Abstract
Large deletions that are associated with insertions of Alu-derived sequence represent a rare, but potentially unique class of alterations. Whether they form by a one-step mechanism or by a primary insertion step followed by an independent secondary deletion step is not clear. We resolved two disease-associated SPAST deletions, which involve distinct exons by long range PCR. Alu-derived sequence was observed between the breakpoints in both cases. The intronic regions that represent the targets of potentially involved Alu retrotransposition events overlapped. Microsatellite- and SNP-based haplotyping indicated that both deletions originated on one and the same founder allele. Our data suggest that the deletions are best explained by two-step insertion–deletion scenarios for which a single Alu retrotransposition event represents the shared primary step. This Alu then mediated one of the deletions by non-homologous end joining and the other by non-allelic homologous recombination. Our findings thus strongly argue for temporal separation of insertion and deletion in Alu insertion-associated deletions. They also suggest that certain Alu integrations confer a general increase in local genomic instability, and that this explains why they are usually not detected during the probably short time that precedes the rearrangements they mediate.
Similar content being viewed by others
Introduction
Genomic deletions have a wide size distribution. Those involving one or a few bases are thought to be caused by local polymerase slippage, whereas recombination between low copy repeats generates deletions that may span several megabases. Deletions of intermediate size can be caused by a variety of mechanisms, including non-homologous end joining, non-allelic homologous recombination, and DNA replication-associated events such as microhomology-mediated break-induced replication.1
None of the above concepts would readily explain insertion of unrelated, retrotransposon-derived sequence at the site of breakpoint fusion. That such events exist was first observed upon induction of retrotransposition in vitro.2, 3 Comparative analysis of the human and chimp genomes revealed that this kind of copy number alteration is also relevant on an evolutionary scale. The authors termed the phenomenon Alu retrotransposition-mediated deletion (AMD), and suggested that a novel, one-step mutational mechanism underlies AMD.4 Meanwhile, a few disease-associated deletions that harbor Alu-derived sequence insertions have been reported, including in the BRCA-1, CHD7, NF1, PMM2, GLA, and LPL genes.5, 6, 7, 8, 9, 10 Mechanistic interpretations referred to the one-step AMD concept,8 or alternatively suggested a two-step scenario in which an Alu insertion precedes the actual deletion event.10
The present study reports on the characterization of two distinct Alu insertion-associated deletions in the SPAST gene, alterations of which cause hereditary spastic paraplegia type SPG4 (OMIM 604277).11 Haplotyping as well as detailed analysis of the sequences involved was used to trace the probable history of the corresponding alleles, and to unravel the likely mutational mechanisms. Our findings enable a better understanding of the formation and evolutionary role of Alu insertion-associated deletions.
Materials and methods
Patient and control samples
One of the SPAST deletions investigated here in detail involves two internal exons (family N-16), whereas the other involves the four immediate 3′ exons (family D-W). These deletions have been reported as ex10–12del and ex13–16del previously, and had been identified by cDNA analysis and by multiplex ligation-dependent probe amplification, respectively.12, 13 DNA from multiple family members, including affected as well as unaffected individuals, was available for family D-W, whereas only the index case could be analyzed for family N-16. A total of 118 anonymised genomic DNAs from unrelated individuals were obtained from the Department of Human Genetics of Jena University Hospital, and, following approval by the local ethics committee, used to estimate the general frequency of a critical haplotype at the SPAST locus.
Long-range PCR and sequencing
The SPAST genomic sequence was downloaded from the UCSC genome browser (https://genome.ucsc.edu/) with common SNPs (dbSNP 142), repeats, and exons highlighted (exons according to NM_014946.3). The introns presumed to harbor the breakpoints were targeted with sets of primers that avoided SNPs and repeats, and that showed a spacing of 1–2 kb. Candidate deletion-specific products were confirmed, narrowed, and eventually sequenced from both sides, with additional internal primers. The two variants resolved at sequence level by the present study have been submitted to a publicly funded variant database (http://databases.lovd.nl/shared/variants/SPAST; patient IDs 00056433 and 00057909). All primer sequences are available upon request.
Haplotyping
The SPAST genomic sequence ±1 MB was searched for potentially polymorphic di- and trinucleotide repeats, using the microsatellite display option of the USCS genome browser. Intragenic SNPs with minor allele frequencies >0.4 were identified in dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). Microsatellite alleles as amplified by PCR were visualized on a sequencing type polyacrylamide gel (LICOR Biosciences, Lincoln, NE, USA); SNPs were investigated by direct sequencing of purified PCR products. Detailed information regarding the markers finally applied is provided in Supplementary Figure 1.
Results
Two intragenic SPAST deletions that involve distinct exons carry distinct insertions at their sites of fusion
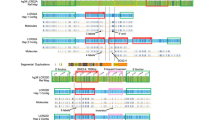
Long-range PCR utilizing appropriate combinations of forward and reverse primers identified sample-specific products that were considered good candidates for harboring the fusion sequences (not shown). Additional primers narrowed the regions of interests to analyzable size (Figure 1a). Direct sequencing revealed that the more 5′ deletion affected 4365 bp (nucleotides 77 798–82 162 in NG_008730.1) and was accompanied by a 15 bp insertion (Figure 1b′). In the more 3′ deletion, 7095 bp were deleted (nucleotides 82 179–89 273 in NG_008730.1) and 255 bp inserted (Figure 1b′′). Conventional interpretation would regard these variants as resulting from distinct, unrelated insertion–deletion events (Figures 1c′–c′′).
Determination of the fusion sequences for two distinct deletions in the SPAST gene. (a) Appropriate primers amplify variant-specific PCR products from samples with an exon 10–12 deletion (index case (IC) from family N-16; middle panel), and from two samples with an exon 13–16 deletion (patients 308 and 316 from family D-W; lower panel). (b ′ , b ′′) Sequencing of the products depicted in a identifies the deletion breakpoints, and reveals the presence of 15 bp (b ′) and 255 bp (b ′′) insertions for the exon 10–12 and the exon 13–16 deletions, respectively (shaded boxes). (c ′ , c ′′) Conventionally, the variants would be interpreted as deriving from independent insertion–deletion events. Note that the 3′ breakpoint of the exon 10–12 deletion (c ′) and the 5′ breakpoint of the exon 13–16 deletion (c ′′) are depicted as distinct, but as mapping in close proximity within an AT-rich region in intron 12 (underlined).
Two-step scenarios that share a primary Alu retrotransposition explain both rearrangements
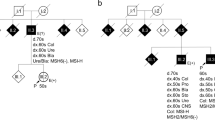
BLAT analysis of the 255 bp insertion revealed numerous hits throughout the human genome; all matches represented the 3′ end of AluYb8 elements. One proper description (of several possible considering the ambiguous origin of the inserted sequence) of this variant would therefore be c.1494-1115_1728+625delinsNC_000006.12:g.117393-117647inv. The 15 bp insertion in c.1246-155_1494-1132delinsCCACCGCGCCCGGCC was too small for BLAT analysis. We noticed, however, that it exactly matches the 5′ end of the general Alu consensus sequence.14 We therefore started to consider more complex events that involve Alu retrotranspositions. The sequences directly neighboring the intact Alu termini suggested overlapping intronic regions as the target for retrotransposition in both cases. The fact that they reside in one and the same AT-rich stretch (Figures 1c′–c′′), together with the general rarity of germline Alu retrotransposition15 made us hypothesize that a single insertion, rather than two independent ones, may be part of an explanation (Figure 2a–a′′). To test this hypothesis we used three highly polymorphic microsatellites to construct haplotypes for the SPAST locus in both families. The availability of many affected as well as unaffected members of family D-W (Figure 2b) allowed unambiguous determination of the haplotype associated with the more 3′ deletion (Figure 2c and d). Strikingly, the same haplotype was also found in the sample that carried the more 5′ deletion (Figure 2c and d), but in none of 118 unrelated controls (data not shown). The idea of both deletions to derive from one and the same founder allele was further supported by investigating a set of local SNPs (not shown). Detailed comparative analysis of all relevant sequences suggested that, as a first step, an AluYb8 element inserted by classical retrotransposition at an L1-endonuclease cleavage site, and thereby created target site duplications 16 bp in size (Figure 2e). The allele harboring the more 5′ deletion would then be derived from non-homologous end joining, involving the novel Alu and unique sequence in an upstream intron (Figure 2e′). The more 3′ deletion, in contrast, would be due to non-allelic homologous recombination between the novel Alu and an AluY in a downstream intron (Figure 2e′′).
Reinterpretation of the two deletions as being derived from one and the same founder allele that carries a non-reference sequence Alu insertion. (a–a ′′) Two-step scenario for formation of the rearrangements. Shared step I: retrotransposition of an AluYb8 into intron 12 (a). Distinct steps II: deletions in which the newly inserted Alu harbors the 3′ breakpoint and gets truncated at its 5′ end (a ′), or harbors the 5′ breakpoint and gets truncated at its 3′ end (a ′′). (b) Affected members of family D-W show a specific band in a PCR that targets the fusion sequence. (c) A PCR that targets a microsatellite near SPAST defines the exon 13–16 deletion-associated repeat allele (arrow). Note that this allele is also present in sample N-016_IC carrying the exon 10–12 deletion (arrow head). (d) Typing of three highly polymorphic microsatellites suggests that both deletions arose on the same haplotype (shaded in gray). (e–e ′′) Sequence level resolution for scenario suggested in a–a ′′. The Alu (italic) inserts at the endonuclease cleavage site 5′-TT/AAAA-3′ (stippled box) as present within the 42 bp AT-rich region (underlined), thereby creating target site duplications (TSDs; underlined bold) (e). The deletion of exons 10–12 is associated with a 1 bp microhomology and only random sequence similarity at the breakpoints (e ′). The deletion of exons 13–16 shows 26 bp microhomology and a generally high-sequence similarity at the breakpoints (e ′′). Note that either the left (e ′) or the right (e ′′) TSD gets removed during the deletion step. Exclamation marks: identity; boxes: microhomology; gray: hypothetical sequence not present in either insertion (derived from the AluYb8 consensus).
Discussion
The present study adds two disease-associated Alu insertion-associated deletions to the few published previously.5, 6, 7, 8, 9, 10 Although we cannot completely rule out the possibility of two independent two-step events, we suggest that both aberrations can be traced back to a shared primary Alu retrotransposition event. This interpretation is based on (i) identity of the region harboring the presumed integration; (ii) involvement of AluYb8 as one of the most active Alu subfamilies;16 (iii) presence of a classical L1-endonuclease site;16 (iv) the sequence representing the potential target site duplication to be of the typical size of 16 bp;16 and (v) the rarity of the haplotype on which both variants reside. The fact that previous genome-wide studies did not identify the corresponding polymorphic Alu insertion17 may be explained by the high genomic instability (mediation of two different deletions!) obviously associated with its presence. The same argument may apply to the lack of alleles that carry only the presumed insertions for other Alu insertion-associated deletions. The absence of such ‘missing links’ has been used to argue for a one-step mutational mechanism,4 whereas our data provide indirect evidence that they can temporarily exist. They would, however, be under strong negative selection due to representing the far end of a spectrum of Alu insertions, with increasing potential to get involved in gene-inactivating rearrangements.
With the availability of MLPA, the fraction of copy number variants in the SPAST gene has been revealed as unusually high, and the gene’s high Alu content has been suggested as an explanation.18 Indeed, the majority of deletions previously resolved at sequence level involve Alus present in the introns, the 3′UTR, and the neighboring intergenic regions.19 The present study reveals that SPAST is still actively targeted by Alu retrotransposition. We show that an AluYb8, which was newly inserted into the intronic sequence, would have been involved in the two distinct deletions. On the basis of the degree of homology for the predicted breakpoints, one deletion would have occurred by non-homologous end joining, whereas the other would resemble the majority of previously resolved rearrangements by being based on non-allelic homologous recombination between corresponding parts of Alus.19
In summary, our findings add to the spectrum of mutational mechanisms responsible for SPAST deletions, confirm the pivotal role played by Alus, and reveal that Alu insertion-associated deletions may form by two temporally separated mutational steps.
References
Hastings PJ, Lupski JR, Rosenberg SM, Ira G : Mechanisms of change in gene copy number. Nat Rev Genet 2009; 10: 551–564.
Gilbert N, Lutz-Prigge S, Moran JV : Genomic deletions created upon LINE-1 retrotransposition. Cell 2002; 110: 315–325.
Symer DE, Connelly C, Szak ST et al: Human l1 retrotransposition is associated with genetic instability in vivo. Cell 2002; 110: 327–338.
Callinan PA, Wang J, Herke SW, Garber RK, Liang P, Batzer MA : Alu retrotransposition-mediated deletion. J Mol Biol 2005; 348: 791–800.
Okubo M, Horinishi A, Saito M et al: A novel complex deletion-insertion mutation mediated by Alu repetitive elements leads to lipoprotein lipase deficiency. Mol Gen Metabol 2007; 92: 229–233.
Udaka T, Okamoto N, Aramaki M et al: An Alu retrotransposition-mediated deletion of CHD7 in a patient with CHARGE syndrome. Amer J Med Gen A 2007; 143A: 721–726.
Schollen E, Keldermans L, Foulquier F et al: Characterization of two unusual truncating PMM2 mutations in two CDG-Ia patients. Mol Gen Metabol 2007; 90: 408–413.
Dobrovolny R, Nazarenko I, Kim J, Doheny D, Desnick RJ : Detection of large gene rearrangements in X-linked genes by dosage analysis: identification of novel alpha-galactosidase A (GLA) deletions causing Fabry disease. Hum Mut 2011; 32: 688–695.
Peixoto A, Pinheiro M, Massena L et al: Genomic characterization of two large Alu-mediated rearrangements of the BRCA1 gene. J Hum Genet 2013; 58: 78–83.
Hsiao MC, Piotrowski A, Callens T et al: Decoding NF1 Intragenic Copy-Number Variations. AJHG 2015; 97: 238–249.
Hazan J, Fonknechten N, Mavel D et al: Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet 1999; 23: 296–303.
Sauter S, Miterski B, Klimpe S et al: Mutation analysis of the spastin gene (SPG4) in patients in Germany with autosomal dominant hereditary spastic paraplegia. Hum Mut 2002; 20: 127–132.
Erichsen AK, Inderhaug E, Mattingsdal M, Eiklid K, Tallaksen CM : Seven novel mutations and four exon deletions in a collection of Norwegian patients with SPG4 hereditary spastic paraplegia. Eur J Neurol 2007; 14: 809–814.
Batzer MA, Deininger PL : Alu repeats and human genomic diversity. Nat Rev Genet 2002; 3: 370–379.
Xing J, Zhang Y, Han K et al: Mobile elements create structural variation: analysis of a complete human genome. Genome Res 2009; 19: 1516–1526.
Stewart C, Kural D, Stromberg MP et al: A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Gen 2011; 7: e1002236.
David M, Mustafa H, Brudno M : Detecting Alu insertions from high-throughput sequencing data. NAR 2013; 41: e169.
Beetz C, Nygren AO, Schickel J et al: High frequency of partial SPAST deletions in autosomal dominant hereditary spastic paraplegia. Neurology 2006; 67: 1926–1930.
Boone PM, Yuan B, Campbell IM et al: The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. AJHG 2014; 95: 143–161.
Acknowledgements
We thank the patients and their families for participating in the study, Kerstin Stein for excellent technical support, and Ingo Kurth (Department of Human Genetics, Jena University Hospital) for providing control DNAs. This study was supported by the Tom-Wahlig-Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Jahic, A., Erichsen, A., Deufel, T. et al. A polymorphic Alu insertion that mediates distinct disease-associated deletions. Eur J Hum Genet 24, 1371–1374 (2016). https://doi.org/10.1038/ejhg.2016.20
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2016.20
This article is cited by
-
Fragile Sites as Drivers of Gene and Genome Evolution
Current Genetic Medicine Reports (2018)