Abstract
Purpose: Mutations in the MECP2 gene are associated with Rett syndrome, an X-linked mental retardation disorder in females. Mutations also cause variable neurodevelopmental phenotypes in rare affected males. Recent clinical testing for MECP2 gene rearrangements revealed that entire MECP2 gene duplication occurs in some males manifesting a progressive neurodevelopmental syndrome.
Methods: Clinical testing through quantitative DNA methods and chromosomal microarray analysis in our laboratories identified seven male patients with increased MECP2 gene copy number.
Results: Duplication of the entire MECP2 gene was found in six patients, and MECP2 triplication was found in one patient with the most severe phenotype. The Xq28 duplications observed in these males are unique and vary in size from approximately 200 kb to 2.2 Mb. Three of the mothers who were tested were asymptomatic duplication carriers with skewed X-inactivation. In silico analysis of the Xq28 flanking region showed numerous low-copy repeats with potential roles in recombination.
Conclusions: These collective data suggest that increased MECP2 gene copy number is mainly responsible for the neurodevelopmental phenotypes in these males. These findings underscore the allelic and phenotypic heterogeneity associated with the MECP2 gene and highlight the value of molecular analysis for patient diagnosis, family members at risk, and genetic counseling.
Similar content being viewed by others
Main
The MECP2 gene located on chromosome Xq28 encodes the methyl-CpG-binding protein 2 (MeCP2), which normally functions as a transcriptional repressor.1 The MeCP2 protein preferentially binds methylated CpG dinucleotides and functions as a co-repressor of target gene transcription.2 MECP2 encodes two isoforms: MeCP2_e2 (exons 2–4) and MeCP2_e1 isoform (exons 1, 3, and 4), which is more abundant in brain tissue.3–5 Mutations in the MECP2 gene cause Rett syndrome (Online Mendelian Inheritance in Man [database] 312750), a progressive neurodevelopmental disorder that affects approximately 1 in 10,000 females.6 Classic Rett syndrome is characterized by apparently normal development in girls 6 to 18 months, followed by regression. Clinical features include loss of purposeful hand use replaced by stereotypic hand wringing, loss of speech, acquired microcephaly, mental retardation, autistic features, seizures, ataxia, and breathing dysrhythmias.7,8 In addition to classic Rett syndrome, MECP2 mutations have been identified in patients with a broader range of phenotypes including severe or mild Rett variants, Angelman syndrome-like phenotype, mental retardation with seizures, autistic phenotypes, or mild learning disabilities.9,10 The clinical phenotype in females can be modulated significantly by nonrandom X-inactivation patterns.11
Molecular testing currently involves sequencing of the entire MECP2 coding region, which detects the majority of mutations in Rett syndrome. Additional tests for MECP2 gene rearrangements, such as deletions and duplications, have been developed to augment overall test sensitivity. Our previous analysis on a cohort of 216 females with classic Rett syndrome identified MECP2 mutations in 96% of cases. Sequencing of MECP2 exons 1 to 4 detected mutations in 86%, and additional testing by dosage-sensitive Southern analysis identified MECP2 partial gene deletions in another 10% of females with classic Rett syndrome.12
Rett syndrome was initially considered to be an X-linked dominant lethal condition in males. However, MECP2 gene mutations were identified in males affected with a broader range of neurodevelopmental phenotypes.13,14 Hemizygous MECP2 point mutations that inactivate the protein are associated with a severe neonatal encephalopathy phenotype in affected males.15 When such mutations occur in males with somatic mosaicism or a 47, XXY karyotype, the presentation is more consistent with a classic Rett phenotype because of a compensating normal X-chromosome.16–18 Hypomorphic mutations, which barely cause a phenotype in females, cause mental retardation, tremors, and a variety of neuropsychiatric features in affected males.13,19,20 More recent clinical testing for MECP2 gene rearrangements revealed that entire MECP2 gene duplications occur in some males manifesting a progressive neurodevelopmental syndrome.21–23
Diagnostic studies at the Baylor Medical Genetics Laboratories have identified seven affected males with increased copy-number of the MECP2 gene. These include two males who were referred specifically for mutation analysis of the MECP2 gene and five males who were referred for microarray comparative genomic hybridization analysis for deletions or duplications of multiple genomic regions involved in clinical genomic disorders.24,25 Confirmatory testing using complementary methods were performed in all seven cases.
MATERIALS AND METHODS
Patient material
Peripheral blood samples from patients were submitted for clinical testing to the Baylor Medical Genetics Laboratories for either MECP2 deletion/duplication analysis (patients 1 and 2) or chromosomal microarray analysis (CMA) (patients 3–7). Patients' clinical features are summarized in Table 1. Genomic DNA was extracted from blood leukocytes using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN), and DNA concentration was measured using the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Rockland, DE).
MECP2 Southern analysis
Genomic DNA (2 μg) was digested with PstI or BanI restriction enzymes (New England Biolabs, Ipswich, MA), electrophoresed on a 0.8% agarose gel, and blotted onto a Hybond XL nylon membrane (Amersham Pharmacia, Buckinghamshire, UK). Blots were hybridized to 32P-labeled probes corresponding to MECP2 exon 2, 3, and 4 polymerase chain reaction (PCR) products. Dosage differences indicative of MECP2 whole gene duplication by Southern analysis were detected by visual inspection of autoradiogram band intensities compared with ethidium bromide-stained agarose gel images.
Quantitative real-time polymerase chain reaction
Quantitative real-time PCR (qRT-PCR) analysis for the MECP2 gene was performed using 20 ng of genomic DNA, and PCR primers and Taqman MGB probes were designed using Primer Express software (Applied Biosystems, Foster City, CA). Reactions were done in triplicate, wherein each target region was coamplified with an internal control (RNaseP) using the ABI 7500 RT-PCR system (Applied Biosystems). Relative gene copy number was determined by the comparative threshold cycle method (ddCt).26 qRT-PCR analysis of multiple loci flanking MECP2 on Xq28 was performed using the SYBR-Green chemistry, which measured the fluorescent signal at the end of the elongation phase. Data were normalized against an endogenous reference gene (GAPDH), and a melting curve analysis was performed to verify PCR product specificity.
Multiplex ligation-dependent probe amplification
Dosage analysis of the MECP2 gene and flanking loci was performed by multiplex ligation-dependent probe amplification (MLPA) analysis using commercially available reagents and instructions from MRC-Holland (Amsterdam, The Netherlands). Ligation products were amplified using a Gene Amp PCR System 9700 (Applied Biosystems). Products were resolved on an ABI-3100 genetic analyzer, and data were analyzed using Genescan software (Applied Biosystems). Patient data were normalized to controls for copy-number differences using the Gene Marker Software (Softgenetics, State College, PA).
Chromosomal microarray analysis
Microarray-based comparative genomic hybridization for selected genomic regions involved in clinical genomic disorders was performed at our institution as previously described.27 Patients 1 and 2 and their respective mothers were analyzed with our version 4 microarray containing 366 fluorescence in situ hybridization (FISH)-verified bacterial artificial chromosome (BAC) clones that cover genomic regions involved in more than 40 deletion/duplication disorders.27 Subsequent patients 3 to 7 were analyzed with an expanded microarray version 5 containing 860 FISH-verified BAC clones, including four clones for MECP2 and flanking loci (http://www.bcm.edu/cma/). Reciprocal dye reversal experiments were performed for each patient sample, and hybridized arrays were scanned into 16-bit tiff image files using an Axon two-color microarray scanner 4000B and quantified by using GenePix Pro 6.0 software (GenePix 4000B from Axon Instruments, Union City, CA). Data analysis was performed by a web-based software platform with the capability of displaying the raw, normalized, and integrated data of all clones to detect genomic copy-number changes for each patient relative to a sex-matched control, as previously described.27
Fluorescence in situ hybridization
FISH experiments on interphase or metaphase nuclei chromosome preparations were performed to confirm increased copy-number changes in the MECP2 or other loci identified by CMA. Miniprep BAC DNA (100 ng) was labeled with Spectrum Orange-dUTP or Spectrum Green-dUTP (Vysis, Downers Grove, IL), according to the manufacturer's instructions, and used as probes for FISH analysis using standard protocols.28
X-inactivation studies
Skewing of X-inactivation patterns in duplication carrier mothers was assessed with the androgen receptor methylation assay.29 One microgram of genomic DNA was digested at 37°C overnight with 10 units of the methylation-sensitive restriction enzyme HpaII (New England Biolabs, Ipswich, MA) followed by heat inactivation at 65°C for 20 minutes. Undigested and digested DNA were used as templates for 32P-labeled PCR amplification of the Androgen Receptor CAGn repeat.29 Resulting PCR products were analyzed on a 6% (39:1 acrylamide/bisacrylamide) denaturing gel, which was dried and exposed to x-ray film overnight at −80°C.
RESULTS
In the course of DNA diagnostic testing for MECP2 mutations, our laboratories received referrals for MECP2 deletion/duplication testing for neurodevelopmentally delayed males. Of 122 males referred specifically for MECP2 gene rearrangement testing within a 17-month period, two patients tested positive for duplication of the entire MECP2 gene. Representative data are shown for patient 1. Our standard test for deletion/duplication consisted of a dosage-sensitive Southern analysis for MECP2 exons 1 to 4. Figure 1A shows the results of MECP2 Southern analysis for patient 1 using a genomic restriction digestion with the enzyme PstI. The resulting hybridization pattern consists of a 7.60-kb PstI fragment containing MECP2 exons 1 and 2, and a second fragment of 2.5 kb containing MECP2 exons 3 and 4. Relative band intensities are internally consistent within this patient lane. However, the collective band intensities are disproportionately higher compared with a normal male and resemble a normal female with two X-chromosomes. Comparison of signal intensities on the PstI autoradiogram (Fig. 1A) versus the corresponding ethidium bromide gel (Fig. 1B) is consistent with duplication of MECP2 exons 1 to 4 in male patient 1. These findings were corroborated by Southern analysis using a second restriction enzyme, BanI. Figure 1C shows the autoradiogram of three BanI fragments of 2.14 kb, 1.74 kb, and 1.2 kb that correspond to MECP2 exons 3, 2, and 4, respectively. Thus, Southern analyses using two restriction enzymes indicate duplication of the entire MECP2 gene. Our laboratories performed confirmatory testing by independent dosage-sensitive methods. MECP2 duplication in these patients was verified by MLPA analysis using commercially available reagents. Figure 1D shows the raw MLPA data for patient 1, and Figure 1E shows patient 1 data normalized to a male control, which indicates a twofold increased copy number of probes corresponding to MECP2 exons 1 to 4, plus flanking loci on Xq28 (SLC6A8, IDH3G, L1CAM and IRAK1). In addition to dosage-sensitive Southern and MLPA analyses, we developed qRT-PCR testing for MECP2 exons 1 to 4 using a Taqman assay. Representative data are shown in Figure 1F, wherein MECP2 exon 4 is coamplified with the autosomal RNaseP gene as an endogenous control. Patient 1 was determined to have a twofold MECP2 gene copy number relative to a normal male using the comparative cycle threshold method on qRT-PCR. Thus, duplication of the entire MECP2 gene was established by several DNA methods in two male patients referred specifically for MECP2 gene rearrangement testing.
Molecular detection of MECP2 gene duplication. A: Southern analysis with PstI-digested genomic DNA from male patient 1 (lane 3), normal male controls (lanes 1 and 2), and normal female controls (lanes 4 and 5). Hybridization with MECP2 exon 2 probe generates a 7.6-kb fragment containing MECP2 exons 1 and 2 and a 2.5-kb fragment containing MECP2 exons 3 and 4. B: Corresponding ethidium bromide-stained gel of the PstI-digested DNA. C: Southern analysis with BanI-digested genomic DNA from male patient 1 (lane 3), normal male controls (lanes 1 and 2), and normal female controls (lanes 4 and 5). Hybridization with probes for MECP2 exons 2 to 4 detects three fragments of 2.14 kb (exon 3), 1.74 kb (exon 2), and 1.2 kb (exon 4). D: MLPA data of male patient 1 with MECP2 duplication. E: The same MLPA data normalized to a male control using Gene Marker Software (Softgenetics, State College, PA). MECP2 duplicated exons appear as outliers (blue) from the data set, as points outside of the normal copy threshold lines. F: qRT-PCR analysis of MECP2 exon 4 (orange) and RNAaseP control (blue) in a duplex assay. Results are shown for a male control and patient 1 with MECP2 duplication (ddCt ratio ∼2.00). ΔRn is plotted on the y-axis, and the cycle number is on the x-axis.
The Baylor Medical Genetics Laboratories have also been performing a highly multiplexed CMA for genetic disorders associated with deletions and duplications of specific chromosomal regions. We developed a microarray containing large-insert genomic DNA clones of clinically relevant regions as targets for comparative genomic hybridization (array-CGH).27 The advantages of CMA include increased sensitivity for subchromosomal deletions or duplications that are likely to go undetected by conventional karyotyping or metaphase FISH, and simultaneous detection of a wide range of duplication and deletion syndromes in one highly multiplexed assay.27,30–32 Our CMA microarray was designed to include BAC clones for MECP2, based on the observation that increased MECP2 levels cause progressive neurologic features in a mouse model.33,34 CMA detection of MECP2 duplications was demonstrated on our initial patients 1 and 2, along with their asymptomatic mothers, using our version 4 microarray (366 FISH-verified BAC clones).27 Figure 2A shows representative CMA data from patient 1, which was cohybridized with a normal male genome; the results show a twofold increased copy number of the two Xq28 genomic clones on the CMA version 4 microarray (i.e., BAC RP11-119A22 including MECP2, and RP11-24410 including the flanking L1CAM gene). Corresponding DNA and CMA studies on the asymptomatic mother of patient 1 were positive for heterozygous carrier status (Fig. 2B). The magnitude of her deviation is relatively smaller, consistent with three copies of MECP2 in the carrier mother versus two copies in the normal female. Similar results were obtained on the healthy carrier mother of male patient 2 with duplication (data not shown).
Molecular cytogenetic detection of increased copy number of the MECP2 gene. A: Chromosomal microarray detection of MECP2 duplication in our male patient 1 using CMA version 4.0 microarray. B: Corresponding CMA data showing heterozygous MECP2 duplication in the mother of patient 1. C and D: FISH detection of MECP2 triplication in male patient 4 using RP11-119A22 clone containing the MECP2 gene. E: FISH detection of a flanking duplication in male patient 4 using the RP11-24410 clone containing the L1CAM gene.
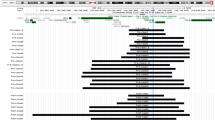
Clinical CMA testing using an updated version 5 microarray identified five additional male patients with increased MECP2 gene copy number of 1380 cases tested by CMA within a 6-month period. Collectively, increased copy number for BAC clones RP11-119A22 (containing MECP2) and RP11-24410 (L1CAM) were seen in all seven patients. A third BAC clone on CMA version 5 (RP11-54120 centromeric to MECP2) was duplicated in patients 1, 3, 4, and 5. A fourth BAC clone RP11-157E12 (more proximally located) was duplicated in patients 3 and 4 (data not shown). Patient 4 had unusual CMA results that suggested triplication of the MECP2 clone. This unexpected finding was confirmed by additional studies, with the most graphic evidence provided by FISH analysis.28 Figure 2C and D show interphase FISH data for patient 4 using clone RP11-119A22, which clearly demonstrate three signals for MECP2 (red) versus one signal for the control probe. In contrast, Figure 2E shows duplication of clone RP11-24410 flanking the MECP2 gene in the same patient. Patient 4 has a complex rearrangement that involves triplication of the MECP2 gene amid duplication of a flanking region of approximately 2 Mb on Xq28. The detection of increased MECP2 gene copy number by CMA analysis in these five additional male patients was also confirmed by quantitative DNA testing methods in our laboratories. Figure 3 shows a composite duplication map based on collective data from Southern, MLPA, CMA, and qRT-PCR studies. Endpoint mapping was facilitated by qRT-PCR analysis performed on 25 loci flanking the MECP2 gene. Individual duplications ranged from approximately 2.2 Mb as the largest to a minimal region of approximately 200 kb containing the L1CAM and MECP2 genes. The genomic rearrangements are unique and vary in size among our seven male patients.
Nonrecurrent duplications involving the MECP2 gene in the Xq28 region among seven affected males determined by quantitative molecular methods. Array CGH BAC clones mapping in the Xq28 region (blue bars). Locations of MLPA probes (red arrows); Xq28 genes tested by qRT-PCR (loci in black text). The extent of the duplicated region based on combined molecular and cytogenetic data for each patient (1–7) (colored horizontal lines). The common duplicated region (vertical dotted lines) (∼200 kb) including the MECP2 and L1CAM genes. CMA, chromosomal microarray analysis; MLPA, multiplex ligation-dependent probe amplification.
The major clinical features in these patients are summarized in Table 1. Predominant features include developmental delay, hypotonia, mild to moderate dysmorphic features, and limited to absent speech. Some patients presented with autistic features, seizures, and progressive loss of ambulation. Four of seven patients had a history of respiratory infections. Involvement of other organ systems was seen in two of seven patients, whereas patient 4 had hydronephrosis and patient 7 had mild aortic dilatation. The most severe, early-onset presentation was seen in patient 4. This is a white male who was 3 months old at the time of testing, with dysmorphic facies, moderate developmental delay, hydronephrosis, tracheomalacia, and recurrent pneumonia. Notably, recurrent respiratory infections have been reported in a number of males with MECP2 gene duplication.35
Our laboratories received samples from several of the patients' mothers, who were reported to be asymptomatic and tested positive as heterozygous duplication carriers. Unexpectedly, the mother of our complex patient 4 has a heterozygous duplication of the MECP2 gene and the surrounding region of approximately 2 Mb (data not shown); this is in contrast with her affected son who has triplication of MECP2 and duplication of the flanking approximately 2-Mb region. These unusual results underscore the complexity of the genomic rearrangement in this family, which has yet to be fully characterized. Given the normal clinical status of these carrier mothers, we performed X-inactivation studies at the androgen receptor locus. Genomic DNA samples were treated with and without predigestion with the methylation-sensitive restriction enzyme HpaII, followed by PCR amplification of the androgen receptor CAGn repeat.29 The X-inactivation results for three cases wherein sufficient material was available are shown in Figure 4. In all three cases, one of two alleles predominates after HpaII digestion. These data are consistent with skewed X-inactivation that presumably inactivates the mutant allele in these unaffected carrier mothers.
X-inactivation studies by methylation analysis of the androgen receptor locus. Data for individual maternal DNA samples (lanes 1–3). PCR amplifications performed on template DNA without prior digestion with the methylation-sensitive HpaII enzyme (lanes 1a–3a) and corresponding results with prior HpaII digestion of template DNA (lanes 1b–3b). All samples are informative, and the observed predominance of one allele after HpaII digestion indicates skewing of X-inactivation in the three tested carrier mothers. Data for female controls DNA with skewed X inactivation and random X inactivation before HpaII digestion (lanes 4a and 5a) and post-HpaII digestion (lanes 4b and 5b).
DISCUSSION
Mutations in the MECP2 gene encompass a wide range of mutation types that include inactivating and hypomorphic point mutations, as well as loss-of-function deletions36 and gain-of-function duplications.21,22 MECP2 mutations are associated with a broad range of neurodevelopmental phenotypes that extend beyond classic Rett syndrome in females. We describe seven male patients with increased copy number of the MECP2 gene and surrounding loci on Xq28 identified by quantitative molecular testing and CMA in our diagnostic laboratory. The phenotypic severity in these male patients did not correlate strictly with their duplication sizes, which ranged from approximately 200 kb to 2.2 Mb. We identified Xq28 duplications that involve MECP2 and flanking genes such as L1CAM and SLC6A8, which are known to play a role in neuronal cell migration or nonspecific mental retardation.37,38 Although flanking gene contribution is a theoretic possibility, these collective data suggest that increased MECP2 gene dosage is specifically responsible for the phenotypes in male patients. Submicroscopic Xq28 duplications containing the MECP2 gene with corresponding increase in RNA expression were reported in multiple male patients.21,22 Those include one male who was duplicated for the entire MECP2 gene but not the flanking L1CAM gene.21 In our study, the most severely affected patient is triplicated for the MECP2 gene. Taken together, the critical duplication region seems to be limited to the MECP2 gene. These patient findings are consistent with data from transgenic mouse models overexpressing the wild-type human MECP2 protein.33,34 These mice appear clinically normal at birth but develop Rett-like progressive neurologic problems including motor dysfunction, hypoactivity, tremors, and ataxia, and die prematurely.33 Higher MECP2 protein levels were found to correlate with more severe phenotypes in these mice.33
Duplications within the human genome are increasingly recognized as a cause of neurodegenerative phenotypes. Such rearrangements are responsible not only for Mendelian traits such as Charcot-Marie-Tooth neuropathy type 1A39 and Pelizaeus-Merzbacher leukodystrophy,40,41 but also for conditions generally thought to be acquired in nature such as Parkinson42 and Alzheimer43 diseases. Recurrent rearrangements in Charcot-Marie-Tooth neuropathy type 1A are mediated by nonallelic homologous recombination,44 wherein genome architectural features seem to lead to genomic instability.44,45 In a similar manner, nonrecurrent rearrangements may also have predisposing features in the local genome.46,47 Segmental duplications or low-copy repeats (LCRs) have been implicated in the cause of chromosomal rearrangements that are associated with several genomic disorders.24,48 Some of these repeats have the potential to form cruciform structures that are highly susceptible to double-strand breaks, which could initiate recombination events leading to duplications within Xq28. To investigate this possibility we performed extensive in silico analysis of an approximately 4-Mb region surrounding the MECP2 gene on Xq28 (149,400,000–154,824,264, Build 35). Figure 5 shows the presence of numerous LCRs in both the direct and inverted orientations, which range from approximately 31 kb to 59 kb in size. Similar analysis was performed by Lee et al.49 on the region surrounding the PLP1 locus, wherein duplications are associated with the X-linked Pelizaeus-Merzbacher disease. Their studies found a statistically significant association between this complex genomic architecture and the various duplication breakpoints. This suggests that the LCRs may stimulate the genomic rearrangements responsible for the majority of Pelizaeus-Merzbacher disease cases and supports an alternative role of genomic architecture in nonrecurrent rearrangements. Along these lines, we hypothesize that LCRs in the Xq28 region may be involved in nonrecurrent rearrangements observed in our male patients with MECP2 duplication. Nonallelic homologous recombination44,48,50 is the usual mechanism for recurrent rearrangements with breakpoints clustering in LCRs. However, nonrecurrent rearrangement breakpoints are also associated with LCRs.41,51 Sequencing of the actual MECP2 duplication breakpoint junctions could confirm the mechanism of nonhomologous end joining,41,51 which has been observed for other nonrecurrent rearrangements. Such experiments are challenged by the highly complex and repetitive nature of this genomic region, and lie beyond the immediate scope of our present study.
In silico analysis using BLAST 255 on repeat-masked sequence reveals LCRs in an ∼4-Mb region surrounding the MECP2 gene. Novel LCRs are displayed centromeric (cen) to telomeric (tel) and labeled A to O. Those with apparent shared identity are shown graphically in like colors; the directions of the block arrows indicate relative orientations. The largest LCRs identified include LCR-MECP2 B1 and B2 (49 and 59 kb, ∼95.9% shared identity), J1 and J2 (40 and 41 kb, ∼99.4% shared identity and highly complex), L1 and L2 (31 and 37 kb, ∼99.6% shared identity), and M2 and M3 (37 and 38 kb, 98.9% shared identity). MECP2 gene (pink circle). Genomic positions (Build 35) are given relative to the MECP2 gene in Megabases.
Males carrying hypomorphic mutations in the MECP2 gene present with a broad and less predictable phenotypic range of mental retardation and diverse neurologic signs and symptoms.52 In addition, MECP2 hypomorphic mutations may be a less common cause of unexplained mental retardation, developmental delay, and associated neurologic features in males than MECP2 duplications,52,53 and to add further complexity, some of the reported MECP2 hypomorphic mutations have been found to be normal genetic variants.54
Table 1 summarizes the clinical findings in our seven male patients and compares their features with literature reports of other males with MECP2 gene duplication. Infantile axial hypotonia was a finding observed in all of our patients. This neurologic finding was also observed by Van Esch et al.22 and Friez et al.35; however, our patients did not exhibit the facial hypotonia described by these two groups. The infantile axial hypotonia in this syndrome leads to progressive spasticity later on in childhood, and this finding was corroborated in our group. A majority of the patients with MECP2 duplication in our study exhibited susceptibility to respiratory infections. Frequent childhood infections, in particular respiratory infections such as pneumonias, have been reported among males with MECP2 duplication.22,23,35 Immune deficiency has been postulated as a possible cause35; however, an extensive immunologic evaluation has not been carried out by other groups or our study to clearly establish immune deficiency as the underlying factor of frequent infections. Although only one of our patients (Table 1) had absent swallowing with aspiration pneumonias, it is not inconceivable to speculate that discoordinated swallowing leading to microaspiration could be responsible for the observed frequent respiratory infections in our group.
The dysmorphic features observed in our patients were heterogeneous. However, brachycephaly, midfacial hypoplasia, and large ears were among the most frequent dysmorphic features in our group. Although the clinical data for some of the patients were not available, Van Esch et al.22 also mentioned prominent ears and flat nasal bridge as some of the most commonly observed mild dysmorphic features among the patients they described. A longitudinal clinical study enrolling a larger number of patients will be required to carefully establish the most characteristic dysmorphic features observed in this syndrome. Multiple congenital anomalies are not commonly found in this condition. The patient with the MECP2 triplication (patient 4, Table 1) exhibited hydronephrosis and tracheomalacia, and another patient with MECP2 duplication presented with mild aortic dilatation (patient 7, Table 1). Two of our patients had genital abnormalities (cryptorchidism and hypospadias). None of the cases described by Van Esch et al.22 presented with hypoplastic genitalia; however, genital abnormalities were commonly reported in the cases described by Sanlaville et al.23 and in the case reported by Meins et al.,21 suggesting that they could be part of the clinical spectrum of this condition. In addition, digital abnormalities such as long, slender fingers were observed in some of our patients. Digital abnormalities have also been described in the published literature of Xq28 disomy.22,23
Three of our patients (Table 1) exhibited autistic-like features, including a limited range of facial expression, gaze avoidance, and repetitive behaviors. Autism or autistic-like features have been associated with Rett syndrome10. The male patient with MECP2 duplication described by Meins et al.21 exhibited autistic features. Autism or autistic-like features were not described in previous studies; however, it is not known whether the patients previously described underwent a neuropsychologic evaluation.22,23 The current findings suggest that autistic-like features could also be found in patients with MECP2 duplication expanding the neuropsychologic phenotype of MECP2-related disorders. Our patients did not exhibit the breathing dysrhythmias commonly observed in females with classic Rett syndrome. Only one patient in our study (patient 5, Table 1) developed stereotypic hand movements; hand stereotypies were only observed in the study described by Meins et al.,21 but not in the studies carried out by other groups.22,23 The carrier females with MECP2 duplication did not manifest any evidence of cognitive dysfunction or abnormal neurologic findings, which could be a reflection of the skewed X-inactivation pattern.
These data collectively highlight the value of comprehensive MECP2 clinical testing in both female and male patients with a diverse range of neurodevelopmental phenotypes, in addition to classic Rett syndrome. This includes sequence analysis for the majority of Rett syndrome mutations, plus quantitative analysis to assess MECP2 deletions and duplications. Although we have a limited dataset to assess recurrence risks associated with MECP2 duplications, our findings of three of three asymptomatic mothers tested to be duplication carriers with skewed X-inactivation raises important issues for carrier testing and genetic counseling in families with identified abnormalities in the MECP2 gene. In conclusion, a large cohort study of patients with MECP2 duplication will be required to establish the frequency of this microduplication in males with mental retardation and associated neurologic features, and to determine the variability in the clinical phenotype observed for this syndrome.
References
Nan X, Campoy FJ, Bird A . MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 1997; 88: 471–481.
Nan X, Ng HH, Johnson CA, Laherty CD, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998; 393: 386–389.
Mnatzakanian GN, Lohi H, Munteanu I, Alfred SE, et al. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat Genet 2004; 36: 339–341.
Kriaucionis S, Bird A . The major form of MeCP2 has a novel N-terminus generated by alternative splicing. Nucleic Acids Res 2004; 32: 1818–1823.
Saxena A, de Lagarde D, Leonard H, Williamson S, et al. Lost in translation: translational interference from a recurrent mutation in exon 1 of MECP2. J Med Genet 2006; 43: 470–477. Epub 2005 Sep 9.
Hagberg B . Rett's syndrome: prevalence and impact on progressive severe mental retardation in girls. Acta Paediatr Scand 1985; 74: 405–408.
Hagberg B . Clinical manifestations and stages of Rett syndrome. Ment Retard Dev Disabil Res Rev 2002; 8: 61–65.
Huppke P, Held M, Laccone F, Hanefeld F, et al. The spectrum of phenotypes in females with Rett Syndrome. Brain Dev 2003; 25: 346–351.
Ylisaukko-Oja T, Rehnstrom K, Vanhala R, Kempas E, et al. MECP2 mutation analysis in patients with mental retardation. Am J Med Genet A 2005; 132: 121–124.
Zoghbi HY . MeCP2 dysfunction in humans and mice. J Child Neurol 2005; 20: 736–740.
Zoghbi HY, Percy AK, Schultz RJ, Fill C, et al. Patterns of X chromosome inactivation in the Rett syndrome. Brain Dev 1990; 12: 131–135.
Fang P, Ward PA, Glaze DG, Van der Veyver IB, et al. Comprehensive clinical testing of the MECP2 gene for Rett syndrome. 2005 ACMG Annual Clin Genet Meeting. March 17–20, Dallas, Texas.
Meloni I, Bruttini M, Longo I, Mari F, et al. A mutation in the Rett syndrome gene, MECP2, causes X-linked mental retardation and progressive spasticity in males. Am J Hum Genet 2000; 67: 982–985.
Orrico A, Lam C, Galli L, Dotti MT, et al. MECP2 mutation in male patients with non-specific X-linked mental retardation. FEBS Lett 2000; 481: 285–288.
Zeev BB, Yaron Y, Schanen NC, Wolf H, et al. Orr-Urtreger. Rett syndrome: clinical manifestations in males with MECP2 mutations. J Child Neurol 2002; 17: 20–24.
Armstrong J, Pineda M, Aibar E, Gean E, et al. Classic Rett syndrome in a boy as a result of somatic mosaicism for a MECP2 mutation. Ann Neurol 2001; 50: 692.
Clayton-Smith J, Watson P, Ramsden S, Black GC, et al. Somatic mutation in MECP2 as a non-fatal neurodevelopmental disorder in males. Lancet 2000; 356: 830–832.
Schwartzman JS, De Souza AM, Faiwichow G, Hercowitz LH, et al. [Rett phenotype in patient with XXY karyotype: case report]. Ar Qneuropsiquiatr 1998; 56: 824–828.
Kleefstra T, Yntema HG, Oudakker AR, Romein T, et al. De novo MECP2 frameshift mutation in a boy with moderate mental retardation, obesity and gynaecomastia. Clin Genet 2002; 61: 359–362.
Cohen D, Lazar G, Couvert P, Desportes V, et al. MECP2 mutation in a boy with language disorder and schizophrenia. Am J Psychiatry 2002; 159: 148–149.
Meins M, Lehmann J, Gerresheim F, Herchenbach J, et al. Submicroscopic duplication in Xq28 causes increased expression of the MECP2 gene in a boy with severe mental retardation and features of Rett syndrome. J Med Genet 2005; 42: e12.
Van Esch H, Bauters M, Ignatius J, Jansen M, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet 2005; 77: 442–453.
Sanlaville D, Prieur M, de Blois MC, Genevieve D, et al. Functional disomy of the Xq28 chromosome region. Eur J Hum Genet 2005; 13: 579–585.
Lupski JR . Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 1998; 14: 417–422.
Lupski JR, Stankiewicz P, editors. Genomic Disorders. Totowa, NJ: Humana Press, 2006; 1–427.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408.
Cheung SW, Shaw CA, Yu W, Li J, et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med 2005; 7: 422–432.
Trask BJ . Fluorescence in situ hybridization: applications in cytogenetics and gene mapping. Trends Genet 1991; 7: 149–154.
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, et al. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992; 51: 1229–1239.
Bejjani BA, Theisen AP, Ballif BC, Shaffer LG, et al. Array-based comparative genomic hybridization in clinical diagnosis. Expert Rev Mol Diagn 2005; 5: 421–429.
Bejjani BA, Saleki R, Ballif BC, Rorem EA, et al. Use of targeted array-based CGH for the clinical diagnosis of chromosomal imbalance: is less more?. Am J Med Genet A 2005; 134: 259–267.
Lugtenberg D, de Brouwer AP, Kleefstra T, Oudakker AR, et al. Chromosomal copy number changes in patients with non-syndromic X linked mental retardation detected by array CGH. J Med Genet 2006; 43: 362–370.
Collins AL, Levenson JM, Vilaythong AP, Richman R, et al. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet 2004; 13: 2679–2689.
Luikenhuis S, Giacometti E, Beard CF, Jaenisch R, et al. Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc Natl Acad Sci U S A 2004; 101: 6033–6038.
Friez MJ, Jones JR, Clarkson K, Lubs H, et al. Recurrent infections, hypotonia and mental retardation caused by duplication of MECP2 and adjacent region in Xq28. Pediatrics 2006, Nov 6; [Epub ahead of print].
Schollen E, Smeets E, Deflem E, Fryns JP, et al. Gross rearrangements in the MECP2 gene in three patients with Rett syndrome: implications for routine diagnosis of Rett syndrome. Hum Mutat 2003; 22: 116–120.
Vits L, Van Camp G, Coucke P, Fransen E, et al. MASA syndrome is due to mutations in the neural cell adhesion gene L1CAM. Nat Genet 1994; 7: 408–413.
Salomons GS, van Dooren SJ, Verhoeven NM, Cecil KM, et al. X-linked creatine-transporter gene (SLC6A8) defect: a new creatine-deficiency syndrome. Am J Hum Genet 2001; 68: 1497–1500.
Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 1991; 66: 219–232.
Inoue K, Osaka H, Thurston VC, Clarke JT, et al. Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet 2002; 71: 838–853.
Woodward KJ, Cundall M, Sperle K, Sistermans EA, et al. Heterogeneous duplications in patients with Pelizaeus-Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am J Hum Genet 2005; 77: 966–987.
Singleton AB, Farrer M, Johnson J, Singleton A, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science 2003; 302: 841.
Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur NAPP, et al. Locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 2006; 38: 24–26.
Stankiewicz P, Lupski JR . Genome architecture, rearrangements and genomic disorders. Trends Genet 2002; 18: 74–82.
Stankiewicz P, Inoue K, Bi W, Walz K, et al. Genomic disorders: genome architecture results in susceptibility to DNA rearrangements causing common human traits. Cold Spring Harb Symp Quant Biol 2003; 68: 445–454.
Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, et al. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am J Hum Genet 2003; 72: 1101–1116.
Lee JA, Madrid RE, Sperle K, Ritterson CM, et al. Spastic paraplegia type 2 associated with axonal neuropathy and apparent PLP1 position effect. Ann Neurol 2006; 59: 398–403.
Emanuel BS, Shaikh TH . Segmental duplications: an ‘expanding' role in genomic instability and disease. Nat Rev Genet 2001; 2: 791–800.
Lee JA, Inoue K, Cheung SW, Shaw CA, et al. Role of genomic architecture in PLP1 duplication causing Pelizaeus-Merzbacher disease. Hum Mol Genet 2006; 15: 2250–2265.
Shaw CJ, Lupski JR . Implications of human genome architecture for rearrangementbased disorders: the genomic basis of disease. Hum Mol Genet 2004; 13 Spec No 1: R57–64.
Shaw CJ, Lupski JR . Non-recurrent 17p11.2 deletions are generated by homologous and non-homologous mechanisms. Hum Genet 2005; 116: 1–7.
Moog U, Smeets EE, van Roozendaal KE, Schoenmakers S, et al. Neurodevelopmental disorders in males related to the gene causing Rett syndrome in females (MECP2). Eur J Paediatr Neurol 2003; 7: 5–12.
Moog U, Van Roozendaal K, Smeets E, Tserpelis D, et al. MECP2 mutations are an infrequent cause of mental retardation associated with neurological problems in male patients. Brain Dev 2006; 28: 305–310.
Laccone F, Zoll B, Huppke P, Hanefeld F, et al. MECP2 gene nucleotide changes and their pathogenicity in males: proceed with caution. J Med Genet 2002; 39: 586–588.
Tatusova TA, Madden TL . BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 1999; 174: 247–250.
Acknowledgements
This work was supported in part by National Institutes of Health Grants P01 HD40301 (to Huda Y. Zogbhi) and HD24064 to the Baylor College of Medicine Mental Retardation and Developmental Disabilities Research Center.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
del Gaudio, D., Fang, P., Scaglia, F. et al. Increased MECP2 gene copy number as the result of genomic duplication in neurodevelopmentally delayed males. Genet Med 8, 784–792 (2006). https://doi.org/10.1097/01.gim.0000250502.28516.3c
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/01.gim.0000250502.28516.3c
This article is cited by
-
Duplication within two regions distal to MECP2: clinical similarity with MECP2 duplication syndrome
BMC Medical Genomics (2023)
-
IRAK1 Duplication in MECP2 Duplication Syndrome Does Not Increase Canonical NF-κB–Induced Inflammation
Journal of Clinical Immunology (2023)
-
A brief history of MECP2 duplication syndrome: 20-years of clinical understanding
Orphanet Journal of Rare Diseases (2022)
-
Reversal of Social Recognition Deficit in Adult Mice with MECP2 Duplication via Normalization of MeCP2 in the Medial Prefrontal Cortex
Neuroscience Bulletin (2020)
-
Distinct patterns of complex rearrangements and a mutational signature of microhomeology are frequently observed in PLP1 copy number gain structural variants
Genome Medicine (2019)