Abstract
Purpose
The purpose of this study was to assess the endothelium of corneal grafts by in vivoconfocal microscopy (IVCM), and to evaluate an automated endothelial software system in comparison with a manual cell count and planimetry.
Patients and methods
Overall, 40 corneal grafts (20 deep anterior lamellar keratoplasties (DALKs) and 20 penetrating keratoplasties (PKs)) were assessed by scanning-slit IVCM. The endothelial cell density (ECD) was estimated with the automated and the manual cell count method of the instrument's Nidek Advanced Vision Information System (NAVIS) software. The results were compared with planimetry as the reference method, and the agreement was assessed.
Results
The mean (±SD) automated ECD was 2278±524 cells/mm2 (range 1167–3192 cells/mm2), whereas the manual cell count method gave significantly lower ECDs with a mean of 1213±677 cells/mm2 (range 218–2440 cells/mm2; P<0.001). The manual cell counts were also significantly lower than those by planimetry, with a mean ECD of 1617±813 cells/mm2 (range 336–2941, P<0.001). Bland–Altman analyses indicated that the limits of agreement (LoA) between the automated and the planimetry method were −671 and +1992 cells/mm2, whereas they were −1000 and +202 cells/mm2 when comparing the manual cell counts with planimetry.
Conclusion
Following keratoplasty, the NAVIS automated method is likely to overestimate endothelial cell counts due to oversegmenting of the cell domains. Automated ECDs are substantially higher than those by the manual counting method or planimetry. The differences are considerably larger post-keratoplasty than for normal corneas, and the methods should not be used interchangeably.
Similar content being viewed by others
Introduction
Penetrating keratoplasty (PK) is an established technique to restore corneal transparency and has a good long-term prognosis. However, studies have shown that a substantial number of endothelial cells are lost from the transplanted donor cornea, which, as a result, is at risk of decompensation. This decompensation, associated with an unwanted increase in corneal thickness that may occur after an initially successful graft, can be attributed to endothelial dysfunction. Deep anterior lamellar keratoplasty (DALK) is an alternative to PK, especially for conditions that primarily affect the anterior corneal layers such as keratoconus and carries no risk of endothelial rejection per se, with a lower risk of post-operative endothelial cell loss.1
The causes of endothelial cell loss after corneal transplantation (PK or DALK) are numerous, including mechanical damage associated with the operation itself and later stage immune reactions that lead to rejection of the donor endothelial cells. An assessment of the endothelial cell density (ECD) is a commonplace method of pre- and post-operative monitoring of a corneal graft by using either specular microscopy2, 3, 4, 5, 6, 7, 8 or, more recently, confocal microscopy.9, 10, 11, 12 Such high-resolution imaging of the corneal endothelium is generally needed to allow for endothelial cell counting and this is done, as it has long been recognised that a very low endothelial cell count may ultimately lead to reduced endothelial function and to subsequent graft failure. For such assessments, it is important that the method of assessment of ECD is reasonably reliable.13, 14, 15 Various semi-automated and automated methods are available on specular and confocal microscopes that are routinely used to assess ECD.11, 16, 17
In vivo confocal microscopy (IVCM) provides highly magnified images of corneal layers, including the corneal endothelium,11, 18, 19, 20, 21 even from corneas with advanced Fuchs’ dystrophy before grafting19 or from previously failed grafts with partial opacities.10 One of the endothelial morphometry programmes available with one confocal microscope (Confoscan CS4, Nidek Technologies, Albignasego, Italy) is the Nidek Advanced Vision Information System software (NAVIS). The automated analysis option of this programme (or versions thereof) has been reported to provide cell density values that show reasonable agreement to those obtained with semi-automated or manual methods.22, 23, 24, 25, 26 Such agreement for ECD values was, however, obtained on relatively normal appearing endothelia with high ECD values and did require manual correction of erroneous cell boundaries created by the automated method.24, 25
The main purpose of this study was to evaluate the agreement of three endothelial analysis methods used on endothelial images obtained with the Confoscan 4 scanning-slit confocal microscope after corneal transplantation. The reliability of ECD estimates for relatively normal and for very abnormal-appearing endothelia was also evaluated.
Materials and methods
This cross-sectional study was undertaken as part of routine post-operative assessments of corneal graft patients attending the ophthalmology department at Gartnavel General Hospital in Glasgow. The study adhered to the tenets of the Declaration of Helsinki, and consent from patients was obtained to use data for analysis before confocal microscopy. All patients received a comprehensive ophthalmic examination including best-corrected visual acuity, slit-lamp biomicroscopy and Goldmann tonometry. Medical case records were reviewed retrospectively, and demographic and surgical information retrieved. In total, 40 corneal grafts (20 with DALK and 20 with PK) from 36 subjects were included. Keratoconus was the main indication for transplantation. The mean (±SD) age of the subjects was 43±17 years, the confocal assessments were made at 57±64 months after surgery (range 1–208 months).
Scanning-slit IVCM was undertaken after the main ophthalmic examination using a scanning-slit Confoscan 4 instrument (Nidek Technologies) under topical ocular anaesthesia (Minims Proxymetacaine eye drops 0.5%, Bausch & Lomb, Kingston-upon-Thames, UK). All confocal assessments were made on the central cornea. To ensure similar central corneal locations were assessed, all subjects were asked to fixate at a small light inside the instrument. The full-thickness mode and the ‘auto-landing’ function were selected; the × 40 objective lens was used without the Z-ring. The clearest endothelial image frame was selected for analysis with the frame size being approximately 0.460 × 0.345 mm, from which a region of interest (ROI) was selected for all subsequent analyses. Efforts were made to include as large a number of cells as possible in the ROI. The mean size of the ROI was 0.05 mm2 (range 0.03–0.09 mm2). A scale marker (50 μm) was placed on each of the image frames using the NAVIS line drawing tool and used for calibration and standardisation of distances on all the images. Cell analysis was first performed using the automated option on the NAVIS software without manual cell border correction, which is part of the Confoscan 4 package (version 3.6.6, Nidek Technologies). Following the automated analysis, a manual cell count was carried out, a technique similar to the ‘frame method’ described in a recent review by McCarey et al,27 also available on the NAVIS system. This analysis was performed including cells overlapping the upper and left border of the ROI. Cells overlapping the lower and right border were not included.
The NAVIS automated software requires that the operator select this option from the menu and its output includes the number of cells identified and the mean cell area from which a cell density value is calculated. The NAVIS manual cell count method requires the operator to select cells; this is done by placing a dot on each cell within the pre-defined ROI using a mouse-based cursor. These dots are visible on the screen and, as each cell is selected, the number of cells is progressively counted along with the mean cell area from the set of cells analysed.
Following the automated and the manual counting, planimetry was carried out and used as the reference method. The planimetry technique involves printing of the endothelial image file following careful visual inspection and manual outlining of the cell-cell borders within the ROI with a pen.28 The area of each of these cells was then measured using a Digipro digitiser pad in stream mode (Elestree Computing Ltd, London, UK), with calibration checked for each image, both against its own internal system and against the scale marker on the printed image.28 Planimetry allows measurements to be made to a precision of ±1%.13 From the set of digitised cells, the mean cell area was calculated and this used to calculate the ECD.
Statistical analysis
Descriptive statistics were generated. The data were compared with a non-parametric statistical test (Wilcoxon signed-rank test), as most data sets were different from a normal distribution, with statistical significance set at P<0.05. Differences in ECD among the three methods were analysed and the agreement between the methods assessed by calculating limits of agreement (LoA, mean difference±1.96 SD) and by plotting differences between measurement techniques against the mean of two respective measurements.29 Linear regression analysis was used to determine any predictable association between the means and the differences.
Results
ECD for 40 grafts
Figure 1 gives an example of a graft with a relatively high ECD (upper panel) and of a graft with a relatively low ECD (lower panel). For the 40 graft endothelia analysed, the manual counting method provided the lowest cell density, followed by planimetry and the automated method, which gave the highest values (Figure 2). Descriptive statistics including the mean, SD and the range of the main outcome measures, as well as the mean number of cells counted with each analysis method are shown in Table 1. The mean differences between the methods including the LoA are provided in Table 2. These differences were statistically significant (P<0.001).
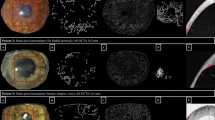
(A) Representative set of IVCM images for a corneal graft with a relatively high cell count. (a) Confocal image showing cells with cell-cell borders marked for planimetry. (b) Image illustrating the NAVIS automated cell count analysis for the same cells. Scale bar=50 μm. (B) Representative set of IVCM images for a corneal graft with a relatively low cell count. (a) Confocal image showing cells with cell-cell borders marked for planimetry. (b) Image illustrating the NAVIS automated cell count analysis for the same cells. Scale bar=50 μm.
Differences between the manual cell count and planimetry
The manual counting method provided lower ECD estimates than planimetry, that is, it effectively underestimated the ECD. The mean (±SD) difference was −405±303 cells/mm2. The difference between manual counting method and planimetry appears to be a systematic effect in that, across the 40 grafts, the ECD from the planimetry was between 48 and 1311 cells/mm2 higher. Method comparison (Bland–Altman) analysis of the differences vs the mean value of the two methods indicate LoA of −1000 and +202 cells/mm2. For lower average ECD values, the difference between the two methods appeared less variable (Figure 3), whereas for higher mean values, the difference between the two methods was slightly greater and also more variable. Linear regression indicated a predictable bias with the difference declining by 189 cells/mm2 per 1000 cells/mm2 change in the mean for the two methods (P=0.003, r=−0.457).
Agreement between the NAVIS manual cell count method and planimetry for 40 corneal grafts. The scatterplot shows the difference vs the mean of the two methods (Bland–Altman analysis). Reference lines are provided for the mean difference (−405 cells/mm2) and at ±1.96 × SD of the difference, which represent the 95% LoA.
However, this effect was no longer evident if DALK and PK grafts were analysed separately (not shown). DALKs had generally higher cell counts with a mean ECD of 1689±519 cells/mm2 (manual cell count) and 2215±610 cells/mm2 (planimetry). The mean difference between these two methods for the DALKs was −525±361 cells/mm2. There was no measurable bias in relation to the mean ECD (linear regression, P>0.05). For the 20 PKs, the group mean ECD was 736±438 cells/mm2 (manual cell count) and 1019±482 cells/mm2 (planimetry) and the mean difference was −284±167 cells/mm2. Again, the difference was independent from the mean ECD (P>0.05).
Differences between the automated method and planimetry
The automated method generated higher ECDs than the planimetry method for 93% of grafts. For all 40 grafts, the mean difference was 661±666 cells/mm2 (Table 2). The LoA were −671 and +1992 cells/mm2. Method comparison (Bland–Altman) analysis of the difference illustrates the poor agreement between the automated method and planimetry (Figure 4). The differences between automated and planimetry counts were greater for grafts with relatively low cell densities and smaller for grafts with a relatively high ECD. This bias was statistically significant and linear regression analysis indicated that the difference between the methods would decline by 541 cells/mm2 for each 1000 cells/mm2 change in the mean ECD for the two methods (P=0.001, r=−0.486).
Agreement between the NAVIS automated method and planimetry for 40 corneal grafts. The scatterplot shows the differences vs the mean of the two methods (Bland–Altman analysis). The output of the automated analysis was not manually corrected. Reference lines are provided for the mean difference (661 cells/mm2) and at ±1.96 × SD of the difference, which represent the 95% LoA.
The mean automated count was 2447±572 cells/mm2 (DALK) and 2108±419 cells/mm2 (PK). Comparison of the automated method and planimetry for the 20 DALKs yielded a smaller mean difference of 232±263 cells/mm2. There was no magnitude-dependent bias across the range of cell density values seen for the DALKs (linear regression, P>0.05). However, the difference for the PKs was larger at 1089±674 cells/mm2, but again the difference was independent from the magnitude of the measurements (P>0.05).
Discussion
In this study, confocal microscopy was used to assess the endothelium after keratoplasty. The output from two different software options for endothelial cell counts was found to be substantially different, with both not being in good agreement with the planimetry technique. For the particular set of images analysed, the automated option of the NAVIS software yields a modest to very substantial overestimation of the graft ECD. The main reason appears to be oversegmenting of endothelial cells by the software. The NAVIS manual counting method provided lower ECD estimates than planimetry, effectively underestimating the cell density. These errors were substantially larger than those reported in previous studies using this confocal microscope. However, these previous studies assessed mostly relatively normal endothelia11, 22, 23, 24, 25, 26, 30 and the automated method was compared with various other methods.
Planimetry is an established technique and was used as the reference method against which the automated and the manual cell count were compared. Planimetry has previously been undertaken on both regular and irregular cell mosaic sets.13, 28, 31
The automated and the manual cell count methods of the NAVIS system are easy to operate and potentially time-saving analysis options. Earlier studies assessing the software have provided conflicting data. Using a manual cell count, ECD estimates on normal endothelia have been reported to be both lower23 and higher24 than those from applying the automated analysis software. Manual editing of the automated Confoscan 3 output was reported to give marginally higher ECD values than a reference method in normal subjects and a slightly better agreement than the automated option.26 In another study on normal corneas, the manual mode of a Confoscan instrument was reported to give slightly higher ECD values than a count from specular microscope images,19 but the authors also note that in cases with Fuchs endothelial dystrophy, the ECD values ‘differed markedly’ (when comparing these two methods). In a very recent study,11 a centre method was compared with two other options, but the Confoscan-acquired images were analysed by another image analysis programme by exporting the images from the confocal microscope. For these comparisons, similar repeatability was reported for 10 normal corneas as well as for 10 graft endothelia. For the 10 grafts studied there was, however, no statistically significant difference in ECD among the three methods used. The ECD differences reported in these various studies11, 19, 22, 23, 24, 25, 26, 30 are notably smaller than the substantial discrepancies found in the present study on corneal grafts.
The mean ROI used in this study was 0.054 mm2 and this is similar to the size of the ROI used in a previous study.32 The minimum ROI in our study was similar in size to the one chosen by the investigators of the aforementioned study (0.03 mm2) for the analysis of images with cell borders that were difficult to visualise. In our study, once the ROI had been selected it was kept constant while each of the three analysis methods were used.
One question that arises when discussing possible reasons for the noted discrepancies in ECD is whether the same cells were analysed with every method. Each set of images had the same scale marker and was cross-checked for calibration. The same ROI was used and every effort was made to see that the number of cells manually outlined for the planimetry matched those marked during the manual cell counting routine. This does not mean, however, that exactly the same cell domains were used in every case and, indeed, the data obtained strongly indicate that these could be rather different between the planimetry and the manual cell count and different again from the automated option. As discussed elsewhere,33 automated and semi-automated image analyses systems differ in the way cell-cell borders are both identified and displayed. The NAVIS software provides the operator with the borders and thus the individual cell domains if the automated technique is used, but not if the manual cell counting method is used. For sets of uniform or slightly irregular cells, slight differences in cell counts would not be expected to result in such a marked discrepancy in ECD, but ECD estimates are clearly more affected in endothelia with very irregular cell mosaics.
Small differences in the manual cell count can arise because of the ambiguity in user instructions ‘…regarding the cells cut by the ROI borders…’, that is, it has yet to be established what constitutes ‘cut’ by the borders (NAVIS Manual). However, the overall effect is small in comparison with errors in the automated counting method (unpublished data).
The number of cells analysed per image was not constant throughout the study. Using the manual cell counting method, the mean (±SD) number of cells counted was 59±31 cells. Slightly fewer cells were counted and analysed during planimetry with a mean of 54±24 cells. Overall, the inter-subject variability in the number of cells that were analysed was unavoidable, as some endothelia had substantially enlarged cell areas and, therefore, fewer cells were available for analysis. In one case, only 14 very large endothelial cells could be counted, despite a relatively large (above average) ROI of 0.064 mm2. For another subject, 10 times as many cells (142) were counted within a ROI of nearly identical size of 0.062 mm2. Assessments of the variability (unpublished data) have indicated that if the ROI contained 40 cells, any error could be expected to be close to ±1%, whereas this increased to around ±4% if 30 cells were included. This is substantially less than the discrepancies noted in the ECD estimates. Owing to the oversegmentation, the mean number of cells as detected by the automated method was substantially higher at 96±29 cells.
An option available on the NAVIS software is manual correction of any inaccurately placed cell borders when using the automated method. Provided there are not too many errors, it should be fairly easy and not too time consuming to correct these cell border errors.11, 24, 25, 34 However, for the majority of grafts with non-uniform and large cells, this requires considerable time and is therefore largely impractical in a busy clinic. The automated option on the NAVIS software is substantially oversegmenting cell domains, thereby creating additional cell-cell borders at numerous locations across the ROI that are simply not present, resulting in an artificially high ECD. This artefact does not seem to be caused by abnormal reflections of material on the endothelial surface (Figure 1) or by individual hyperreflective nuclei.32 The oversegmentation makes the automated method essentially unusable, because it may lead to an overestimation of the health of a graft. Similarly, the automated method should not be used for research purposes either, specifically when comparing the outcome of different keratoplasty techniques. The detailed analyses of the differences between the methods strongly indicate that the chance of errors is much more substantial for any grafts with lower cell counts. Of interest in this context is what the best method is for assessment of ECD in busy clinical settings. As noted previously, the perspective in the present study is that a planimetry-based method should be the ‘gold standard’ and used for morphometry, and this is in line with recent recommendations.35 It is however acknowledged that planimetry may be too time consuming for routine use in clinical settings.
Over- (automated method) or underestimation (manual cell counting method) of ECD can have clinical consequences. For the very grafts for which accurate and reliable ECD estimates are needed, the automated software overestimates the ECD and the results could be misinterpreted as indicating that the post-operative cell density was acceptable. This is important when there is really a precipitous loss of endothelial cells as in the early post-operative period.1, 36 In our clinic, endothelial imaging is commonly undertaken on graft patients undergoing further ocular surgery (eg, late cataract extraction). Patients with normal corneal grafts, but with a cell count that is already lower than in normal eyes may have an increased risk for corneal decompensation due to the potential endothelial damage inflicted by cataract surgery.37, 38 Counselling on the basis of the endothelial examination is likely to influence a patient's decision whether or not to consent to cataract surgery and is therefore of importance to the surgeon. Underestimation of the ECD by the manual counting method might prompt concern even if the graft appeared clear, whereas an overestimation using the fully automated method might lead to a decision that the particular graft was not at risk. A further consequence of underestimating ECD by the manual counting method is that a poorer graft outcome is assumed than has really occurred.
In addition to the methods described in this study, other methods that could be used to assess the ECD in corneal endothelia include exporting images to either a standard computer or a tablet-style processing device and then using compatible software programmes to trace cell borders and to quantify the cell density.
Conclusions
The NAVIS automated method on the Confoscan is likely to grossly overestimate endothelial cell counts in corneal grafts due to substantial oversegmenting of endothelial cells, and should not routinely be used in clinic or research settings. Both, the NAVIS manual cell count and the planimetry method provides lower cell counts, and the three methods described in this paper should not be used interchangeably. Clinicians need to be cautious when extrapolating results from assessment of reliability of endothelial morphometry on normal endothelia to post-keratoplasty endothelia.

References
Reinhart WJ, Musch DC, Jacobs DS, Lee WB, Kaufman SC, Shtein RM . Deep anterior lamellar keratoplasty as an alternative to penetrating keratoplasty: a report by the American Academy of Ophthalmology. Ophthalmology 2011; 118 (1): 209–218.
Bourne WM . Cellular changes in transplanted human corneas. Cornea 2001; 20 (6): 560–569.
Inoue K, Kimura C, Amano S, Oshika T, Tsuru T . Corneal endothelial cell changes twenty years after penetrating keratoplasty. Jpn J Ophthalmol 2002; 46 (2): 189–192.
Patel SV, Hodge DO, Bourne WM . Corneal endothelium and postoperative outcomes 15 years after penetrating keratoplasty. Am J Ophthalmol 2005; 139 (2): 311–319.
Cornea-Donor-Study-Investigator-Group. Donor age and corneal endothelial cell loss 5 years after successful corneal transplantation. Ophthalmology 2008; 115: 627–632.
Borderie VM, Boelle PY, Touzeau O, Allouch C, Boutboul S, Laroche L . Predicted long-term outcome of corneal transplantation. Ophthalmology 2009; 116 (12): 2354–2360.
Jensen LB, Hjortdal J, Ehlers N . Long term follow-up of penetrating keratoplasty for keratoconus. Acta Ophthalmol 2010; 88: 347–351.
Lass JH, Sugar A, Benetz BA, Beck RW, Dontchev M, Gal RL et al. Endothelial cell density to predict endothelial graft failure after penetrating keratoplasty. Arch Ophthalmol 2010; 128 (1): 63–69.
Niederer RL, Perumal D, Sherwin T, McGhee CN . Corneal innervation and cellular changes after corneal transplantation: an in vivo confocal microscopy study. Invest Ophthalmol Vis Sci 2007; 48 (2): 621–626.
Ramamurthi S, Cornish KS, Steeples L, Ramaesh K . Deep anterior lamellar keratoplasty on a previously failed full-thickness graft. Cornea 2009; 28 (4): 456–457.
Patel SV, McLaren JW, Bachman LA, Bourne WM . Comparison of Flex-center, center, and corner methods of corneal endothelial cell analysis. Cornea 2010; 29 (9): 1042–1047.
Sarnicola V, Toro P, Gentile D, Hannush SB . Descemetic DALK and predescemetic DALK: outcomes in 236 cases of keratoconus. Cornea 2010; 29: 53–59.
Doughty MJ, Muller A, Zaman ML . Assessment of the reliability of human corneal endothelial cell-density estimates using a noncontact specular microscope. Cornea 2000; 19 (2): 148–158.
Prinz A, Varga J, Findl O . Reliability of a video-based noncontact specular microscope for assessing the corneal endothelium. Cornea 2007; 26 (8): 924–929.
Thuret G, Deb-Joardar N, Zhao M, Gain P, Gavet Y, Nguyen F . Agreement between two non-contact specular microscopes: Topcon SP2000P versus Rhine-Tec. Br J Ophthalmol 2007; 91 (7): 979–980.
Doughty MJ . Could the coefficient of variation (COV) of the corneal endothelium be overestimated when a centre-dot method is used? Clin Exp Optom 2008; 91 (1): 103–110.
Doughty MJ, Oblak E . A comparison of two methods for estimating polymegethism in cell areas of the human corneal endothelium. Ophthalmic Physiol Opt 2008; 28 (1): 47–56.
Patel S, McLaren J, Hodge D, Bourne W . Normal human keratocyte density and corneal thickness measurement by using confocal microscopy in vivo. Invest Ophthalmol Vis Sci 2001; 42 (2): 333–339.
Hara M, Morishige N, Chikama T, Nishida T . Comparison of confocal biomicroscopy and noncontact specular microscopy for evaluation of the corneal endothelium. Cornea 2003; 22 (6): 512–515.
Weed KH, MacEwen CJ, Cox A, McGhee CN . Quantitative analysis of corneal microstructure in keratoconus utilising in vivo confocal microscopy. Eye (Lond) 2007; 21 (5): 614–623.
Hillenaar T, Weenen C, Wubbels RJ, Remeijer L . Endothelial involvement in herpes simplex virus keratitis: an in vivo confocal microscopy study. Ophthalmology 2009; 116 (11): 2077–2086.e1-2.
Imre L . Elsó hazai tapasztalok konfokális corneamikroszkópiával. Szemeszet 1999; 136: 97–102.
Imre L, Nagymihaly A . Reliability and reproducibility of corneal endothelial image analysis by in vivo confocal microscopy. Graefes Arch Clin Exp Ophthalmol 2001; 239 (5): 356–360.
Klais CM, Buhren J, Kohnen T . Comparison of endothelial cell count using confocal and contact specular microscopy. Ophthalmologica 2003; 217 (2): 99–103.
Kitzmann AS, Winter EJ, Nau CB, McLaren JW, Hodge DO, Bourne WM . Comparison of corneal endothelial cell images from a noncontact specular microscope and a scanning confocal microscope. Cornea 2005; 24 (8): 980–984.
Sheng H, Parker EJ, Bullimore MA . An evaluation of the ConfoScan3 for corneal endothelial morphology analysis. Optom Vis Sci 2007; 84 (9): 888–895.
McCarey BE, Edelhauser HF, Lynn MJ . Review of corneal endothelial specular microscopy for FDA clinical trials of refractive procedures, surgical devices, and new intraocular drugs and solutions. Cornea 2008; 27 (1): 1–16.
Doughty MJ, Aakre BM . Further analysis of assessments of the coefficient of variation of corneal endothelial cell areas from specular microscopic images. Clin Exp Optom 2008; 91 (5): 438–446.
Bland JM, Altman DG . Measuring agreement in method comparison studies. Stat Methods Med Res 1999; 8 (2): 135–160.
Popper M, Morgado AM, Quadrado MJ, Van Best JA . Corneal cell density measurement in vivo by scanning slit confocal microscopy: method and validation. Ophthalmic Res 2004; 36 (5): 270–276.
Doughty MJ . Concerning the symmetry of the ‘hexagonal’ cells of the corneal endothelium. Exp Eye Res 1992; 55 (1): 145–154.
Patel DV, Phua YS, McGhee CN . Clinical and microstructural analysis of patients with hyper-reflective corneal endothelial nuclei imaged by in vivo confocal microscopy. Exp Eye Res 2006; 82 (4): 682–687.
Oblak E, Doughty MJ, Oblak L . A semi-automated assessment of cell size and shape in monolayers, with optional adjustment for the cell-cell border width-application to human corneal endothelium. Tissue Cell 2002; 34 (4): 283–295.
Patel HY, Patel DV, McGhee CN . Identifying relationships between tomography-derived corneal thickness, curvature, and diameter and in vivo confocal microscopic assessment of the endothelium in healthy corneas of young adults. Eye (Lond) 2009; 23 (2): 270–278.
Hirneiss C, Schumann RG, Gruterich M, Welge-Luessen UC, Kampik A, Neubauer AS . Endothelial cell density in donor corneas: a comparison of automatic software programs with manual counting. Cornea 2007; 26 (1): 80–83.
Jones MN, Armitage WJ, Ayliffe W, Larkin DF, Kaye SB . Penetrating and deep anterior lamellar keratoplasty for keratoconus: a comparison of graft outcomes in the United Kingdom. Invest Ophthalmol Vis Sci 2009; 50 (12): 5625–5629.
Edelhauser HF . The resiliency of the corneal endothelium to refractive and intraocular surgery. Cornea 2000; 19 (3): 263–273.
Kim EC, Kim MS . A comparison of endothelial cell loss after phacoemulsification in penetrating keratoplasty patients and normal patients. Cornea 2010; 29 (5): 510–515.
Acknowledgements
This clinical study was supported, in part, by a grant from The Royal College of Surgeons, Edinburgh (KR, MJD, SJ).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Presentation: Part of this work was presented at the World Ophthalmology Congress 2010, Berlin, Germany.
Rights and permissions
About this article
Cite this article
Jonuscheit, S., Doughty, M. & Ramaesh, K. In vivo confocal microscopy of the corneal endothelium: comparison of three morphometry methods after corneal transplantation. Eye 25, 1130–1137 (2011). https://doi.org/10.1038/eye.2011.121
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/eye.2011.121
Keywords
This article is cited by
-
Potential applications of artificial intelligence in image analysis in cornea diseases: a review
Eye and Vision (2024)
-
Fully automatic evaluation of the corneal endothelium from in vivo confocal microscopy
BMC Medical Imaging (2015)
-
Objective assessment of changes in nuclear morphology and cell distribution following induction of apoptosis
Diagnostic Pathology (2014)
-
Assessment of central corneal thickness and corneal endothelial morphology using ultrasound pachymetry, non-contact specular microscopy, and Confoscan 4 confocal microscopy
BMC Ophthalmology (2013)
-
Assessment of a variable frame (polygonal) method to estimate corneal endothelial cell counts after corneal transplantation
Eye (2012)