as phPIV2ΔF. The EGFP gene was amplified by PCR using pEGFP-N1 (Clontech, Palo Alto, CA, USA) as a template and a pair of NotI-tagged primers of 5′-ATTGATTGCGGCCGCGGTCGCCACCATGGTGAGCAAGGGCGAGGAGC-3′ and 5′-ATTGATTGCGGCCGCCTAACCCGTCCGGGCCTATGATTTTTTCTTAAATTATGAGAGTTACTTGTACAGCTCGTCCATG-3′ containing hPIV2 NP (R2)-intergenic-hPIV2 P (R1) sequences, cloned into the NotI site just upstream of the NP gene of phPIV2ΔF, and was designated as pEGFP-hPIV2ΔF. The influenza A virus M2 gene tagged with an intervening sequence of hPIV2 was amplified by the one-step RT-PCR (Qiagen, Hilden, Germany) by using 2 μg of purified genomic RNA of PR8, which was obtained from supernatants of MDCK cell cultures after infection at an MOI of 0.1 by using a High Pure Viral RNA Kit (Roche, Indianapolis, IN, USA). The primers used were as follows: 5′-AAAAAGCGGCCGCATAGGTCGCCACCATGAGTCTTCTAACCGAGGTCGAAAC GCCTATCAGAAACGAATGG-3′ (a forward primer containing a NotI site and fusion sequence of 1–26 (underlined) and 715–733 (italic) of matrix (M) gene) and 5′-AAAAAGCGGCCGCCTAACCCGTCCGGGCCTATGATTTTTTCTTAAATTATGAGAGTTATTACTCCAGCTCTATGC-3′ (a reverse primer containing a NotI site and hPIV2 NP (R2)-intergenic-hPIV2 P (R1) sequence array). A NotI fragment of the amplified M2 gene was inserted into a NotI site of phPIV2ΔF, which was designated as pM2-hPIV2ΔF.
hPIV2 NP, P and L genes were amplified from pPIV2 by PCR and each gene was cloned into pCAGGS.17 pcDL-SRα296 (pSRα),18 pSRα-F and pSRα-HN were described previously.19 The gene encoding T7 RNA polymerase was inserted into an EcoRI site of pSRα to generate pSRα-T7 RNA polymerase. An M2 gene fragment tagged with EcoRI and XhoI sequences, which was amplified with PCR by using pM2-hPIV2ΔF DNA as a template, was inserted into the corresponding sites of pcDNA to generate pcDNA-M2 for a positive control experiment.
Establishment of the cells constitutively expressing hPIV2 F protein
Vero cells were transfected with 2 μg of pCXN2-hPIV2F DNA with Cell Line Nucleofector Kit V (Lonza, Basel, Switzerland) by using the Amaxa Nucleofector type II (Lonza) according to the manufacturer’s instructions and cultured in MEM supplemented with 10% heat-inactivated FBS and 1.0 mg ml−1 of G418 (Geneticin) (Thermo Fisher Scientific Inc.) with replacement of the medium two times per week. Cells were cultured for 2–4 weeks and G418-resistant colonies were isolated. To purify single-cell-derived clones, 1 × 104 cells were diluted to 2−1 to 2−12 with the medium on 96-well plates. A single cell in a well was confirmed by microscopy and was proliferated.
To examine mRNA of the hPIV2 F gene, total RNA was extracted with ISOGEN (Nippon Gene, Tokyo, Japan) from 6 × 105 cells of the Vero transfectants with G418 resistance, as well as control Vero cells with and without infection of wild-type hPIV2 at an MOI of 0.2 on the 6-well plates. Total RNA was dissolved in nuclease-free water and each 2 μg of the total RNA was subjected to one-step RT-PCR to amplify the F gene using a pair of primers described in Table 1 (i).
For syncytial formation by the F and HN proteins, 1.0 × 106 of the F-stable clone and control Vero cells were cultured in 6-well plates. Control Vero cells were transfected with 1 μg of pSRα-HN and 1 μg of pSRα-F, and Vero cells stably expressing F protein were transfected with 2 μg of pSRα-HN, by using FuGENE HD (Roche) and X-tream GENE HP (Roche). Numbers of syncytia (defined as more than three nuclei aggregation) were counted 20, 24 and 28 h after transfection.
For the cell growth assay, 5.0 × 104 of the control Vero cells and the F-stable clone candidates were initially prepared in 6-well plates and cultured in MEM supplemented with 10% heat-inactivated FBS (growth medium). The cell numbers were counted on various days.
To detect the F protein on the surface, 6 × 105 of control Vero cells, Vero cells infected with wild-type hPIV2 at an MOI of 0.2 and the Vero/BC-F cells were cultured in 6-well plates. Twenty-four hours later, the cells were harvested in a FACS buffer (phosphate-buffered saline (PBS) containing 2% heat-inactivated FBS) and fixed with 4% paraformaldehyde in PBS. Then, the cells were incubated with the anti-F (144-1A) mAb,20 washed with a FACS buffer three times, labeled with Alexa Flour 488 (Invitrogen, Eugene, OR, USA) and analyzed on a FACSCalibur (BD Biosciences, San Jose, CA, USA) using CellQuest software (BD Biosciences).
To investigate cyototoxicity of overexpressed F protein in the cells, 1.0 × 106 Vero/BC-F cells were transfected with 2 μg of pSRα-F or control pSRα by using X-tream GENE HP. After 2 days, the cells were stained with PI (BioLegend, San Diego, CA, USA) without fixation to evaluate cell damage, and were then analyzed by FACS.
To compare the expression levels of hPIV2 F mRNA between the Vero/BC-F cells cultured for a few months and those cultured for more than 2.5 years, an RT-qPCR was carried out. Total RNA was isolated from 1 × 106 cells by using a High Pure RNA Isolation Kit (Roche). The cDNA was synthesized from 1 μg of the total RNA with an oligo (dT)20 primer and SuperScript II reverse transcriptase (Invitrogen), according to the manufacturer’s instructions. Subsequently, qPCR was performed in triplicate with specific primers shown in Table 1 (ii) for detecting the hPIV2 F transcripts. Each reaction contained 10 μl of 2 × Power SYBR green master mix (Applied Biosystems/Invitrogen, Foster City, CA, USA), forward and reverse primers and 1 μl of the cDNA products, giving a final reaction volume of 20 μl. qPCR assay was performed on a StepOnePlus real-time PCR system (Applied Biosystems/Invitrogen), and StepOne software v.2.1 (Applied Biosystems/Invitrogen) was used to analyze the qPCR data, according to the manufacturer’s instructions. Cycle conditions were set as follows: initial template denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 1 min.
Recovery of a recombinant F-defective hPIV2 vector
Vero/BC-F cells were transfected with pEGFP-hPIV2ΔF (10 μg), altogether with each plasmid encoding the hPIV2 NP (2 μg), P (0.9 μg), L (2 μg) and T7 RNA polymerase (3 μg) by using X-tremeGENE HP according to the manufacturer’s instructions. One week later, the supernatant was centrifuged to remove cells and cell debris, and was then transferred to the fresh Vero/BC-F cell culture in MEM with 1% heat-inactivated FBS (maintenance medium) for virus propagation.
To examine the recovery of the replicon particles, the Vero/BC-F and Vero cells were infected with the EGFP-hPIV2ΔF viruses produced by reverse genetics and wild-type hPIV2, respectively, at an MOI of 0.1. Seven days later, the supernatant of the transfectants was centrifuged to remove cell debris and viral RNA was isolated with High Pure Viral RNA Kit (Roche). Two μg of purified RNA was subjected to one-step RT-PCR to generate various regions of an hPIV2 genome using the pair of primers shown in Table 1 (iii–vi).
Vero cells (1.0 × 106 cells) were infected with either wild-type hPIV2 or EGFP-hPIV2ΔF at an MOI of 5. The cell surface proteins were biotinylated as described previously.24,25 The proteins were immunoprecipitated from the cell lysates with the anti-F (144-1A) mAb or the anti-HN (108-S1) mAb.20 Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane. For detection of biotinylated proteins, the membrane was treated with a streptavidin–biotin–peroxidase complex (Vector Laboratories, Burlingame, CA, USA) and enhanced chemiluminescence reagent, followed by exposure to an X-ray film.
To test the propagation of the EGFP-hPIV2ΔF, 1.0 × 106 cells of the Vero/BC-F or control Vero were inoculated with the EGFP-hPIV2ΔF at an MOI of 0.1. The inoculated cells were incubated in a maintenance medium. The appearance of the GFP fluorescence on the cells was examined 2, 4 and 6 days after inoculation under fluorescence microscopy. Flow cytometry analysis was carried out in the same condition. The Vero/BC-F and Vero cells cultured with EGFP-hPIV2ΔF in various periods were harvested in the FACS buffer, washed two times with the buffer and subjected to FACS analysis.
Also, to test the viral propagation, control Vero cells were infected with the EGFP-hPIV2ΔF or wild-type hPIV2, and the Vero/BC-F cells were infected with the EGFP-hPIV2ΔF at an MOI of 0.1, and incubated in the medium. At appropriate time points, the cultures were examined for production of infectious viruses. The culture supernatant was collected on various days after infection and the virus titer was calculated by the TCID50 method. TCID50 was determined on 96-well plates as follows: the culture supernatants collected at various time points were diluted to 10−4–10−7 with maintenance medium, and each 100 μl was infected to the cells and incubated at 37 °C for 1 week. Virus-positive wells were determined by the presence of cytopathic effects under microscopy.
Expression of M2 protein of the influenza A virus by M2-hPIV2ΔF
A549 cells (1.0 × 106) were infected with the M2-hPIV2ΔF or the EGFP-hPIV2ΔF at an MOI of 5 and incubated with a maintenance medium for 2 days. Cell lysates were subjected to western blot analyses with an anti-M2 (14C2) mAb (Abcam, Cambridge, MA, USA) and an anti-PIV2 NP (20A) mAb,16 respectively.
Infection of EGFP-hPIV2ΔF to the respiratory tract of hamsters and detection of GFP expression in vivo
EGFP-hPIV2ΔF viruses were propagated with Vero/BC-F cells. Cultured supernatants were cleared of the cell debris by centrifugation (2000 g, 4 °C, 10 min), followed by ultracentrifugation (141 000 g, 4 °C, 30 min) to concentrate the virions. Pellets were dissolved in PBS. Virus titer was determined by TCID50. Four-week-old female Syrian hamsters were purchased from Japan SLC (Shizuoka, Japan). Hamsters were anesthetized by intraperitoneal injection of pentobarbital, and EGFP-hPIV2ΔF viruses (7.5 × 108 cell infectious units) in 100 μl of PBS were administered intranasally. Three days after administration, tracheas and lungs were extirpated, and observed under fluorescence microscopy to evaluate the transduction efficiency. All animal studies were approved by the Animal Care Committee of Mie University.
References
Karron RA Collins PL . Parainfluenza viruses. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA et al. Fields Virology, 6th edn. Lippincott Williams & Wilkins: Philadelphia PA, USA, 2013, pp 996–1023.
Lamb RA Parks GD . Paramyxoviridae. Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA et al. Fields Virology, 6th edn Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013, pp 957–995.
Henrickson KJ . Parainfluenza viruses. Clin Microbiol Rev 2003; 16: 242–264.
Ito Y, Tsurudome M, Hishiyama M . The polypeptides of human parainfluenza type 2 virus and their synthesis in infected cells. Arch Virol 1987; 95: 211–224.
Hara K, Fukumura M, Ohtsuka J, Kawano M, Nosaka T . Human parainfluenza virus type 2 vector induces dendritic cell maturation without viral RNA replication/transcription. Hum Gene Ther 2013; 24: 683–691.
Schnell MJ, Mebatsion T, Conzelmann KK . Infectious rabies viruses from cloned cDNA. EMBO J 1994; 13: 4195–4203.
Tang RS, MacPhail M, Schickli JH, Kaur J, Robinson CL, Lawlor HA et al. Parainfluenza virus type 3 expressing the native or soluble fusion (F) protein of respiratory syncytial virus (RSV) confers protection from RSV infection in African green monkeys. J Virol 2004; 78: 11198–11207.
Tompkins SM, Lin Y, Leser GP, Kramer KA, Haas DL, Howerth EW et al. Recombinant parainfluenza virus 5 (PIV5) expressing the influenza A virus hemagglutinin provides immunity in mice to influenza A virus challenge. Virology 2007; 362: 139–150.
Zhan X, Slobod KS, Krishnamurthy S, Luque LE, Takimoto T, Jones B et al. Sendai virus recombinant vaccine expressing hPIV-3 HN or F elicits protective immunity and combines with a second recombinant to prevent hPIV-1, hPIV-3 and RSV infections. Vaccine 2008; 26: 3480–3488.
DiNapoli JM, Yang L, Samal SK, Murphy BR, Collins PL, Bukreyev A . Respiratory tract immunization of non-human primates with a Newcastle disease virus-vectored vaccine candidate against Ebola virus elicits a neutralizing antibody response. Vaccine 2010; 29: 17–25.
Manuse MJ, Parks GD . Role for the paramyxovirus genomic promoter in limiting host cell antiviral responses and cell killing. J Virol 2009; 83: 9057–9067.
Yasumura Y, Kawakita Y . Studies on SV40 in tissue culture-preliminary step for cancer research ‘in vitro’. Nihon Rinsho 1963; 21: 1201–1215; in Japanese.
Desmyter J, Melnick JL, Rawls WE . Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J Virol 1968; 2: 955–961.
Sheets R . History and characterization of the vero cell line. The Vaccines and Related Biological Products Advisory Committee Meeting, Silver spring, MD, USA, 2000.
Montagnon BJ, Fanget B, Nicolas AJ . The large-scale cultivation of VERO cells in micro-carrier culture for virus vaccine production. Preliminary results for killed poliovirus vaccine. Dev Biol Stand 1981; 47: 55–64.
Montagnon BJ . Polio and rabies vaccines produced in continuous cell lines: a reality for Vero cell line. Dev Biol Stand 1989; 70: 27–47.
Niwa H, Yamamura K, Miyazaki J . Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991; 108: 193–199.
Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K et al. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of the human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol 1988; 8: 466–472.
Tsurudome M, Kawano M, Yuasa T, Tabata N, Nishio M, Komada H et al. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 1995; 213: 190–203.
Tsurudome M, Nishio M, Komada H, Bando H, Ito Y . Extensive antigenic diversity among human parainfluenza type 2 virus isolates and immunological relationships among paramyxoviruses revealed by monoclonal antibodies. Virology 1989; 171: 38–48.
Kawano M, Kaito M, Kozuka Y, Komada H, Noda N, Nanba K et al. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology 2001; 284: 99–112.
Calain P, Roux L . The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol 1993; 67: 4822–4830.
Skiadopoulos MH, Vogel L, Riggs JM, Surman SR, Collins PL, Murphy BR . The genome length of human parainfluenza virus type 2 follows the rule of six, and recombinant viruses recovered from non-polyhexameric-length antigenomic cDNAs contain a biased distribution of correcting mutations. J Virol 2003; 77: 270–279.
Tsurudome M, Nishio M, Ito M, Tanahashi S, Kawano M, Komada H et al. Effects of hemagglutinin-neuraminidase protein mutations on cell-cell fusion mediated by human parainfluenza type 2 virus. J Virol 2008; 82: 8283–8295.
Tsurudome M, Ito M, Nishio M, Nakahashi M, Kawano M, Komada H et al. Identification of domains on the fusion (F) protein trimer that influence the hemagglutinin-neuraminidase specificity of the F protein in mediating cell–cell fusion. J Virol 2011; 85: 3153–3161.
Palese P . RNA virus vectors: where are we and where do we need to go? Proc Natl Acad Sci USA 1998; 95: 12750–12752.
Tao T, Skiadopoulos MH, Davoodi F, Surman SR, Collins PL, Murphy BR . Construction of a live-attenuated bivalent vaccine virus against human parainfluenza virus (PIV) types 1 and 2 using a recombinant PIV3 backbone. Vaccine 2001; 19: 3620–3631.
Roberts A, Buonocore L, Price R, Forman J, Rose JK . Attenuated vesicular stomatitis viruses as vaccine vectors. J Virol 1999; 73: 3723–3732.
Rose NF, Marx PA, Luckay A, Nixon DF, Moretto WJ, Donahoe SM et al. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 2001; 106: 539–549.
Jones SM, Feldmann H, Ströher U, Geisbert JB, Fernando L, Grolla A et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat Med 2005; 11: 786–790.
Kapadia SU, Rose JK, Lamirande E, Vogel L, Subbarao K, Roberts A . Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology 2005; 340: 174–182.
Schwartz JA, Buonocore L, Roberts A, Suguitan A Jr, Kobasa D, Kobinger G et al. Vesicular stomatitis virus vectors expressing avian influenza H5 HA induce cross-neutralizing antibodies and long-term protection. Virology 2007; 366: 166–173.
Lamb RA, Zebedee SL, Richardson CD . Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 1985; 40: 627–633.
Sugrue RJ, Hay AJ . Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology 1991; 180: 617–624.
Shim BS, Choi YK, Yun CH, Lee EG, Jeon YS, Park SM et al. Sublingual immunization with M2-based vaccine induces broad protective immunity against influenza. PLoS One 2011; 6: e27953.
Park KS, Lee J, Ahn SS, Byun Y-H, Seong BL, Baek YH et al. Mucosal immunity induced by adenovirus-based H5N1 HPAI vaccine confers protection against a lethal H5N2 avian influenza virus challenge. Virology 2009; 395: 182–189.
Song K, Bolton DL, Wei C-J, Wilson RL, Camp JV, Bao S et al. Genetic immunization in the lung induces potent local and systemic immune responses. Proc Natl Acad Sci USA 2010; 107: 22213–22218.
Le T-vL, Mironova E, Garcin D, Compans RW . Induction of influenza-specific mucosal immunity by an attenuated recombinant Sendai virus. PLoS One 2011; 6: e18780.
Li HO, Zhu YF, Asakawa M, Kuma H, Hirata T, Ueda Y et al. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J Virol 2000; 74: 6564–6569.
Kuypers J, Wright N, Ferrenberg J, Huang M-L, Cent A, Corey L et al. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J Clin Microbiol 2006; 44: 2382–2388.
Nolan SM, Skiadopoulos MH, Bradley K, Kim OS, Bier S, Amaro-Carambot E et al. Recombinant human parainfluenza virus type 2 vaccine candidates containing a 3′ genomic promoter mutation and L polymerase mutations are attenuated and protective in non-human primates. Vaccine 2007; 25: 6409–6422.
Acknowledgements
We thank Dr Yasuhiro Yasutomi for providing valuable discussions. This work was supported by a Grant-in-Aid for the Regional Innovation R&D Program by the Ministry of Economy, Trade and Industry of Japan.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
JO, MF, MK and TN are patent applicants for Vero/BC-F cells. MF is a founder of Biocomo Inc. MF, MK and TN have shares of stock in Biocomo Inc. JO is an employee of Biocomo Inc.
Rights and permissions
About this article
Cite this article
Ohtsuka, J., Fukumura, M., Tsurudome, M. et al. Vero/BC-F: an efficient packaging cell line stably expressing F protein to generate single round-infectious human parainfluenza virus type 2 vector. Gene Ther 21, 775–784 (2014). https://doi.org/10.1038/gt.2014.55
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2014.55