Abstract
Lynch syndrome is an autosomal dominant hereditary cancer syndrome in which many cancers develop, the main one being colorectal cancer. Germline pathogenic variants in one of four mismatch repair (MMR) genes are known to be causative of this disease. Accurate diagnosis using genetic testing can greatly benefit the health of those affected. Recently, owing to the improvement of sequence techniques, complicated variants affecting the functions of MMR genes were discovered. In this study, we analyzed insertions of a retrotransposon-like sequence in exon 5 of the MSH6 gene and exon 3 of the MSH2 gene found in Japanese families suspected of having Lynch syndrome. Both of these insertions induced aberrant splicing, and these variants were successfully identified by mRNA sequencing or visual observation of mapping results, although a standard DNA-seq analysis pipeline failed to detect them. The insertion sequences were ~2.5 kbp in length and were found to have the structure of an SVA retrotransposon (SVA). One SVA sequence was not present in the hg19 or hg38 reference genome, but was in a Japanese-specific reference sequence (JRGv2). Our study illustrates the difficulties of identifying SVA insertions in disease genes, and that the possibility of polymorphic insertions should be considered when analyzing mobile elements.
Similar content being viewed by others
Introduction
Lynch syndrome is an autosomal dominant hereditary cancer syndrome in which colon cancer is particularly prominent. It is caused by a pathogenic variant in one of the mismatch repair genes (MMR genes; MLH1, MSH2, MSH6, or PMS2) [1,2,3,4,5]. In recent years, owing to the advent of next-generation sequencing (NGS) and the progression of analytical techniques, many variants affecting the functions of MMR genes have been revealed [6]. Lynch syndrome accounts for about 3% of all colon cancer cases [7, 8] and its prevalence in the general population has been estimated at 1:440 [9]. Because various cancers besides colon cancer occur in those affected by this syndrome, determination of the responsible variant and appropriate surveillance are important to the healthcare of patients [10].
Retrotransposons are “mobile genetic elements” that move in the genome. Transposon insertion causes a change in the gene at or near the insertion point. Such a change is considered to provide room for the evolution of the genome [11]. Retrotransposons occupy ~40% of the human genome sequence and are classified into two groups: long terminal repeat (LTR) and non-LTR. LTR retrotransposons do not have transposable element activity, while non-LTR retrotransposons, including LINE-1 (constitute 16.9% of the human genome), Alu (10.6%), and SVA (0.2%) elements, have such activity; this latter group covers approximately one-third of the human genome and causes various hereditary diseases via its insertion [12]. SVA retrotransposons, hominid-specific retrotransposons, consist of SINE (short interspersed repetitive elements), VNTR (variable number of tandem repeats), and Alu, and are ~3 kb in length. This type of retrotransposon is rare, with ~2700 copies of it in the human genome [13]. Regarding its relationship with disease, it was reported that a cause of Fukuyama-type congenital muscular dystrophy was splice abnormality by the insertion of an SVA-type retrotransposon [14]. With regard to Lynch syndrome, an Alu insertion variant in MSH2 was reported [15, 16], while a report of a single case of SVA insertion in PMS2 was also published [17].
In this study, insertion of an SVA-type retrotransposon was found in exonic regions of the MSH2 or MSH6 gene. One of the inserted sequences aligned only with JRGv2 (Japanese Reference Genome V2) provided by the Tohoku Medical Megabank but not with standard human reference genomes. RNA sequencing revealed an aberrant alternative splicing event associated with these variants.
Materials and methods
Patients
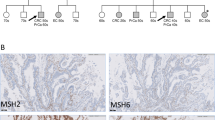
Patients enrolled in Study for the Establishment of Effective Screening and Diagnosis of Lynch Syndrome (Dial study) were analyzed. Case 1 (Fig. 1a) had never been diagnosed with cancer. However, his father had developed three metachronous Lynch syndrome-associated cancers (colorectal, ureteral, and bladder cancers). Microsatellite instability (MSI) and immunohistochemical testing revealed that the ureteral and bladder cancers showed microsatellite instability-high (MSI-H) and loss of MSH6. Case 2 (Fig. 1b) was diagnosed with metachronous multiple cancers and his older brother showed a similar phenotype. In both siblings, the colorectal cancer was MSI-H, and loss of MSH2/MSH6 was observed (Fig. 1b, III.2, 3). All procedures were performed in accordance with the ethical standards of the responsible committee on human experimentation and with the 1964 Helsinki Declaration, as revised in 2013, as well as the Japanese ethical guidelines for human genome/gene analysis research. This study was approved by the Institutional Review Boards of Saitama Cancer Center (no. 729). Written consent was obtained from the patient before inclusion in the study.
a Family of Case 1. Patient III.2 was Case 1 (arrow). b Family of Case 2. Patient III.3 was Case 2 (arrow) (d died, dx diagnosed at, P proband, E evaluation, Bla bladder, CNS central nervous system, Col colorectum, Eso esophagus, Ile ileocecum, Pro prostate, Sto stomach, Ure ureter, Ute uterus, each organ name indicates the primary site of cancer). No personal information (including the type of cancer) was shown for family members who had not undergone any kind of molecular testing, except for a history of cancer (indicated black square). The information for the generation number Ι in both families was unknown.
DNA and RNA extraction from peripheral blood mononuclear cells (PBMCs)
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood collected in heparinized vacutainer tubes using Ficoll®-Paque Premium (GE Healthcare, Chicago, IL, USA). These cells were resuspended in KBM502 (KOHJIN BIO, Sakado, Japan) supplemented with 10% FBS (GE Healthcare) and penicillin–streptomycin (FUJIFILM Wako Pure Chemical, Osaka, Japan), and plated and cultured in tissue culture tubes (TPP, Trasadingen, Switzerland) at 37 °C in a 5% CO2 humidified atmosphere. After 1 week, cells were divided into culture tubes for DNA extraction and RNA extraction with or without puromycin (Thermo Fisher Scientific, Waltham, MA, USA) treatment, as reported previously [18]. DNA and RNA (with/without puromycin treatment) were extracted using AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany), in accordance with the manufacturer’s instructions.
Next-generation sequencing analysis for genetic testing
Genetic testing for LS was conducted with both DNA and RNA. DNA was sequenced using QIAseq Targeted DNA Custom Panel (Qiagen) including MMR genes (MLH1, MSH2, MSH6, PMS2, and EPCAM), in accordance with the manufacturer’s instructions. Transcripts of MMR genes were amplified by PCR with cDNA synthesized from RNA and sequenced using Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, USA), in accordance with the manufacturer’s instructions with slight modifications. [The library amplification was carried out using KAPAHiFi DNA Polymerase (Kapa Biosystems, Wilmington, DE, USA), not NPM.] Sequencing was performed on Miseq (Illumina). The sequence reads were analyzed with CLC Genomics Workbench (Qiagen, RRID: SCR_011853) using hg19 as a reference. The Japanese reference genome was obtained from JMORP (https://jmorp.megabank.tohoku.ac.jp/, [19]). The accession numbers for the MSH2 and MSH6 genes (transcripts) were NG_007110.2 (LRG218t1) and NG_007111.1 (LRG219t1), respectively. Exons are numbered according to the accession number of LRG.
To confirm the inserted sequences, amplified PCR products were sequenced using the same method as in the RNA sequencing. Because there was difficulty amplifying the insertion sequence of Case 2 using standard PCR enzyme, we used PrimeSTAR GXL DNA Polymerase (Takara Bio, Kusatsu, Japan). The list of primers designed for the amplification of MMR transcripts and confirmation of the inserted sequences is given in Table 1. Visualization of mapping results was performed using Integrative Genomics Viewer software (http://software.broadinstitute.org/software/igv/, RRID: SCR_011793). Sequence data resulted from this study is already submitted to the DNA DataBank of Japan (DDBJ) repository, accession DRA009831 and DRA009891.
Results
Finding of insertion in exonic regions of MSH6
Multigene panel testing on the DNA sample of Case 1 using our standard sequencing pipeline detected neither a pathogenic single-nucleotide variant nor copy number variation. However, sequencing of the MSH6 transcript revealed a deletion of the 5′ region of exon 5 in 27% of the reads of transcripts isolated from puromycin treated PBMCs (23% in untreated cells) (Fig. 2a), suggesting that aberrant splicing was induced by the use of a cryptic 3′ splice site. Via careful evaluation of the mapping of the DNA sequence around this region, we found a soft-clipped sequence in the middle of exon 5 consisting of repetitive “GGGAGA” units and accounted for 304 out of a total of 749 reads covering this site (Fig. S2a). These results suggested the presence of a larger inserted sequence with the sequence at the 3′ end although the 5′ end of the insertion had not been detected. According to this assumption, we attempted to amplify the inserted DNA fragment using a forward primer (MSH6 F) located in exon 4 together with an insertion-specific reversed primer (MSH6 R) (Table 1, Fig. 3a). Since the wild-type sequence between this primer pair is 2.6 kb, amplification of an ~5 kb PCR fragment revealed that the inserted sequence is ~2.4 kb (Fig. 3c).
a RNA mapping result of Case 1. Colored area shows sequence coverage (blue, Case 1; orange, control). Arrow indicates low-coverage area compared with control. b RNA mapping result of Case 2. Blue area is Case 2, and orange is control. No significant change was observed in sequence coverage. In exon 3, low-frequency deletion was detected (magnified in box). Both cases of RNA was derived from puromycin treated cells.
a Schematic illustration of the primer design for Case 1. Forward primer was designed in exon 4 (F) and the reverse primer was in a border region with a soft-clipped sequence (R). In the reference genome, the distance between the two primers was 2572 bp. b Schematic illustration of the primer design for Case 2. In this case, two forward primers were designed in exon 3 (F1) and in the 5′ soft-clipped sequence manually defined from a fastq file (F2). And two reverse primers were also designed in exon 3 (R1) and in the border region with a 3′ soft-clipped sequence (R2). c Agarose gel electrophoresis of PCR products. In Case 1, specific amplification was observed compared with control. d Electrophoretic image of Case 2. Here, 1 and 2 represent two-step and three-step PCR, respectively. Using F1/R1 primer with the three-step condition and F2/R2 with the two-step condition, good amplification occurred.
Insertion of SVA-type retrotransposon causes aberrant splicing
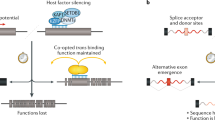
To examine the inserted fragment, we sequenced the whole 5 kbp amplicon as described in “Materials and methods” section. The results revealed the presence of an insertion sequence in exon 5 with a target site duplication starting at a poly-T tract (Fig. S1a). Together with the “GGAGA” repeats at the 3′ end, this suggested that the insertion resulted from retrotransposition of an SVA element into exon 5 (Fig. S1b). The characteristics of this sequence are reminiscent of an SVA-type retrotransposon (SVA). However, our standard mapping method failed to pinpoint the position in the reference genome, probably because of the repetitive sequence. Using de novo assembly software, we obtained the whole sequence of the insert. By re-mapping on the reference genome, the sequence starting from poly-T turned out to be unique to chr12: 96233959–96236309 and this region was annotated as SVA E by Repeat Masker (http://www.repeatmasker.org/). This variant was considered to be represented as NC_000002.11: g.48030698_48030699ins[SVA;48030684_48030698]. In addition, detailed RNA analysis revealed the deletion of the initial 174 bp sequence of exon 5, which caused an in-frame deletion of the MSH6 protein. Taking these findings together, we concluded that the insertion of a ~2.4 kbp SVA E retrotransposon into exon 5 changes its splicing acceptor site (Fig. 4a, Table 2).
The alternative patterns of splicing caused by the insertions are indicated by red lines. a In Case 1, the upper reaches of MSH6 exon 5 from the inserted part were spliced out. A new splicing acceptor site was created downstream of the insertion sequence. b In Case 2, new splicing donor and acceptor sites were created upstream and downstream from the inserted part. Via these splicing sites, exon 3 of MSH2 was divided into two exons.
SVA insertion may be a more frequent cause of Lynch syndrome than we assumed
As described above, aberrant splicing induced by SVA insertion has been reported for PMS2. Case 1 in this study led us to the assumption that SVA insertion is not a very rare causes of Lynch syndrome. Thus, we performed a thorough investigation of cases in which one of the Lynch syndrome genes was apparently mutated, but the nature of the variant had not been determined. The RNA sequence of MSH2 from Case 2 showed an aberrant transcript lacking 88 bp in the middle of exon 3 (Fig. 2b). Carefully evaluating the read mapping exon 3 revealed soft-clipped sequence similar to Case 1 consisting of repetitive “GGGAGA” (234 out of a total of 406 reads) and poly-T tract (234 out of 394 reads) (Fig. S2b). We assumed that this insertion sequence was also an SVA retrotransposon, but this insertion was not amplified by our standard PCR reactions. Therefore, we employed PrimeSTAR GXL DNA polymerase (Takara Bio) instead of our standard PCR polymerase EX Taq (Takara Bio) to achieve better extension of “difficult to replicate” regions such as AT- or GC-rich regions and attempted both two-step and three-step PCR cycles with two kinds of primer pairs, MSH2 F1/R1 and MSH2 F2/R2 (Table 1, Fig. 3b). Then, the insert was successfully amplified (Fig. 3d) and the product was subjected to NGS. Unfortunately, our de novo assembly program failed to construct a single sequence as in Case 1. Therefore, we manually created contigs from adjacent reads and merged them into a single sequence. However, we could not map the merged sequence on either hg19 or hg38. Only a slightly similar sequence in chromosome 16 of hg19 was present (Fig. S3a). We thus assumed that the sequence is specific to the Japanese. By searching GGGenome (https://gggenome.dbcls.jp/en/), the sequence was revealed to be unique to chr6: 111170450–111172854 of JRGv2, a Japanese-specific reference sequence [19] (Fig. S3b). Coverage analysis using the JRGv2 decoy sequence also suggested that this insertion sequence may be specific to the sequence (Fig. S4). Furthermore, this sequence was thought to be inserted into NC_000006.11:g.111278198_111278199 in hg19 (Fig. S5). This sequence was annotated as SVA F by Repeat Masker. Thus, in Case 2, an SVA F retrotransposon was integrated into exon 3 of MSH2 and created an extra intron in the middle of this exon (Fig. 4b). This variant was considered to be represented as NC_000002.11: g.47637427_47637428ins[SVA;47637413_47637427]. As a result, shorter mRNA causing a frameshift was formed (Table 2).
Discussion
In the genetic testing of patients suspected of having Lynch syndrome in a multi-facility collaborative investigation, we found two novel exonic insertions of SVA in MSH2 and MSH6, and showed the splice effect of these variants. Insertion of SVA has been reported only in an intron of the PMS2 gene [17], so this study is the first to identify it in other MMR genes and in an exonic region of these four genes.
As to the final evaluation of the pathogenicity of the two variants against Lynch syndrome, that in Case 1 is a variant of uncertain significance (VUS; PM2: absent in population data, PM4: protein-length-changing variant) and that in Case 2 is likely pathogenic (PVS1: predicted null variant, PM2: absent in population data), according to ACMG criteria [20]. Regarding Case 1, we tried to determine that the variant was pathogenic or likely pathogenic because it is a long deletion variant of 58 amino acids, but the variants registered in InSiGHT (http://insight-database.org/) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) did not have an in-frame deletion variant that was determined to be pathogenic or likely pathogenic within these 58 amino acids. For the diagnosis of Lynch syndrome, molecular tests, such as MSI testing and immunohistochemistry (IHC), are widely used, but those results are not included in the criteria. In recent years, evaluation methods that include the results of MMR gene variants of a tumor and the results of IHC have been advocated [21, 22].
Our results indicate the difficulty in finding SVA insertions (and possibly any other insertions of DNA sequence larger than a few kilobases) by standard methods of genetic testing. We had problems detecting SVA insertion in both Case 1 and Case 2. In Case 1, our standard DNA analysis failed to detect the abnormality. Only a special tool, such as Scramble (https://github.com/GeneDx/scramble), could detect this change (Table S1). However, we were able to notice the abnormality by analyzing RNA, which prompted us to re-examine the result of DNA sequencing. The DNA change was barely detectable by visual inspection of the mapping results. This supports the idea that RNA analysis is often useful to discover variants that are difficult to detect and evaluate using DNA [23, 24]. However, RNA analysis does not always work well. In Case 2, quite a few reads (6%) supportive of aberrant splicing were detected in the RNA derived from PBMCs with puromycin treatment (no reads in untreated cells). The ratio of aberrant-splicing reads was also similar in a relative (Fig. 1b, III-2), but was not recognized as an aberrant change by our standard computational analysis. Because the loss of MSH2 protein was observed in IHC, we examined the whole region of this gene by manually viewing the mapping and eventually discovered SVA insertion. It is unclear why the ratio of variant transcripts was low, but here are a various possible causes, including technical problems [25, 26]. In view of the difficulty of detection, it is possible that there are a significant number of hidden Lynch syndrome patients who harbor an insertion of SVA (or any fragment extending a few kilobases) in any of the genes causative of this condition. In support of this idea, variants involving the insertion of a mobile element such as Alu and LINE-1 sequences have been reported in genes causative of the hereditary cancer-predisposing syndrome including NF1 and BRCA [27, 28].
Another problem that we encountered is that the insertion sequence of Case 2 did not exist in hg19 or hg38, and existed only in JRGv2, the Japanese reference genome. Active retrotransposons are known to generate polymorphic insertions by themselves [29, 30]. In addition, it has been reported that an Alu sequence moves to another locus once per 20 births [31]. Our data and these studies suggested that the consideration of ethnic and individual differences is important for identifying the origin of inserted mobile elements.
In this study, we detected insertions of SVA in exons of the MSH2 and MSH6 genes. To date, we have identified 137 pathogenic or likely pathogenic variants and 65 variants of uncertain significance (VUS) from 580 probands suspected of Lynch syndrome based on family history and molecular testing. In light of the difficulty in detecting them, the insertion of mobile elements including SVA may not be a rare cause of Lynch syndrome. In addition, our results indicate that RNA analysis helps to increase the possibility of detection, although it is not sufficient to detect all kinds of structural variants. To achieve precise diagnoses of genetic disorders and provide appropriate surveillance/treatment to patients, technological advances to detect currently undetectable (or hardly detectable) variants are awaited.
References
Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–25.
Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–61.
Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80.
Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–9.
Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–2.
Espenschied CR, LaDuca H, Li S, McFarland R, Gau CL, Hampel H, et al. Multigene panel testing provides a new perspective on Lynch syndrome. J Clin Oncol. 2017;35:2568–75.
Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801–18.
Cunningham JM, Kim CY, Christensen ER, Tester DJ, Parc Y, Burgart LJ, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69:780–90.
Chen S, Wang W, Lee S, Nafa K, Lee J, Romans K, et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA. 2006;296:1479–87.
Dominguez-Valentin M, Sampson JR, Seppälä TT, Ten Broeke SW, Plazzer JP, Nakken S, et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database. Genet Med. 2020;22:15–25.
Kazazian HH Jr, Moran JV. Mobile DNA in Health and Disease. N. Engl J Med. 2017;377:361–70.
Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703.
Hancks DC, Kazazian HH Jr. SVA retrotransposons: evolution and genetic instability. Semin Cancer Biol. 2010;20:234–45.
Taniguchi-Ikeda M, Kobayashi K, Kanagawa M, Yu CC, Mori K, Oda T, et al. Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature. 2011;478:127–31.
Kloor M, Sutter C, Wentzensen N, Cremer FW, Buckowitz A, Keller M, et al. A large MSH2 Alu insertion mutation causes HNPCC in a German kindred. Hum Genet. 2004;115:432–8.
Baert-Desurmont S, Coutant S, Charbonnier F, Macquere P, Lecoquierre F, Schwartz M, et al. Optimization of the diagnosis of inherited colorectal cancer using NGS and capture of exonic and intronic sequences of panel genes. Eur J Hum Genet. 2018;26:1597–602.
van der Klift HM, Tops CM, Hes FJ, Devilee P, Wijnen JT. Insertion of an SVA element, a nonautonomous retrotransposon, in PMS2 intron 7 as a novel cause of Lynch syndrome. Hum Mutat. 2012;33:1051–5.
Nomura S, Sugano K, Kashiwabara H, Taniguchi T, Fukayama N, Fujita S, et al. Enhanced detection of deleterious and other germline mutations of hMSH2 and hMLH1 in Japanese hereditary nonpolyposis colorectal cancer kindreds. Biochem Biophys Res Commun. 2000;271:120–9.
Nagasaki M, Kuroki Y, Shibata TF, Katsuoka F, Mimori T, Kawai Y, et al. Construction of JRG (Japanese reference genome) with single-molecule real-time sequencing. Hum Genome Var. 2019;6:27. https://doi.org/10.1038/s41439-019-0057-7.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Gray PN, Tsai P, Chen D, Wu S, Hoo J, Mu W, et al. TumorNext-Lynch-MMR: a comprehensive next generation sequencing assay for the detection of germline and somatic mutations in genes associated with mismatch repair deficiency and Lynch syndrome. Oncotarget. 2018;9:20304–22.
Shirts BH, Konnick EQ, Upham S, Walsh T, Ranola JMO, Jacobson AL, et al. Using somatic mutations from tumors to classify variants in mismatch repair genes. Am J Hum Genet. 2018;103:19–29.
Pagenstecher C, Wehner M, Friedl W, Rahner N, Aretz S, Friedrichs N, et al. Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants. Hum Genet. 2006;119:9–22.
Brandão RD, Mensaert K, López-Perolio I, Tserpelis D, Xenakis M, Lattimore V, et al. Targeted RNA-seq successfully identifies normal and pathogenic splicing events in breast/ovarian cancer susceptibility and Lynch syndrome genes. Int J Cancer. 2019;145:401–14.
Thakur PK, Rawal HC, Obuca M, Kaushik S. Bioinformatics Approaches for Studying Alternative Splicing. In: Ranganathan S, Gribskov M, Nakai K, Schönbach, editors. Encyclopedia of Bioinformatics and Computational Biology (volume 2). Amsterdam, Netherland: Elsevier B.V.; 2019, p. 221–34.
Tajaddod M, Tanzer A, Licht K, Wolfinger MT, Badelt S, Huber F, et al. Transcriptome-wide effects of inverted SINEs on gene expression and their impact on RNA polymerase II activity. Genome Biol. 2016;17:220.
Katharina W, Tom C, Annekatrin W, Ludwine M. The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLos Genet. 2011;7:e1002371.
Qian Y, Mancini-DiNardo D, Judkins T, Cox HC, Brown K, Elias M, et al. Identification of pathogenic retrotransposon insertions in cancer predisposition genes. Cancer Genet. 2017;216–217:159–69.
Wang L, Norris ET, Jordan IK. Human retrotransposon insertion polymorphisms are associated with health and disease via gene regulatory phenotypes. Front Microbiol. 2017;8:1418. https://doi.org/10.3389/fmicb.2017.01418.
Puurand T, Kukuškina V, Pajuste FD, Remm M. AluMine: alignment-free method for the discovery of polymorphic Alu element insertions. Mob DNA. 2019;10:31.
Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–26.
Acknowledgements
We thank Edanz (www.edanzediting.co.jp) for editing the English text of a draft of this manuscript. IM received generous support from M. Kagawa, T. Ito, A. Yamamoto, and N. Kamae for providing clinical information.
Funding
This research was supported by Japan Agency for Medical Research and Development (AMED) under grant JP 18kk0205004, JSPS KAKENHI Grant Number JP18K07339 and National Cancer Center Research and Development Found Grant Number 31-A-2.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yamamoto, G., Miyabe, I., Tanaka, K. et al. SVA retrotransposon insertion in exon of MMR genes results in aberrant RNA splicing and causes Lynch syndrome. Eur J Hum Genet 29, 680–686 (2021). https://doi.org/10.1038/s41431-020-00779-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-020-00779-5
This article is cited by
-
Incidence and molecular characteristics of deficient mismatch repair conditions across nine different tumors and identification of germline variants involved in Lynch-like syndrome
International Journal of Clinical Oncology (2024)
-
Diagnostic and therapeutic challenges of glioblastoma as an initial malignancy of constitutional mismatch repair deficiency (CMMRD): two case reports and a literature review
BMC Medical Genomics (2023)
-
A 39 kb structural variant causing Lynch Syndrome detected by optical genome mapping and nanopore sequencing
European Journal of Human Genetics (2023)
-
Retrotransposon insertion as a novel mutational cause of spinal muscular atrophy
Human Genetics (2023)
-
Splicing analyses for variants in MMR genes: best practice recommendations from the European Mismatch Repair Working Group
European Journal of Human Genetics (2022)