Abstract
Advanced gene delivery techniques can be combined with rational gene design to further improve the efficiency of plasmid DNA (pDNA)-mediated transgene expression in vivo. Herein, we analyzed the influence of intragenic sequence modifications on transgene expression in vitro and in vivo using murine erythropoietin (mEPO) as a transgene model. A single electro-gene transfer of an RNA- and codon-optimized mEPOopt gene into skeletal muscle resulted in a 3- to 4-fold increase of mEPO production sustained for >1 year and triggered a significant increase in hematocrit and hemoglobin without causing adverse effects. mEPO expression and hematologic levels were significantly lower when using comparable amounts of the wild type (mEPOwt) gene and only marginal effects were induced by mEPOΔCpG lacking intragenic CpG dinucleotides, even at high pDNA amounts. Corresponding with these observations, in vitro analysis of transfected cells revealed a 2- to 3-fold increased (mEPOopt) and 50% decreased (mEPOΔCpG) erythropoietin expression compared with mEPOwt, respectively. RNA analyses demonstrated that the specific design of the transgene sequence influenced expression levels by modulating transcriptional activity and nuclear plus cytoplasmic RNA amounts rather than translation. In sum, whereas CpG depletion negatively interferes with efficient expression in postmitotic tissues, mEPOopt doses <0.5 μg were sufficient to trigger optimal long-term hematologic effects encouraging the use of sequence-optimized transgenes to further reduce effective pDNA amounts.
Similar content being viewed by others
Introduction
Among the prevalent delivery systems evaluated for in vivo gene transfer, naked plasmid DNA (pDNA) provides some crucial advantages over viral vectors regarding safety, tolerance and lack of vector-specific immune responses.1, 2 However, to overcome the rather poor transduction efficiency of pDNA in vivo,3, 4 either large doses or multiple applications of the respective transgene are required for the induction of long-term therapeutic effects. Whereas the former strategy bears the risk of inducing inflammatory responses,5, 2 the latter approach would be rather costly and intricate. Further progress in boosting transduction efficiencies has been achieved by complexing pDNA with cationic lipids and gold particles or by using improved transfer protocols, such as hydrodynamic injection, sono- or electroporation (reviewed in6). Especially, the latter method referred to as electro-gene transfer (EGT) has improved transgene expression in muscle cells of various species by 10- to 1 000-fold.7, 8, 9
Apart from sophisticated delivery techniques, vector-intrinsic features, such as DNA structure, stability and size10, as well as the strength and tissue specificity of promoters11, 12 may critically affect the level and longevity of therapeutic transgenes in vivo. The most widely applied strategy to improve in vitro protein production13 or in vivo expression of DNA vaccines14 is the adaptation of the codon bias of a gene of interest to the codon preference of highly expressed host genes (reviewed in15). Gene optimization routinely involves the elimination of repetitive or inhibitory sequence elements, such as AU-rich elements, RNA secondary structures, UpA dinucleotides or cryptic splice sites, all of which have been shown to substantially affect transgene expression.12, 16, 17
Codon adaptation for expression in mammalian hosts is mostly associated with an elevated guanine–cytosine content and consequently, with an increase in CpG dinucleotides (CpGs) known to impact in vivo transgene expression by different mechanisms. First, unmethylated CpG motifs in microbial DNA were shown to stimulate proinflammatory immune responses leading to rapid clearing of transgene expressing cells.18, 19 Second, the de novo methylation of CpG-enriched sequences induced epigenetic gene silencing, thereby impeding persistent expression.20, 21 Apart from promoter-associated CpG-islands, which play a major role in epigenetic regulation, the CpG-content of the transgene open reading frame was also reported to contribute to gene silencing.22, 23, 24 Thus, to avoid methylation-induced shut off of transgenes in vivo, the current credo strives to remove CpGs from both, the vector backbone and the expression cassette.11, 25, 26 Contrary to popular belief, we have recently reported that the depletion of CpG dinucleotides from a green fluorescent protein (GFP) gene results in a significant reduction of in vitro expression in transiently and stably transfected cells suggesting a methylation-independent effect of intragenic CpGs on gene expression.27
In the present study, we have evaluated the influence of rational gene design on the expression level, longevity and bioactivity of the model gene murine erythropoietin (mEPO) in electroporated mice. Herein, we have demonstrated that a codon-optimized mEPO gene containing 20 CpGs induced a significant and constant increase of mEPO expression and the associated hematologic parameters for more than 1 year following single EGT of low pDNA amounts. In contrast, the depletion of all intragenic CpG dinucleotides from the wild type gene resulted in a clear reduction of mEPO production and bioactivity in vivo. Transient expression analysis in cell culture revealed a direct correlation of transgene CpG content with de novo transcription suggesting that codon-optimized CpG-containing transgenes might be preferable over CpG-depleted genes to achieve high-level and long-term expression in skeletal muscle.
Results
Rational design of mEPO genes for high-level and long-term in vivo expression
As previous reports have assigned an important role to pDNA CpG content in compromising long-term gene expression in vivo by methylation-dependent gene silencing, we aimed to clarify to what extent CpG-specific modifications in the coding region of a therapeutic transgene influence the level and longevity of in vivo expression. To exclude unspecific effects on long-term expression by transgene-directed immune responses,28 we chose the autologous murine erythropoietin gene (mEPOwt). Furthermore, EPO allows for direct quantification of secreted protein from mouse blood and functional analysis via correlated hematologic parameters.
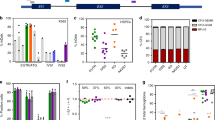
To scrutinize the common belief that the removal of CpG dinucleotides avoids (i) methylation-dependent gene silencing21, 23, 24, 25, 29, 30 and (ii) CpG-motif-mediated inflammation31 both of which hamper prolonged in vivo expression, we designed a CpG-free mEPO gene (mEPOΔCpG) by eliminating all 14 intragenic CpGs without changing the encoded amino acid sequence. Alternatively, we followed the standard strategy to improve effective protein expression by adapting the encoding sequence to the codon usage of highly expressed mammalian genes, resulting in mEPOopt gene with a total of 20 CpGs (Figure 1). Both synthetic sequences were carefully designed to avoid inhibitory DNA- and RNA-sequence motifs known to impair transgene expression. Optimization of the mEPO gene yielded an enhanced guanine–cytosine content (62%) compared with the wild type (54% guanine–cytosine) and CpG-depleted (51% guanine–cytosine) genes. Whereas the elimination of intragenic CpGs (mEPOΔCpG) from the mEPOwt gene did not significantly influence the codon adaptation index (0.75 and 0.73), codon optimization clearly increased the codon adaptation index of mEPOopt (0.98) as expected (Figure 1).
Comparison of sequence-specific parameters of the various mEPO genes. The codon quality distribution plot shows the percentage of codons falling into a certain quality class. The quality value of the codon used most often for a given amino acid in the desired expression system is set to 100 and the remaining codons are scaled according to Sharp and Li.60 The codon quality plot shows the quality of the used codon at the indicated codon position. The GC-content plot displays the GC content in a 40-bp window centered at the indicated nucleotide position. Abbreviation: CAI, codon adaptation index; GC, guanine–cytosine.
For expression studies, the various mEPO genes were inserted into a eukaryotic expression vector under control of the strong human cytomegalovirus (CMV) promoter shown to trigger long-term transgene expression in the skeletal muscle of mice without being silenced.8, 28 To exclude transgene-unrelated effects on mEPO expression, no further CpG modifications were introduced into the vector backbone.
EGT of mEPOopt induces high level and long-term mEPO expression in mice associated with a sustained increase in bioactivity
The longevity of postmitotic muscle cells promoting durable transgene expression from naked pDNA in the absence of chromosomal integration32 and the capability of skeletal muscle to secrete protein like an endocrine tissue33, 34 provide optimal conditions for analysis of long-term EPO expression. For maximum gene transfer efficiency, BALB/c mice were injected intramuscularly with 2.5 μg of the various mEPO pDNAs per leg followed by electroporation. The mEPO levels in mice sera were monitored for more than 12 months by mEPO-specific enzyme-linked immunosorbent assay (ELISA). As indicated in Figure 2a, mEPO concentrations in mEPOwt-injected mice were only slightly increased above endogenous mEPO levels as determined from the negative control group. Surprisingly, the deletion of all CpGs from the coding region yielded mEPO amounts at background level, which were not further augmented in the long-term. In contrast, a significant and persistent increase of mEPO expression was induced following single pDNA injection of the mEPOopt gene with an average serum concentration of ∼37 mU ml−1 maintained for at least 12 months. The longevity of high-level mEPOopt expression further indicates that the CpG-containing codon-optimized sequence was not prone to methylation-dependent gene silencing.
Influence of transgene design on long-term mEPO expression and bioactivity in electroporated BALB/c mice. BALB/c mice were assigned to groups of six animals each and injected intramuscularly into musculus tibialis anterior with 5 μg of the mEPO-encoding vectors followed by in vivo electroporation. Mouse blood was collected from the tail vein at depicted times and sera were obtained by centrifugation. (a) mEPO levels in sera were determined by a commercial mEPO enzyme-linked immunosorbent assay. *P<0.001 (SPSS; univariant analysis). HCT levels of mice treated with (b) 5 μg, (c) 15 μg or (d) 50 μg of pDNA were measured by microcapillary centrifugation. Upward triangles, mEPOopt; squares, mEPOwt; diamonds, mEPOΔCpG; downward triangles, negative (neg) group. *P<0.001 (SPSS; Statistical Package for the Social Sciences, IBM, Chicago, IL, USA; univariant analysis). All data are presented as mean values obtained from six mice per group. Abbreviation: HCT, hematocrit.
We next investigated the bioactivity of the mEPO species encoded by various genes by monitoring the long-term hematocrit (HCT) levels in mouse blood. As depicted in Figure 2b, the observed mEPO expression pattern directly correlated with HCT levels of corresponding mice sera. Accordingly, maximum HCT values were achieved in mEPOopt-injected mice, with ∼72% measured 56 days after EGT and remaining constant for at least 1 year. As expected, HCT levels in mEPOwt-injected mice were significantly lower with an average long-term value of ∼53%, whereas mEPOΔCpG application did not even result in a noticeable HCT increase. Differences in mEPO protein expression and HCT values were also nicely reflected by long-term hemoglobin levels with maximum values above 25 g per 100 ml measured in mEPOopt-treated mice (data not shown).
To clarify, whether the poor effect of the mEPOwt and mEPOΔCpG genes on mEPO and HCT levels could be further boosted at higher pDNA dose, mice were electroporated with 15 or 50 μg of the various pDNAs. Indeed, mEPOwt showed a clear increase in HCT at 15 μg (Figure 2c) and reached HCT levels comparable to those induced by mEPOopt at 50 μg (Figure 2d). In contrast, even 50 μg of the CpG-depleted gene were not sufficient to induce a maximum HCT level. As mEPOwt and mEPOΔCpG genes only differ in their CpG content with otherwise similar sequence parameters, these findings clearly point to a CpG-specific effect on mEPO production.
Interestingly, the maximum mEPO concentration (data not shown) and HCT induced by mEPOopt were not further improved at higher pDNA amounts, suggesting that 5 μg of the optimized gene were sufficient to saturate mEPO production in muscle cells.
Albeit inducing higher initial HCT values of almost 80%, the 50 μg dose of mEPOopt and mEPOwt pDNA resulted in a slight long-term HCT decrease in corresponding mice, which might either be due to tissue damage induced by high pDNA amounts35 or negative feedback regulation of the endogenous mEPO level.36 Taken together, these data demonstrate that a single injection of a codon-optimized mEPO gene efficiently triggers long-term expression of functional mEPO resulting in significantly enhanced HCT and hemoglobin levels in mice for more than 12 months. It is noteworthy that the long-term increase in hematologic parameters was not accompanied by adverse effects and the blood parameters determined from animals in the mEPOopt group at day 171 after EGT were in the normal range (data not shown).
Exogenous mEPO DNA is limited to the electroporated muscle and is still detectable at day 467 after EGT
Next, we aimed to identify those tissues and cells that harbored exogenous mEPO DNA to exclude a horizontal transfer of EGT-administered pDNA to tissues other than the treated muscle. For this purpose, we isolated total DNA at day 30 after EGT from the tibialis anterior (electroporated) and the quadriceps femoris (non-electroporated) muscles, kidney and liver, and amplified mEPO DNA by qualitative PCR using mEPO-specific primers. Unlike endogenous mEPO DNA present in all analyzed tissues (Figure 3a), exogenous mEPO DNA was exclusively detected in the electroporated muscle cells (Figure 3b). The sensitivity of mEPO-specific pDNA detection by PCR was determined to be limited to 10 copies per tissue cell number (Figure 3e). These findings indicate that the injected mEPO DNA was not transferred to neighboring or remote tissues following EGT. When repeating the experiment at day 467 after EGT (Figure 3c), all exogenous mEPO genes were still detected in the electroporated muscle, suggesting that the rather poor expression profiles of mEPOwt and mEPOΔCpG were not due to loss or degradation of the corresponding pDNAs.
Detection of exogenous mEPO DNA in various mouse tissues. Total DNA from electroporated muscles (M+), non-electroporated muscles (musculus quadriceps femoris; (M−), kidney (K) and liver (L) were isolated, and equal amounts of DNA (200 ng) were analyzed for (a) endogenous mEPO (170 bp), (b, c) exogenous mEPO (200 bp) and (d) β-actin (370 bp) using specific primer pairs. 1, mEPOwt; 2, mEPOΔCpG; 3, mEPOopt; 4, control group; 5, PCR negative control; 6, PCR positive control; M, marker. (e) The PCR detection limit was determined using dilution series of mEPO-specific pDNA copies and 200 ng of total DNA prepared from muscle injected with reference vector (neg) as templates. Data are presented for one representative mouse of six per group.
Codon optimization and CpG content strongly affect mEPO expression in cell culture
As the above in vivo studies did not allow discrimination between expression- and secretion-based effects on differential mEPO production, we further analyzed the underlying molecular mechanisms in in vitro cell culture. For this purpose, human HEK-293T and murine 3T3-NIH cells were transiently transfected with the different mEPO pDNAs and protein production was analyzed by western blot using an mEPO-directed antibody (Figures 4a and b). Two different mEPO species were detected in cells and supernatants, whereas a smaller protein of ∼27 kDa predominated in the cell lysates, only the larger fully glycosylated 34 kDa mEPO variant was found to be secreted into cell supernatants.37 In accordance with the in vivo data, only faint amounts of mEPO were detected in human (Figure 4a, top) or murine (Figure 4b, bottom) cells transfected with mEPOwt and mEPOΔCpG genes. In contrast, transfection with the codon-optimized gene resulted in a strong mEPO expression in both cell lines, indicating that the observed differences were not cell type specific. However, larger amounts of the glycosylated mEPO species were found in supernatants of 3T3-NIH cells (compare Figures 4a and b, middle) suggesting that post-translational modifications of mEPO might be more efficient in murine than in human cells with a possible impact on secretion efficiency.
Transient expression analysis of modified mEPO genes in mammalian cells. (a) HEK-293T and (b) 3T3-NIH cells were transiently transfected with 15 μg of the indicated mEPO pDNA vectors. After 48 h, cell lysates and supernatants were collected, and 40 μg of total protein were analyzed by western blot using a mEPO-specific antibody. For quantification of mEPO expression, cell lysates (white bars) and supernatants (black bars) of HEK-293T (c) and 3T3-NIH (d) cells were measured with a Quantikine mouse EPO immunoassay using equal amounts of total protein (1 μg). Data are presented as means of triplicates of one representative experiment. *P<0.001 for both, lysates and supernatants (Student’s t-test).
The subsequent quantification of mEPO expression by ELISA basically confirmed the western blot data, although some relative differences between the western blot signal intensities and the ELISA values were observed, which might be due to differential recognition of the folded mEPO variants by the used antibodies. The ELISA data revealed a strong ∼6- to 7-fold increase in mEPOopt levels in HEK-293T (Figure 4c) and 3T3-NIH (Figure 4d) cells and corresponding supernatants, which confirms previous reports about the expression boost of a codon-optimized human EPO gene in cell culture.13 In contrast, mEPOΔCpG expression in lysates and supernatants of both cell lines was found to be reduced by approximately 50% compared with mEPOwt-specific amounts. Basically, total mEPO levels obtained from HEK-293T cells were 4- to 7-fold higher than those from 3T3-NIH cells, which might be due to better transfection efficiencies. In sum, these findings are consistent with the above in vivo data and suggest that the intragenic CpG content has a direct impact on mEPO protein production.
Intragenic sequence modifications do not affect mEPO translation efficiency
To further assess whether the observed differences in mEPO expression correlate with translation efficiency, we performed a cell-based translation assay. To avoid any sequence-specific effects on post-transcriptional regulation in the nucleus, HEK-293T cells were infected with an MVA virus expressing a T7-RNA polymerase, which allowed for cytoplasmic transcription of the mEPO genes under the control of the T7 promoter. At 24 h after transfection of the MVA/T7-infected cells with the various mEPO genes, cell-associated mEPO amounts were analyzed by western blot (Figure 5a) and quantified by ELISA (Figure 5b). Transfected cells that were not MVA/T7 infected were used as negative controls to demonstrate that the transcription of the mEPO genes strictly depended on the T7 polymerase. Relative glycosylation ratios were comparable for all mEPO species following cytoplasmic transcription, which implies that exclusion of the nuclear compartment did not affect glycosylation efficiency. Interestingly, similar amounts of mEPO were produced from either construct, as also verified by ELISA (Figure 5b) indicating that neither CpG modifications nor codon optimization significantly affected translation in this assay. Taken together, these results suggest that a decreased translation rate was not responsible for the poor expression of the CpG-depleted mEPO gene.
Influence of mEPO sequence modifications on translation efficiency. HEK-293T cells were infected with recombinant MVA/T7 virus and 1 h later transfected with pDNA encoding the indicated mEPO variants. (a) After 24 h cell lysates were collected, and 40 μg of total protein were analyzed by western blot using an anti-mouse erythropoietin (upper panel) or a β-actin-specific (lower panel) antibody. (b) For quantification of in vitro mEPO expression, a Quantikine mouse EPO immunoassay was performed with 1 μg of cell lysates. White bars, MVA/T7-infected cells; black bars, uninfected controls. Data are presented as means of triplicates of one representative experiment.
Intragenic CpG content directly correlates with de novo transcription of mEPO constructs
As translation was not substantially affected by mEPO gene design, we next scrutinized the possible impact on cellular RNA distribution. For this purpose, HEK-293T cells were transiently transfected with the various mEPO constructs, and mEPO-specific mRNAs obtained from nuclear and cytoplasmic cell fractions were analyzed by northern blot. As indicated in Figure 6a, both fractions contained large amounts of mEPOopt transcripts, whereas mEPOwt mRNA was only present in the cytoplasm and at significantly lower amounts. However, only a faint signal was detected in the cytoplasmic fraction of mEPOΔCpG-transfected cells. As revealed by densitometric quantification of mRNA-specific signals (Figure 6b), CpG depletion resulted in a ∼50% reduction of cytoplasmic mRNA levels compared with mEPOwt transcripts.
Analysis of mEPO-specific mRNA distribution and de novo transcription. Human HEK-293T cells were transfected with the indicated constructs and nuclear and cytoplasmic RNA species were harvested 48 h after transfection. (a) mEPO-specific RNAs were detected by northern blot analysis using a DIG-labeled bovine growth hormone-specific RNA probe. 18S and 28S rRNA (middle panel) and β-actin-specific transcripts (lower panel) were analyzed for RNA integrity and as loading control, respectively. (b) Integrated optical bands density was determined using a densitometer. Black bars, nuclear fractions; white bars, cytoplasmic fractions. For relative quantification of mEPO transcripts, nuclear (c) and cytoplasmic (d) RNA fractions as well as (e) de novo synthesized mEPO-specific RNAs were retranscribed into complementary DNA by reverse transcriptase PCR and measured in a Light Cycler 3.5 (Roche). Relative Quantification Software was used for analysis, and target genes (mEPO) were normalized against endogenous β-actin. Data are presented as means of triplicates of one representative experiment.
These findings were confirmed by quantification of mRNAs with the more sensitive real-time PCR (Figures 6c and d). Accordingly, CpG depletion reduced mRNA levels by ∼50% in both, the nuclear and cytoplasmic fractions, which corresponds to the respective protein amounts measured in mEPO ELISA (Figures 4c and d). In contrast, mEPOopt-transfected cells showed a ∼17-fold and ∼7-fold increase of mEPO transcripts in the nucleus and cytoplasm, respectively, as compared with mEPOwt, explaining the ∼6-fold increased protein amounts (Figure 4). Comparable mRNA patterns were obtained from a murine cell line (data not shown). These results thus clearly indicate that the differences in mEPO expression became manifested at the nuclear RNA level.
Next, we asked whether the observed differences in nuclear RNA were based on transcriptional effects. To determine de novo transcription rates, a nuclear run on assay was performed and the obtained RNA amounts were quantified by real-time PCR. As shown in Figure 6e, the de novo-synthesized transcript rates are consistent with the relative amounts of steady-state nuclear mRNA (Figure 6c). Accordingly, de novo transcription of the mEPOΔCpG gene was reduced by almost 50%, whereas mEPOopt transcripts were increased by ∼2-fold compared with mEPOwt. Although an additional impact of codon optimization on RNA stability cannot be excluded, as suggested by the overly increase of mEPOopt steady-state mRNA (Figures 6c and d), these findings propose that the expression strength of the various mEPO genes directly correlates with transcriptional activities. Hence, the above data led to conclude that the depletion of intragenic CpG dinucleotides negatively affects de novo transcription resulting in a loss of transgene expression in vitro and in vivo.
Minimal amounts of the mEPOopt transgene are sufficient to trigger optimal mEPO effects in electroporated mice
The above in vivo studies have demonstrated that EGT of 5 μg of the mEPOopt gene resulted in a long-term HCT saturation of >70% in electroporated mice. However, with regard to human gene therapy, a prolonged maximum HCT increase is not desirable and anemic patients were reported to benefit from a yet moderate HCT increase without risking long-term side effects.38, 39 Thus, to further adjust the mEPO expression to a therapeutically relevant level and analyze gene-specific dose effects, BALB/c mice were injected with decreasing amounts of the mEPOopt and the mEPOwt genes (10–0.1 μg). For comparison, 5 and 2.5 μg doses of the less efficient mEPOΔCpG gene were applied in parallel studies. HCT values were determined at day 56 after EGT, the time point where the average long-term HCT levels had been reached in previous in vivo studies (Figure 2). As outlined in Figure 7a, HCT levels of >70% were still induced by 0.5 μg of mEPOopt pDNA but were rapidly declining at lower doses and reached background levels at 0.1 μg suggesting an efficiency threshold of gene transfer at very low DNA concentrations. In contrast, HCT levels were reduced comparatively fast at mEPOwt-pDNA doses below 5 μg with only a marginal HCT increase obtained with 0.5 μg. The mEPOΔCpG gene did not show any effect at 2.5 μg. The measured HCT levels directly correlated with the corresponding hemoglobin values (Figure 7b), which illustrates that both parameters were equally modulated by the different EPO genes. In sum, these data indicate that the codon-optimized mEPO gene was significantly more efficient than the wild type or CpG-depleted transgenes, allowing for further reduction of the applied pDNA amounts to achieve optimal EPO-related effects.
Efficiency of low dose application of the various mEPO genes on hematocrit and hemoglobin levels. BALB/c mice were assigned to groups of five animals each and were electroporated as described above either with 10, 5, 2.5, 1, 0.5 or 0.1 μg of mEPOopt or mEPOwt constructs or with 5 and 2.5 μg of mEPOΔCpG. Mouse blood was analyzed for HCT levels (a) by microcapillary centrifugation before (filled symbols) or 56 days after EGT (empty symbols) of pDNAs. **P<0.001 (SPSS; univariant analysis) for 10–0.5 μg. (b) Hemoglobin (Hgb) levels were measured from a drop of mouse blood using the HemoCue Hb201+. **P<0.001 (SPSS; univariant analysis) for 10–0.5 μg. The number of animals exhibiting Hgb levels >30 g per 100 ml are indicated by asterisks. Open upward triangles, mEPOopt; open squares, mEPOwt; open diamonds, mEPOΔCpG; open downward triangles, negative group; filled symbols, preimmune sera. All data are presented as mean values obtained from five mice per group.
Discussion
pDNA-based gene therapy has received serious attention as a powerful and cost efficient alternative to recombinant protein applications. Notably, EGT of EPO pDNA into skeletal muscle of small animals has been reported to boost EPO expression up to 100-fold compared with pDNA alone.8, 33, 40 However, clinical application of EGT in larger animals or humans might be more challenging due to large pDNA quantities required to achieve therapeutic effects,41 thus necessitating further improvement of the delivered pDNA.
Most work published to date regarding pDNA-based EPO gene transfer has been focused on the optimization of EPO secretion,41 promoter choice,39 DNA dose effects39, 40, 42 or application routes,38, 39, 41, 42, 43 whereas the contribution of gene design has received less attention. Although several in vivo studies have been performed using either wild type8, 28, 43, 44 or codon-optimized EPO genes41 for EGT, no direct comparison of gene-specific long-term EPO expression has been reported.
In this study, we have provided evidence that in vivo production of murine EPO was significantly enhanced by codon and RNA optimization of the mEPO-coding sequence compared with the wild type gene. Furthermore, a single intramuscular injection of 5 μg of the mEPOopt gene into BALB/c mice followed by electroporation induced a sustained mEPO increase for more than 1 year, associated with a long-term boost of HCT and hemoglobin values proving persistent mEPO bioactivity. Although HCT values of >70% were still obtained with only 0.5 μg of mEPOopt pDNA, even 5 μg of the wild type gene were not sufficient to achieve comparable effects, which demonstrates the potential of gene optimization in achieving maximum effects at low dose.
In contrast to the wild type and CpG-depleted constructs showing a correlation between pDNA dose and physiological response, the long-term HCT levels were not further augmented when applying 15 or even 50 μg of mEPOopt. A dose saturation at high pDNA amounts, in particular in EPO gene therapy, has been described previously4, 45 and might have several reasons. As EPO is a hormone-like protein with multiple systemic functions, EPO-mediated effects are expected to be amplified following expression increase.45 Furthermore, we assume that the codon-optimized gene expresses so efficiently that an EPO-specific production threshold is reached at relatively low pDNA dose. In this context, it has been shown that serum EPO content is rather limited by the saturation of the cellular secretion machinery than by expression limitations, as cell-associated protein levels were still increased at elevated pDNA amounts, although serum EPO levels reached a plateau.41 Even a further increase of EPO secretion, which can be achieved using improved leader sequences, would not boost HCT or hemoglobin values beyond a biologically acceptable level. Finally, one must bear in mind that the EPO-specific regulation is highly complex and overexpression of exogenous EPO has been shown to downregulate endogenous EPO via a negative feedback loop,40 which might contribute to the lack of dose response at high pDNA amounts.
A long-term increase of HCT following single EGT of mEPO pDNA into mice lasting for at least 14 months has been reported previously, but was associated with an early death of the mEPO-injected mice.28 In contrast, we have achieved a comparable long-term HCT increase by applying significantly lower amounts of a codon-optimized gene and an equal number of animals in each transgene group survived for at least 460 days after EGT, which demonstrates that no substantial toxic or adverse effects were induced in the treated animals. This aspect is of paramount importance for safe gene therapy. EGT-specific tissue effects have been shown to be small and transient, which paved the way for this method to the clinical setting.8, 45 The observed damage of muscle cells associated with EGT has rather been ascribed to pDNA injection46, 47 and large amounts of pDNA have been reported to cause infiltration of inflammatory cells leading to a rapid clearing of transfected cells (reviewed in Smith and Nordstrom1; Taylor et al35). In particular, the accumulation of immunostimulatory CpG motifs (CpG in a specific sequence context) within vector backbones has been suggested to trigger the production of inflammatory cytokines, which derogate transgene expression.48 These findings might explain the slight long-term decline of HCT observed for all constructs at the 50 μg dose, whereas a constant high-level HCT value was measured using 5 or 15 μg of the mEPOopt gene. Such pDNA-dependent toxic side effects of EGT could, therefore, be minimized by applying smaller amounts of sequence-improved transgenes.
In view of alternative gene therapy strategies, several groups have demonstrated long-term expression of EPO in mice and non-human primates using recombinant adeno-associated viral vectors for gene delivery.49, 50, 51 However, the utilization of recombinant adeno-associated viral vectors for clinical treatment is still scrutinized owing to induction of transgene-specific immune responses.51, 52, 53 As recently reported, supraphysiological expression of EPO via AAV-based gene transfer elicited EPO-specific antibodies and caused autoimmune anemia in macaques.52, 53 In contrast, we have implemented a long-lasting high-level expression of the biologically active protein implying that no antibodies directed towards the endogenous and pDNA-derived mEPO proteins were generated within the monitored period.
Notwithstanding the widely held view that the deletion of CpGs is beneficial for prolonged transgene expression by preventing methylation-induced gene silencing,20, 21 our data clearly show that both, in vitro and in vivo expression of mEPO was significantly decreased following removal of intragenic CpGs. Furthermore, the durable expression of the mEPOopt gene containing 20 intragenic CpGs argues against CpG-free gene design to achieve a high-level and long-term expression in muscle cells. In fully differentiated tissues such as skeletal muscle, pDNA vectors remain stable as unintegrated episomes and are, therefore, less prone to the methylation-induced silencing mechanisms exerted on genome-integrated transgenes when reorganized into packed chromatin (reviewed in van Gaal et al12; Lufino et al54). The CpG-mediated silencing effects reported for tissues other than skeletal muscle might, therefore, be overcome by using improved vectors engineered to keep the transgene in an extrachromosomal and uncondensed state.54
Although we did not analyze the ratio of integrated versus episomal pDNA over time, all mEPO gene variants were detectable in the injected muscles even 467 days after transfection, which demonstrates the long-term efficacy of EGT. It was also part of this experiment to show that exogenous mEPO DNA was solely detectable in the electroporated myocytes without further translocation to adjacent or remote tissues, thereby confirming previous reports that the transfer of DNA beyond the electrodes is minimal.55
The early and late detection of all mEPO genes did, however, not help to explain the differences in sequence-specific mEPO expression. A more in-depth analysis of the molecular backgrounds in cell culture revealed that sequence modifications did not influence the translation efficiency of the various mEPO transcripts. However, codon and RNA optimization had a strong influence on the steady-state level of mEPO-specific mRNA in transfected cells correlating with a 2-fold increased transcription rate. In contrast, CpG depletion clearly diminished de novo transcription by ∼50% compared with the wild type gene. The molecular mechanism underlying CpG-mediated transcriptional regulation is currently unknown, but has been reproducible for a diverse panel of genes and might be associated with sequence-specific differences in nucleosomal positions, as we have recently reported.27
The negative effects of CpG depletion on mEPO expression in vivo, which were particularly evident at higher pDNA doses might have future implications for rational transgene design. Interestingly, our in vitro experiments have indicated that the expression of codon-optimized genes can be further augmented by inserting additional CpGs into the coding region. CpG-enriched sequence-optimized constructs might, therefore, be optimal tools to further evaluate the thresholds of CpG-mediated effects in future in vivo studies.
The modulatory potential of intragenic CpG content and codon usage may, for example, serve to individually adjust transgene expression to a therapeutically relevant level and should be combined with an efficient regulatory system, allowing for the precise control of transgene expression to avoid long-term side effects in treated individuals. Apart from endogenous regulation via, for example, tissue-specific promoters, several drugs, including doxycycline, ecdysone, mifepristone42 or rapamycin have been successfully tested in vivo for ligand-induced transgene expression (reviewed in Clackson56).
By using the model transgene mEPO, we have shown that sequence optimization combined with efficient gene transfer can significantly increase the level of long-term in vivo transgene expression, thereby further decreasing the threshold of effective pDNA amounts as a major prerequisite to render gene therapy approaches safer, more economic and competitive.
Materials and methods
Generation of modified mEPO genes
Based on the wild type murine erythropoietin gene (mEPOwt) (GenBank accession number NM 007942), a sequence depleted of CpG dinucleotides (mEPOΔCpG) was constructed. Furthermore, a variant adapted to the codon usage of highly expressed mammalian genes (mEPOopt) was designed using the proprietary GeneOptimizer software (Geneart AG, Regensburg, Germany).57 All mEPO sequences were generated by a PCR-based method and were cloned into pPCRScript (Stratagene, La Jolla, CA, USA) via KpnI and SacI restriction sites. The eukaryotic expression vector pRS was generated by deleting the FRT/hygromycin cassette from the commercial expression vector pcDNA5 (Invitrogen, Karlsruhe, Germany) via PCR using oligonucleotides 5′-T CAGATGCATCCGTACGTTAACATGTGAGCAAAAGGCCAGCA-3′ and 5′-AGTCATGCATCCATAGAGCCCACCGCATCCCCA-3′. Synthetic mEPO genes were inserted into pRS via BamHI and HindIII restriction sites. All pDNAs were prepared in supercoiled conformation using Qiagen Endofree Plasmid Kit (Qiagen, Hilden, Germany). For in vivo application, pDNAs were resuspended in sterile saline solution. Endotoxin levels were determined by Limulus amebocyte lysate assay (QCL1000, Cambrex, Walkersville, USA) according to the manufacturer's instructions and were <0.01 U μg−1 DNA in all plasmid preparations.
In vivo electro-gene transfer
Female BALB/c mice (4- to 6-week old, Charles River, Sulzfeld, Germany) were housed at the central animal facility of the University of Regensburg. All animal procedures were conducted in compliance with the national and international laws and policies. Mice were anesthetized with isofluran (Abbott, Wiesbaden, Germany), and mEPO-encoding pDNA constructs (diluted in 100 μl phosphate buffered saline) were injected into both tibialis anterior muscles (50 μl each). Electric pulses were delivered 30 s after injection using 2-Needle Array Handle and steel electrodes (3 cm long, 5 mm apart) (MP Biomedicals, Solon, USA) in parallel orientation with respect to the muscle fibers. Eight long width (50 ms) and low voltage (<50 V cm−1) square bipolar pulses were delivered in a 1 s pulse interval through Electro Square Porator ECM 830 (BTX, Harvard Apparatus, Holliston, MA, USA) with reversal of polarity after four pulses.
Quantification of mEPO, hematocrit and hemoglobin
For detection of mEPO in mouse serum, blood was collected from the tail vein and mEPO concentration was measured using Quantikine ELISA (R&D Systems, MN, USA) as recommended by the manufacturer. Hematocrit levels were determined by high-speed centrifugation of whole blood at 10 000 g in heparinized capillary tubes (Hermle, Wehingen, Germany). Murine hemoglobin levels were determined from a drop of blood using the HemoCue Hb201+ (HemoCue AB, Sweden) as recommended by the manufacturer.
Detection of exogenous and endogenous mEPO DNA
Total DNA from mouse tissues (muscle, kidney and liver) was isolated using QIA amp DNA Mini Kit (Qiagen, Hilden, Germany), tested for RNA contamination and used as PCR templates for the detection of exogenous and endogenous mEPO. For amplification of the exogenous mEPO genes, a forward primer annealing to the CMV promoter (5′-GACGCAAATGGGCGGTAGGCG-3′) was used. Reverse primers were designed to bind within the various mEPO genes: 5′-CAAGGAGAGTAAAAGCAGCAGGGTGG-3′ for mEPOwt and mEPOΔCpG, and 5′-GCAGCAGCAGCAGGGTAGGTCTC-3′ for mEPOopt. Endogenous mEPO was amplified using primers 5′-AAGAACAGGCCATAGAAGTTTG-3′ and 5′-AGCTCCCAGTACCCGAAG-3′ binding within exon 4. To determine the PCR detection limit, 200 ng of total DNA isolated from muscle injected with reference vector (negative group) plus dilution series of mEPO-specific pDNA were used as a template. PCR products were analyzed on a 1% agarose gel.
Cell culture, in vitro transfections and infections
Human embryonic kidney HEK-293T cells and mouse embryonic fibroblast 3T3-NIH cells were maintained in Dulbecco's modified Eagle's medium (Gibco BRL, Grand Island, NY, USA) supplemented with 10% heat-inactivated fetal calf serum (PAN Aidenbach, Germany), 50 IU ml−1 penicillin, 50 μg/ml streptomycin and 1% L-glutamine (3T3-NIH cells), and were cultured in a humidified atmosphere at 5% CO2 and 37 °C. For analysis of mEPO expression, 2 × 105 (HEK-293T) or 2.5 × 105 (3T3-NIH) cells were seeded into six-well plates and were transfected 24 h later with 15 μg of pDNA by calcium phosphate precipitation. To quantify mEPO-specific RNA levels, 3 × 105 HEK-293T cells were seeded into petridishes (∅ 8.5 cm) and transfected with 45 μg of pDNA. For cytoplasmic mEPO expression under control of the T7 promoter, 4 × 105 HEK-293T cells were grown in six-well plates and infected at a multiplicity of infection of 1 with modified vaccinia Ankara virus providing a T7-RNA polymerase (MVA-T7).57 After 1 h, infected cells were washed and transfected with pPCRScript vectors containing the various mEPO genes by using Fugene 6 transfection reagent (Roche, Mannheim, Germany) as recommended by the manufacturer.
Analysis of mEPO expression in vitro
At 24 or 48 h after transfection, cell lysates and supernatants were harvested, and total protein content was determined as described previously.27 For western blot analysis, 40 μg of total protein from both fractions were separated by 12.5% sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis. Erythropoietin was detected using a specific mouse monoclonal antibody (1:500; R&D Systems) and a streptavidin alkaline-phosphatase-conjugated antibody (1:10 000; Roche) followed by staining with the chromogenic substrate NBT/BCIP (Roche). Equal loading of proteins was ensured by β-actin detection using a specific antibody (Millipore, Schwalbach, Germany). For quantification of mEPO proteins in cell lysates and supernatants, Quantikine ELISA was used as described above.
Northern blot analysis
Total RNA was isolated 48 h after transfection using the RNeasy Mini Isolation Kit (Qiagen) according to the manufacturer's protocol. 2 μg of total RNA were electrophoresed on a 1% formaldehyde-agarose denaturing gel and transferred on a nylon membrane. The membrane was prehybridized for 2 h (NaCl 3 M, NaH2PO4 × H2O 0.2 M, Na2EDTA 0.02 M, 50% formamid, 5 × denhard, 0.5% SDS) at 68 °C, followed by a 16 h hybridization at 68 °C with a denatured DIG-labeled RNA-probe specific for the bovine growth hormone polyadenylation signal present in all mEPO constructs. After applying increasingly stringent wash conditions, the membrane was incubated for 30 min with an anti-DIG antibody (Roche, 1:10 000), followed by a 15 min exposition to Hyperfilm ECL (Amersham-Bioscience, Buckinghamshire, UK) and was analyzed using an Optimax X-ray film processor (Protec, Oberstenfeld, Germany). For control of RNA loading and integrity, a β-actin-specific probe was used.
Quantitative real-time PCR
For quantification of mEPO-specific mRNA amounts, 1 μg of purified mRNA was reverse transcribed into complementary DNA using an Oligo (dT)12−18 primer (Invitrogen) and Omniscript reverse transcriptase (Roche). Quantitative real-time PCR was performed in a Light Cycler System 1.2 (Roche) using SYBR-Green as described in the instruction manual for DyNAmo Capillary SYBR Green qPCR Kit (Finnzymes, Espoo, Finland), together with specific primers 5′-CTGCGACAGTCGAGTTCTGGAG-3′ and 5′-CTTCTGAGAGCAGGGACAGGCC-3′ for mEPOwt, 5′-CTGTGACAGTAGAGTTCTGGAG-3′ and 5′-CTTCTGAGAGCAGGGACAGGCC-3′ for mEPOΔCpG, 5′-CTGTGACAGCAGGGTGCTGGAG-3′ and 5′-CCTCGGACAGCAGGCTCAGGCC-3′ for mEPOopt, and 5′-GGTGGGCATGGGCCAGAAGG-3′ and 5′-GATGGGCACAGTGTGGGTGAC-3′ for β-actin complementary DNA. All runs including negative controls were designed and adapted for relative quantification analysis as described by Pfaffl,59 and the relative expression ratio was calculated as follows:
Ratio=((Esample)CPcontrol−CPsample)(target)/((Ereference)CPcontrol−CPsample)(reference);E=efficiency, CP=crossing point. Each reaction was performed in triplicates. Expression levels of mEPO variants were normalized against endogenous β-actin and quantified relative to mEPOwt RNA amounts.
Nuclear run-on
Total RNA from nuclei was prepared 48 h after transfection of HEK-293T cells. Nuclear transcription rates of the various mEPO genes were determined with a non-radioactive nuclear run-on assay as described earlier27 using biotin labeling, magnetic bead capture and analysis by fluorescence-based real-time PCR as outlined above.
Statistical analyses
Statistical analyses were performed using the SigmaPlot 8.0 Student's t-test and SPSS univariant analysis for Windows version 15.0. Differences were considered significant at a P-value of <0.01 and <0.001.
Accession codes
References
Smith LC, Nordstrom JL . Advances in plasmid gene delivery and expression in skeletal muscle. Curr Opin Mol Ther 2000; 2: 150–154.
Nishikawa M, Hashida M . Nonviral approaches satisfying various requirements for effective in vivo gene therapy. Biol Pharm Bull 2002; 25: 275–283.
Glover DJ, Lipps HJ, Jans DA . Towards safe, non-viral therapeutic gene expression in humans. Nat Rev Genet 2005; 6: 299–310.
Fattori E, La Monica N, Ciliberto G, Toniatti C . Electro-gene-transfer: a new approach for muscle gene delivery. Somat Cell Mol Genet 2002; 27: 75–83.
Richard-Fiardo P, Payen E, Chevre R, Zuber J, Letrou-Bonneval E, Beuzard Y et al. Therapy of anemia in kidney failure, using plasmid encoding erythropoietin. Hum Gene Ther 2008; 19: 331–342.
Al Dosari MS, Gao X . Nonviral gene delivery: principle, limitations, and recent progress. AAPS J 2009; 11: 671–681.
Mir LM, Bureau MF, Gehl J, Rangara R, Rouy D, Caillaud JM et al. High-efficiency gene transfer into skeletal muscle mediated by electric pulses. Proc Natl Acad Sci USA 1999; 96: 4262–4267.
Rizzuto G, Cappelletti M, Maione D, Savino R, Lazzaro D, Costa P et al. Efficient and regulated erythropoietin production by naked DNA injection and muscle electroporation. Proc Natl Acad Sci USA 1999; 96: 6417–6422.
Aihara H, Miyazaki J . Gene transfer into muscle by electroporation in vivo. Nat Biotechnol 1998; 16: 867–870.
Mairhofer J, Grabherr R . Rational vector design for efficient non-viral gene delivery: challenges facing the use of plasmid DNA. Mol Biotechnol 2008; 39: 97–104.
Yew NS . Controlling the kinetics of transgene expression by plasmid design. Adv Drug Deliv Rev 2005; 57: 769–780.
van Gaal EV, Hennink WE, Crommelin DJ, Mastrobattista E . Plasmid engineering for controlled and sustained gene expression for nonviral gene therapy. Pharm Res 2006; 23: 1053–1074.
Kim CH, Oh Y, Lee TH . Codon optimization for high-level expression of human erythropoietin (EPO) in mammalian cells. Gene 1997; 199: 293–301.
Graf M, Deml L, Wagner R . Codon-optimized genes that enable increased heterologous expression in mammalian cells and elicit efficient immune responses in mice after vaccination of naked DNA. Methods Mol Med 2004; 94: 197–210.
Gustafsson C, Govindarajan S, Minshull J . Codon bias and heterologous protein expression. Trends Biotechnol 2004; 22: 346–353.
Duan J, Antezana MA . Mammalian mutation pressure, synonymous codon choice, and mRNA degradation. J Mol Evol 2003; 57: 694–701.
Graf M, Schödl T, Wagner R . Rationals of gene design and de novo gene constructions. In: Fu P, Latterich M, Panke S (eds). Systems Biology and Synthetic Biology. John Wiley: New York, 2009, pp 411–438.
Kako K, Nishikawa M, Yoshida H, Takakura Y . Effects of inflammatory response on in vivo transgene expression by plasmid DNA in mice. J Pharm Sci 2008; 97: 3074–3083.
Scheule RK . The role of CpG motifs in immunostimulation and gene therapy. Adv Drug Deliv Rev 2000; 44: 119–134.
Hodges BL, Taylor KM, Joseph MF, Bourgeois SA, Scheule RK . Long-term transgene expression from plasmid DNA gene therapy vectors is negatively affected by CpG dinucleotides. Mol Ther 2004; 10: 269–278.
Yew NS, Zhao H, Przybylska M, Wu IH, Tousignant JD, Scheule RK et al. CpG-depleted plasmid DNA vectors with enhanced safety and long-term gene expression in vivo. Mol Ther 2002; 5: 731–738.
Mitsui M, Nishikawa M, Zang L, Ando M, Hattori K, Takahashi Y et al. Effect of the content of unmethylated CpG dinucleotides in plasmid DNA on the sustainability of transgene expression. J Gene Med 2009; 11: 435–443.
Chevalier-Mariette C, Henry I, Montfort L, Capgras S, Forlani S, Muschler J et al. CpG content affects gene silencing in mice: evidence from novel transgenes. Genome Biol 2003; 4: R53.
Dalle B, Rubin JE, Alkan O, Sukonnik T, Pasceri P, Yao S et al. eGFP reporter genes silence LCRbeta-globin transgene expression via CpG dinucleotides. Mol Ther 2005; 11: 591–599.
de Wolf HK, Johansson N, Thong AT, Snel CJ, Mastrobattista E, Hennink WE et al. Plasmid CpG depletion improves degree and duration of tumor gene expression after intravenous administration of polyplexes. Pharm Res 2008; 25: 1654–1662.
Ill CR, Chiou HC . Gene therapy progress and prospects: recent progress in transgene and RNAi expression cassettes. Gene Ther 2005; 12: 795–802.
Bauer AP, Leikam D, Krinner S, Notka F, Ludwig C, Langst G et al. The impact of intragenic CpG content on gene expression. Nucleic Acids Res 2010; 38: 3891–3908.
Muramatsu T, Arakawa S, Fukazawa K, Fujiwara Y, Yoshida T, Sasaki R et al. In vivo gene electroporation in skeletal muscle with special reference to the duration of gene expression. Int J Mol Med 2001; 7: 37–42.
Chen ZY, He CY, Meuse L, Kay MA . Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther 2004; 11: 856–864.
Hong K, Sherley J, Lauffenburger DA . Methylation of episomal plasmids as a barrier to transient gene expression via a synthetic delivery vector. Biomol Eng 2001; 18: 185–192.
Hyde SC, Pringle IA, Abdullah S, Lawton AE, Davies LA, Varathalingam A et al. CpG-free plasmids confer reduced inflammation and sustained pulmonary gene expression. Nat Biotechnol 2008; 26: 549–551.
Wolff JA, Ludtke JJ, Acsadi G, Williams P, Jani A . Long-term persistence of plasmid DNA and foreign gene expression in mouse muscle. Hum Mol Genet 1992; 1: 363–369.
Kreiss P, Bettan M, Crouzet J, Scherman D . Erythropoietin secretion and physiological effect in mouse after intramuscular plasmid DNA electrotransfer. J Gene Med 1999; 1: 245–250.
MacColl GS, Goldspink G, Bouloux PM . Using skeletal muscle as an artificial endocrine tissue. J Endocrinol 1999; 162: 1–9.
Taylor J, Babbs CF, Alzghoul MB, Olsen A, Latour M, Pond AL et al. Optimization of ectopic gene expression in skeletal muscle through DNA transfer by electroporation. BMC Biotechnol 2004; 4: 11.
Osborne WR, Ramesh N, Lau S, Clowes MM, Dale DC, Clowes AW . Gene therapy for long-term expression of erythropoietin in rats. Proc Natl Acad Sci USA 1995; 92: 8055–8058.
Dubé S, Fisher JW, Powell JS . Glycosylation at specific sites of erythropoietin is essential for biosynthesis, secretion, and biological function. J Biol Chem 1988; 263: 17516–17521.
Hojman P, Gissel H, Gehl J . Sensitive and precise regulation of haemoglobin after gene transfer of erythropoietin to muscle tissue using electroporation. Gene Ther 2007; 14: 950–959.
Sebestyen MG, Hegge JO, Noble MA, Lewis DL, Herweijer H, Wolff JA . Progress toward a nonviral gene therapy protocol for the treatment of anemia. Hum Gene Ther 2007; 18: 269–285.
Maruyama H, Ataka K, Gejyo F, Higuchi N, Ito Y, Hirahara H et al. Long-term production of erythropoietin after electroporation-mediated transfer of plasmid DNA into the muscles of normal and uremic rats. Gene Ther 2001; 8: 461–468.
Fattori E, Cappelletti M, Zampaglione I, Mennuni C, Calvaruso F, Arcuri M et al. Gene electro-transfer of an improved erythropoietin plasmid in mice and non-human primates. J Gene Med 2005; 7: 228–236.
Terada Y, Tanaka H, Okado T, Shimamura H, Inoshita S, Kuwahara M et al. Ligand-regulatable erythropoietin production by plasmid injection and in vivo electroporation. Kidney Int 2002; 62: 1966–1976.
Payen E, Bettan M, Rouyer-Fessard P, Beuzard Y, Scherman D . Improvement of mouse beta-thalassemia by electrotransfer of erythropoietin cDNA. Exp Hematol 2001; 29: 295–300.
Fabre EE, Bigey P, Beuzard Y, Scherman D, Payen E . Careful adjustment of Epo non-viral gene therapy for beta-thalassemic anaemia treatment. Genet Vaccines Ther 2008; 6: 10.
Hojman P, Zibert JR, Gissel H, Eriksen J, Gehl J . Gene expression profiles in skeletal muscle after gene electrotransfer. BMC Mol Biol 2007; 8: 56.
Hartikka J, Sukhu L, Buchner C, Hazard D, Bozoukova V, Margalith M et al. Electroporation-facilitated delivery of plasmid DNA in skeletal muscle: plasmid dependence of muscle damage and effect of poloxamer 188. Mol Ther 2001; 4: 407–415.
Durieux AC, Bonnefoy R, Busso T, Freyssenet D . In vivo gene electrotransfer into skeletal muscle: effects of plasmid DNA on the occurrence and extent of muscle damage. J Gene Med 2004; 6: 809–816.
Yew NS, Cheng SH . Reducing the immunostimulatory activity of CpG-containing plasmid DNA vectors for non-viral gene therapy. Expert Opin Drug Deliv 2004; 1: 115–125.
Tripathy SK, Goldwasser E, Lu MM, Barr E, Leiden JM . Stable delivery of physiologic levels of recombinant erythropoietin to the systemic circulation by intramuscular injection of replication-defective adenovirus. Proc Natl Acad Sci USA 1994; 91: 11557–11561.
Rivera VM, Gao GP, Grant RL, Schnell MA, Zoltick PW, Rozamus LW et al. Long-term pharmacologically regulated expression of erythropoietin in primates following AAV-mediated gene transfer. Blood 2005; 105: 1424–1430.
Maione D, Wiznerowicz M, Delmastro P, Cortese R, Ciliberto G, La Monica N et al. Prolonged expression and effective readministration of erythropoietin delivered with a fully deleted adenoviral vector. Hum Gene Ther 2000; 11: 859–868.
Chenuaud P, Larcher T, Rabinowitz JE, Provost N, Cherel Y, Casadevall N et al. Autoimmune anemia in macaques following erythropoietin gene therapy. Blood 2004; 103: 3303–3304.
Gao G, Lebherz C, Weiner DJ, Grant R, Calcedo R, McCullough B et al. Erythropoietin gene therapy leads to autoimmune anemia in macaques. Blood 2004; 103: 3300–3302.
Lufino MM, Edser PA, Wade-Martins R . Advances in high-capacity extrachromosomal vector technology: episomal maintenance, vector delivery, and transgene expression. Mol Ther 2008; 16: 1525–1538.
Miklavcic D, Semrov D, Mekid H, Mir LM . A validated model of in vivo electric field distribution in tissues for electrochemotherapy and for DNA electrotransfer for gene therapy. Biochim Biophys Acta 2000; 1523: 73–83.
Clackson T . Regulated gene expression systems. Gene Ther 2000; 7: 120–125.
Raab D, Graf M, Notka F, Schoedl T, Wagner R . The GeneOptimizer Algorithm: using a sliding window approach to cope with the vast sequence space in multiparameter DNA sequence optimization. Syst Synth Biol 2010; doi:10.1007/s11693-010-9062-3.
Wyatt LS, Moss B, Rozenblatt S . Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 1995; 210: 202–205.
Pfaffl MW . A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45.
Sharp PM, Li WH . The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 1987; 15: 1281–1295.
Acknowledgements
This study was supported in part by research funding from the Bayerische Forschungsstiftung (grant number 605/05) to RW.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Kosovac, D., Wild, J., Ludwig, C. et al. Minimal doses of a sequence-optimized transgene mediate high-level and long-term EPO expression in vivo: challenging CpG-free gene design. Gene Ther 18, 189–198 (2011). https://doi.org/10.1038/gt.2010.134
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2010.134
Keywords
This article is cited by
-
Minicircle DNA electrotransfer for efficient tissue-targeted gene delivery
Gene Therapy (2013)