Abstract
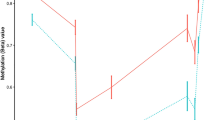
Genetic studies have demonstrated association between single-nucleotide polymorphisms within the IL2RA (interleukin-2 receptor α-subunit) gene and risk of developing multiple sclerosis (MS); however, these variants do not have obvious functional consequences. DNA methylation is a source of genetic variation that could impact on autoimmune disease risk. We investigated DNA methylation of the IL2RA promoter in genomic DNA obtained from peripheral blood mononuclear cells and neural tissue using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry. A differential methylation profile of IL2RA was identified, suggesting that IL2RA expression was regulated by DNA methylation. We extended our analysis of DNA methylation to peripheral blood mononuclear cell (PBMC) of MS cases and controls using MALDI-TOF and Illumina HumanMethylation450 arrays. Analyses of CpG sites within the proximal promoter of IL2RA in PBMC showed no differences between MS cases and controls despite an increase in IL2RA expression. In contrast, we inferred significant DNA methylation differences specific to particular leukocyte subtypes in MS cases compared with controls by deconvolution of the array data. The decrease in methylation in patients correlated with an increase in IL2RA expression in T cells from MS cases in comparison with controls. Our data suggest that differential methylation of the IL2RA promoter in T cells could be an important pathogenic mechanism in MS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Kim HP, Imbert J, Leonard WJ . Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev 2006; 17: 349–366.
Godfrey DI, Kennedy J, Suda T, Zlotnik A . A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8− triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol 1993; 150: 4244–4252.
Kmieciak M, Gowda M, Graham L, Godder K, Bear HD, Marincola FM et al. Human T cells express CD25 and Foxp3 upon activation and exhibit effector/memory phenotypes without any regulatory/suppressor function. J Transl Med 2009; 7: 89.
Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M . Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol 1995; 155: 1151–1164.
Kim HP, Kelly J, Leonard WJ . The basis for IL-2-induced IL-2 receptor alpha chain gene regulation: importance of two widely separated IL-2 response elements. Immunity 2001; 15: 159–172.
Vella A, Cooper JD, Lowe CE, Walker N, Nutland S, Widmer B et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet 2005; 76: 773–779.
Brand OJ, Lowe CE, Heward JM, Franklyn JA, Cooper JD, Todd JA et al. Association of the interleukin-2 receptor alpha (IL-2Ralpha)/CD25 gene region with Graves' disease using a multilocus test and tag SNPs. Clin Endocrinol (Oxf) 2007; 66: 508–512.
Lowe CE, Cooper JD, Brusko T, Walker NM, Smyth DJ, Bailey R et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet 2007; 39: 1074–1082.
Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med 2007; 357: 851–862.
Weber F, Fontaine B, Cournu-Rebeix I, Kroner A, Knop M, Lutz S et al. IL2RA and IL7RA genes confer susceptibility for multiple sclerosis in two independent European populations. Genes Immun 2008; 9: 259–263.
Consortium TSG. Refining genetic associations in multiple sclerosis. Lancet Neurol 2008; 7: 567–569.
Rubio JP, Stankovich J, Field J, Tubridy N, Marriott M, Chapman C et al. Replication of KIAA0350, IL2RA, RPL5 and CD58 as multiple sclerosis susceptibility genes in Australians. Genes Immun 2008; 9: 624–630.
ANZgene. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet 2009; 41: 824–828.
Sharfe N, Dadi HK, Shahar M, Roifman CM . Human immune disorder arising from mutation of the alpha chain of the interleukin-2 receptor. Proc Natl Acad Sci USA 1997; 94: 3168–3171.
Sakaguchi S, Setoguchi R, Yagi H, Nomura T . Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in self-tolerance and autoimmune disease. Curr Top Microbiol Immunol 2006; 305: 51–66.
Laurie KL, Van Driel IR, Gleeson PA . The role of CD4+CD25+ immunoregulatory T cells in the induction of autoimmune gastritis. Immunol Cell Biol 2002; 80: 567–573.
McHugh RS, Shevach EM . Cutting edge: depletion of CD4+CD25+ regulatory T cells is necessary, but not sufficient, for induction of organ-specific autoimmune disease. J Immunol 2002; 168: 5979–5983.
Maier LM, Lowe CE, Cooper J, Downes K, Anderson DE, Severson C et al. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet 2009; 5: e1000322.
Maier LM, Anderson DE, Severson CA, Baecher-Allan C, Healy B, Liu DV et al. Soluble IL-2RA levels in multiple sclerosis subjects and the effect of soluble IL-2RA on immune responses. J Immunol 2009; 182: 1541–1547.
Garg G, Tyler JR, Yang JH, Cutler AJ, Downes K, Pekalski M et al. Type 1 diabetes-associated IL2RA variation lowers IL-2 signaling and contributes to diminished CD4+CD25+ regulatory T cell function. J Immunol 2012; 188: 4644–4653.
Ansel KM, Lee DU, Rao A . An epigenetic view of helper T cell differentiation. Nat Immunol 2003; 4: 616–623.
Wilson CB, Makar KW, Shnyreva M, Fitzpatrick DR . DNA methylation and the expanding epigenetics of T cell lineage commitment. Semin Immunol 2005; 17: 105–119.
Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA . T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 2006; 24: 369–379.
Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY . A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol 2005; 6: 1142–1151.
Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY . Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity 2005; 22: 329–341.
Hori S, Nomura T, Sakaguchi S . Control of regulatory T cell development by the transcription factor Foxp3. Science 2003; 299: 1057–1061.
Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol 2007; 5: e38.
Janson PC, Winerdal ME, Marits P, Thorn M, Ohlsson R, Winqvist O . FOXP3 promoter demethylation reveals the committed Treg population in humans. PLoS One 2008; 3: e1612.
Baron U, Floess S, Wieczorek G, Baumann K, Grutzkau A, Dong J et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol 2007; 37: 2378–2389.
Januchowski R, Prokop J, Jagodzinski PP . Role of epigenetic DNA alterations in the pathogenesis of systemic lupus erythematosus. J Appl Genet 2004; 45: 237–248.
Zhou Y, Lu Q . DNA methylation in T cells from idiopathic lupus and drug-induced lupus patients. Autoimmun Rev 2008; 7: 376–383.
Nile CJ, Read RC, Akil M, Duff GW, Wilson AG . Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum 2008; 58: 2686–2693.
Belot MP, Fradin D, Mai N, Le Fur S, Zelenika D, Kerr-Conte J et al. CpG methylation changes within the IL2RA promoter in type 1 diabetes of childhood onset. PLoS One 2013; 8: e68093.
Graves MC, Benton M, Lea RA, Boyle M, Tajouri L, Macartney-Coxson D et al. Methylation differences at the HLA-DRB1 locus in CD4+ T-cells are associated with multiple sclerosis. Mult Scler 2014; 20: 1033–1041.
Bos SD, Page CM, Andreassen BK, Elboudwarej E, Gustavsen MW, Briggs F et al. Genome-wide DNA methylation profiles indicate CD8+ T cell hypermethylation in multiple sclerosis. PLoS One 2015; 10: e0117403.
Maltby VE, Graves MC, Lea RA, Benton MC, Sanders KA, Tajouri L et al. Genome-wide DNA methylation profiling of CD8+ T cells shows a distinct epigenetic signature to CD4+ T cells in multiple sclerosis patients. Clin Epigenetics 2015; 7: 118.
Kuhn A, Thu D, Waldvogel HJ, Faull RL, Luthi-Carter R . Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat Methods 2011; 8: 945–947.
Cerosaletti K, Schneider A, Schwedhelm K, Frank I, Tatum M, Wei S et al. Multiple autoimmune-associated variants confer decreased IL-2R signaling in CD4+ CD25(hi) T cells of type 1 diabetic and multiple sclerosis patients. PLoS One 2013; 8: e83811.
Butter F, Davison L, Viturawong T, Scheibe M, Vermeulen M, Todd JA et al. Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding. PLoS Genet 2012; 8: e1002982.
Bruniquel D, Schwartz RH . Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol 2003; 4: 235–240.
Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J 2006; 25: 1081–1092.
Malek TR, Bayer AL . Tolerance, not immunity, crucially depends on IL-2. Nat Rev Immunol 2004; 4: 665–674.
Rubin LA, Galli F, Greene WC, Nelson DL, Jay G . The molecular basis for the generation of the human soluble interleukin 2 receptor. Cytokine 1990; 2: 330–336.
Robb RJ, Kutny RM . Structure–function relationships for the IL 2-receptor system. IV. Analysis of the sequence and ligand-binding properties of soluble Tac protein. J Immunol 1987; 139: 855–862.
Rubin LA, Snow KM, Kurman CC, Nelson DL, Keystone EC . Serial levels of soluble interleukin 2 receptor in the peripheral blood of patients with rheumatoid arthritis: correlations with disease activity. J Rheumatol 1990; 17: 597–602.
Rose-John S, Heinrich PC . Soluble receptors for cytokines and growth factors: generation and biological function. Biochem J 1994; 300: 281–290.
Rubinstein MP, Kovar M, Purton JF, Cho JH, Boyman O, Surh CD et al. Converting IL-15 to a superagonist by binding to soluble IL-15R{alpha}. Proc Natl Acad Sci USA 2006; 103: 9166–9171.
Venken K, Hellings N, Broekmans T, Hensen K, Rummens JL, Stinissen P . Natural naive CD4+CD25+CD127low regulatory T cell (Treg) development and function are disturbed in multiple sclerosis patients: recovery of memory Treg homeostasis during disease progression. J Immunol 2008; 180: 6411–6420.
Venken K, Hellings N, Thewissen M, Somers V, Hensen K, Rummens JL et al. Compromised CD4+ CD25(high) regulatory T-cell function in patients with relapsing-remitting multiple sclerosis is correlated with a reduced frequency of FOXP3-positive cells and reduced FOXP3 expression at the single-cell level. Immunology 2008; 123: 79–89.
Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA 2005; 102: 15785–15790.
Maksimovic J, Gordon L, Oshlack A, SWAN . Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol 2012; 13: R44.
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014; 30: 1363–1369.
Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform 2010; 11: 587.
Gagnon-Bartsch JA, Speed TP . Using control genes to correct for unwanted variation in microarray data. Biostatistics 2012; 13: 539–552.
Team RDCR: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2008.
Acknowledgements
We thank all the people with MS and control persons who participated in this study, and are grateful to Jennifer Eckholdt and John Carey for their assistance with the collection of blood samples for this study. This work was supported by NHMRC Project Grants (nos 257518 and 509184) and JF was supported by a Multiple Sclerosis Research Australia Fellowship. This work was also funded by Multiple Sclerosis Research Australia, National Health and Medical Research Council Australia, Lions International and Rebecca L Cooper Foundation. MAJ is supported by an NHMRC/MSRA Betty Cuthbert fellowship. AGB is supported by an Australian National Health and Medical Research Council (NHMRC) Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Field, J., Fox, A., Jordan, M. et al. Interleukin-2 receptor-α proximal promoter hypomethylation is associated with multiple sclerosis. Genes Immun 18, 59–66 (2017). https://doi.org/10.1038/gene.2016.50
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2016.50
This article is cited by
-
Aberrant DNA methylation profile exacerbates inflammation and neurodegeneration in multiple sclerosis patients
Journal of Neuroinflammation (2020)
-
Clinical epigenetics: seizing opportunities for translation
Nature Reviews Genetics (2019)
-
Cell death in cancer in the era of precision medicine
Genes & Immunity (2019)