Abstract
The oral-facial-digital (OFD) syndromes comprise a group of related disorders with a combination of oral, facial and digital anomalies. Variants in several ciliary genes have been associated with subtypes of OFD syndrome, yet in most OFD patients the underlying cause remains unknown. We investigated the molecular basis of disease in two brothers with OFD type II, Mohr syndrome, by performing single-nucleotide polymorphism (SNP)-array analysis on the brothers and their healthy parents to identify homozygous regions and candidate genes. Subsequently, we performed whole-exome sequencing (WES) on the family. Using WES, we identified compound heterozygous variants c.[464G>C];[1226G>A] in NIMA (Never in Mitosis Gene A)-Related Kinase 1 (NEK1). The novel variant c.464G>C disturbs normal splicing in an essential region of the kinase domain. The nonsense variant c.1226G>A, p.(Trp409*), results in nonsense-associated alternative splicing, removing the first coiled-coil domain of NEK1. Candidate variants were confirmed with Sanger sequencing and alternative splicing assessed with cDNA analysis. Immunocytochemistry was used to assess cilia number and length. Patient-derived fibroblasts showed severely reduced ciliation compared with control fibroblasts (18.0 vs 48.9%, P<0.0001), but showed no significant difference in cilia length. In conclusion, we identified compound heterozygous deleterious variants in NEK1 in two brothers with Mohr syndrome. Ciliation in patient fibroblasts is drastically reduced, consistent with a ciliary defect pathogenesis. Our results establish NEK1 variants involved in the etiology of a subset of patients with OFD syndrome type II and support the consideration of including (routine) NEK1 analysis in patients suspected of OFD.
Similar content being viewed by others
Introduction
The oral-facial-digital (OFD) syndromes comprise a group of related disorders with a specific combination of oral, facial and digital anomalies. To date, fourteen phenotypically different subtypes have been reported, of which nine have been well characterized.1, 2 These subtypes show an autosomal recessive inheritance pattern, except for OFD type I, type VII and type VIII, which are X-linked forms of OFD.1, 2 Common characteristic findings associated with these syndromes include: hamartomas of the tongue and a lobulated tongue, midline cleft of the upper lip and/or palate, gingival frenulae, dental anomalies, brachydactyly, syndactyly, pre- or post-axial polydactyly and cystic renal disease.1, 2
In 2001, the causative gene for OFD type 1 (OMIM 300170), OFD1, was identified.3 Functional studies have shown that the encoded protein has an important role in the formation of primary cilia.4, 5 Because cilia have an essential role in the morphogenesis and tissue homeostasis of multiple organ systems, gene variants interrupting cilia function manifest systemically and result in a diverse phenotypic picture.6, 7, 8 In the following years, variants in several other genes involved in ciliogenesis or ciliary signaling have been implicated in specific subtypes of OFD, namely TCTN3 for OFD type IV (OMIM 258860), TMEM216 and C5orf42 for OFD VI (OMIM 277170), DDX59 for an unclassified autosomal recessive form of OFD with intellectual disability, TBC1D32/C6orf170 or SCLT1 for OFD IX (OMIM 258865) and C2CD3 for an atypical type of OFD with severe microcephaly and cerebral malformations.9, 10, 11, 12, 13 Variants in two of these genes, TCTN3 and TMEM216, are also found in other ciliopathies such as Joubert syndrome and Meckel-Gruber syndrome.13, 14 These findings indicate that multiple OFD subtypes can be classified as ciliopathies. However, in the majority of patients with an autosomal recessive form of OFD the underlying genetic cause remains to be identified.
In the present study, we performed genetic analysis on two brothers from consanguineous parents who have features compatible with OFD type II (Mohr syndrome, OMIM 252100). As several other OFD subtypes have been shown to be ciliary diseases, we presumed that the brothers would also have a ciliopathy. The extreme genetic heterogeneity of ciliopathies and the large number of genes involved in ciliary pathways, as well as the large potential number of yet unknown ciliary proteins, prompted us to perform whole-exome sequencing (WES) to identify the disease-related gene in the family.15, 16
Subjects and methods
Patient material collection and consent
Blood was obtained from both brothers and their parents after informed consent. Skin fibroblasts of the eldest brother were obtained during a scheduled knee operation. Informed consent was obtained for whole-genome single-nucleotide polymorphism (SNP)-array analysis and WES in the diagnostics laboratory. The medical ethical committee of the University Medical Center Utrecht (UMCU) approved the WES patient information and informed consent form.
SNP-array analysis
Genomic DNA was extracted from peripheral leukocytes according to standard protocols. SNP-array copy number profiling and analysis of regions of homozygosity were performed according to standard procedures using the Infinium Human Omni Express Exome BeadChip (Illumina, San Diego, CA, USA). Subsequently, visualizations of SNP-array results and data analysis were performed using Nexus 7.5 software (BioDiscovery, Los Angeles, CA, USA). Human genome build Feb. 2009 GRCh37/hg19 was used. Results were classified with Cartagenia BENCH software (Cartagenia, Leuven, Belgium).
Whole-exome sequencing
Fragment libraries of the DNA samples of the two brothers and their parents were prepared (Kapa Biosystems, Wilmington, MA, USA), enriched using Agilent Sureselect V5 (Agilent, Santa Clara, CA, USA) and sequenced as a Rapid Run on the Illumina HiSeq 2500 Sequencer (Illumina, San Diego, CA, USA) at the Utrecht Sequencing Facility (Utrecht, The Netherlands). A phased vcf was created and imported into Cartagenia BENCHlab NGS module (Cartagenia) for variant interpretation. Due to the consanguineous nature of the parents, homozygous variants were initially investigated, and subsequently compound heterozygote and X chromosome variants were also examined (Supplementary Methods S1). Candidate variants for both brothers were examined to detect the variant(s) that both brothers shared (Supplementary Table S2). Variants that cosegregated in both brothers were confirmed by Sanger sequencing (primer sequences available on request). Subsequently, a literature search on the relevant variants was performed. Variants have been submitted to the Leiden Open Variation Database (LOVD) at http://databases.lovd.nl/shared/genes/NEK1 (LOVD DB-IDs NEK1_000002 and NEK1_000003).
Fibroblast RNA isolation and cDNA conversion
Patient and control fibroblasts were cultured with Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal calf serum (FCS) in standard cell culture conditions. Cells were washed with phosphate-buffered saline (PBS), harvested and RNA extracted (RNeasy Mini, Qiagen, Germany). RNA was converted to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA).
Variant screening of NEK1 in an OFD cohort
To evaluate whether NEK1 variants were causal for OFD type II, NEK1 was screened in a cohort of four patients suspected of having OFD and showing overlapping features with the presently described brothers (Supplementary Table S3). The full coding regions and exon–intron boundaries were analyzed by Sanger sequencing. Primer sequences are listed in Supplementary Methods S4.
Immunocytochemistry
To assess primary cilia length and morphology, control- and patient-derived fibroblasts were cultured on glass slides under standard cell culture conditions in DMEM containing 10% FCS. The formation of primary cilia was stimulated using serum-depleted medium (DMEM containing 0% FCS) for 24 h before immunostaining. Cells were put on ice for 5 min before fixation with 4% paraformaldehyde (PFA) in PBS, and permeabilization with 0.1% Triton X-100 in PBS. Cells were blocked with 3% bovine serum albumin (BSA) in PBS minimizing non-specific binding, followed by a 2.5-h incubation with primary antibodies and 1-h incubation with secondary antibodies at room temperature. For a full list of antibodies used, see Supplementary Methods S5. Coverslips were washed with PBS and mounted in Fluormount G (Cell Lab, Beckman Coulter, Brea, CA, USA). Confocal imaging was performed using Zeiss Confocal laser microscope LSM700 (Carl Zeiss AG, Oberkochen, Germany) and images were processed with ZEN 2011 software (Carl Zeiss AG) and Volocity 6.3 software (PerkinElmer, Coventry, UK).
Results
Patient description
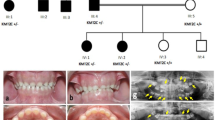
The brothers showed similar features, summarized in Table 1 and Figures 1a–h. They are the only children of healthy, consanguineous parents (fourth-degree relatives), as is depicted in Figure 1i. The elder brother (LOVD individual id 00064295) was born after a gestation of 39 weeks with a birth weight of 3540 g (0.03SD), a birth length of 49 cm (−1.44SD) and a head circumference of 37.5 cm (+1.37SD). The CT scan of the brain shortly after birth did not show any abnormalities. Histopathological examination of the tongue hamartomas showed polyp-like non-keratinizing stratified squamous epithelium. The underlying stroma contained mucoserous acini, fat and muscle cells and the tissue had a benign aspect. At the age of 21 years, he had progressive, mainly conductive hearing loss, his right ear being more severely affected than his left one. A CT scan of the mastoid showed bilateral plump vestibulae and lateral semicircular canals.
Clinical features of both brothers* and pedigree of the family. (a) Partial midline cheilognathoschisis present in the younger brother at the age of 1 month. (b) Bilobulated tongue with tongue hamartoma and short frenulum of the younger brother at the age of 1 month. (c) The intraoral situation of the younger brother (age 3.5 years old) shows a fusion of the two deciduous incisors 5.1 and 5.2. Also, the deciduous upper left lateral incisor (6.2) is congenitally missing. Furthermore, the typical hyperplastic frenula can be seen in the upper jaw. (d) The orthopantomogram of the younger brother at the age of 5 shows that the permanent and deciduous central incisors of the lower jaw (3.1, 4.1, 7.1 and 8.1) are missing. The molar roots are conical and seem to be fused, while the pulp chambers are taurodontic. (e) Incomplete midline cheilognathoschisis and mild protruding ears of the elder brother at the age of 8 months. (f) Broad right hallux of the elder brother at the age of 8 months. (g) Brachydactyly with partial cutaneous syndactyly present in the younger brother. Picture was taken at the age of 9.5 years. (h) Tortuosity of the retinal veins of younger brother at the age of 16 years. (i) Pedigree of the family. Circles represent females and squares represent males. Solid symbols indicate compound heterozygosity for the c.[464G>C];[1226G>A] variants and half solid symbols indicate heterozygosity. Carriership was confirmed in both parents. *We obtained informed consent and approval of both parents and patients to include the present pictures.
The younger brother (LOVD individual id 00064296) was born after a gestation of 39 weeks with a birth weight of 3390 g (−0.37SD), a birth length of 44 cm (−3.59SD) and a head circumference of 37.5 (+1.37SD). A CT scan of the brain that was performed shortly after birth did not show abnormalities. The tongue hamartomas were resected shortly after birth and histopathological examination showed non-keratinizing stratified squamous epithelium, with the underlying stroma containing a mucoserous acinus and fat cells. At the age of 9.5 years, a mild bilateral conductive hearing loss of 15–20 dB was detected. Both brothers had a normal psychomotor development and normal intelligence. An ultrasound of the kidneys was performed at 20 years in the elder brother and at 16 years in the younger brother; these did not show abnormalities. The elder brother had normal serum creatinine level when he was tested at age 11 years and both brothers do not have polyuria. Ophthalmological examination was also performed at the age of 20 years in the eldest brother and 16 years in the younger. Both brothers had bilateral tortuosity of the retinal veins. There were no signs of retinopathy. X-rays of the thorax, humerus, femur, ankle and feet were made at different ages and showed mild shortening of the tibia and broad halluces in both brothers, with a bony duplication of the right hallux in the elder patient. Several dental panoramic X-rays were performed and showed the anomalies described in Table 1. Both brothers had orthodontic treatment before a surgically assisted prosthodontic rehabilitation. Both brothers developed velopharyngeal insufficiency for which they underwent a pharyngoplasty.
Based on the clinical features, specifically the congenital malformations in combination with the recessive inheritance pattern and the absence of cerebral malformations, intellectual disability and severe tibia defects, OFD type II (Mohr syndrome) was diagnosed by two clinical geneticists (MJB and PAT).
SNP-array analysis
SNP-array analysis showed a normal male karyotype in both brothers and several homozygous regions indicative of distant consanguinity of the parents.
WES identifies biallelic variants in NEK1
Subsequently, the WES data of the parents and the two affected brothers were investigated. The genes implicated in the various OFD types were initially analyzed (OFD1, TCTN3, TMEM216, C5orf42, DDX59, TBC1D32/C6orf170, SCLT1, C2CD3) but no variants that could be considered causal were detected. Subsequently, the consanguineous relationship prompted us to investigate the shared homozygous regions in both boys for candidate homozygous variants but no plausible candidate genes were identified. After expanding our search in the whole exome and filtering for autosomal recessive and X-linked inheritance models, several gene variants were identified in candidate genes (Supplementary Table S2). Based on population frequency, predicted effect of the variant on gene function and previous literature, variants in NEK1 were the most likely candidates. Both brothers share the same novel compound heterozygous variants in NEK1, comprising an exonic transversion NM_012224.2:c.464G>C at the last base of exon 6 inherited from the mother and a transition variant NM_012224.2:c.1226G>A inherited from the father, c.[464G>C];[1226G>A] (Figure 2; exons are numbered similarly as in Thiel et al.17). Variants were validated and segregation was confirmed by Sanger sequencing. Both variants were novel and not present in the population frequency reference data sets of NCBI dbSNP Build 137 for Human, the Exome Variant Server (EVS), the 1000Genomes, the Genome of the Netherlands (GoNL) or our in-house data set. The variant c.1226G> A results in a nonsense variant p.(Trp409*) in exon 15 and the resulting shortened mRNA transcript includes the full coding sequence for a functional kinase domain but lacks part of the coiled-coil domain. The effect of the exonic variant c.464G>C appeared to be a non-synonymous coding change from serine to threonine, but we postulated that alternative splicing may also occur due to the position of this base so near to the canonical splice site. This position is conserved and classified as disease-causing by Alamut Visual (Alamut Interactive Biosoftware, Rouen, France) and predicted to result in abnormal splicing. Further Sanger sequencing of NEK1 in four other patients with OFD did not reveal any additional variants in NEK1 that could be considered causal.
NEK1 genomic and protein structure. (a) NEK1 genomic structure including the kinase domain, basic domain, coiled-coil domains (cc1-cc4), nuclear localization signal (NLS) and nuclear exportation signals (NES1-2). Variants found in the patients presented in this study with OFD type II are indicated above, namely c.464G>C in the kinase domain (exon 6) and c.1226G>A in the coiled-coil domain 1 (CCD1, exon 15). NCBI reference transcript NM_012224.2 is used. (b) NEK1 protein structure including the kinase, basic and coiled-coil domains, the NLS and NES.
cDNA sequencing confirms that both variants result in alternative splicing
To assess the splicing impact of the variant c.464G>C, we designed primers covering several exons upstream and downstream of the variant in the cDNA of the elder brother (Supplementary Methods S6). Analysis of the resulting PCR products on 1% agarose gel identified a smaller product in addition to the expected product size, indicating an alternatively spliced product. Subsequent Sanger sequencing of the smaller product confirmed that the variant c.464G>C resulted in alternative splicing and exon skipping of exon 6 (Figures 3a–d). The removal of exon 6 results in shift of the reading frame and introduces an early stop codon. The resulting transcript, r.397_464del, lacking exon 6, most likely undergoes nonsense-mediated decay due to the position of the premature stop codon. We observed that the correctly spliced product including exon 6 has no visible alternative allele C at position 464, demonstrating that this variant has no or minimal effect on coding changes from serine to threonine at this position. Additionally, any transcripts escaping nonsense-mediated decay would lack exon 6 and the critical DFG motif (comprising the amino acids Asp-Phe-Gly) essential for the correct conformation of the ATP-binding site of the kinase domain of NEK1 (Figure 3c).18
NEK1 cDNA analysis of variant c.464G>C (a–d) and nonsense variant c.1226G>A, p.(Trp409*) (e–h). (a) Primers flanking the patient variant c.464G>C were used for PCR on patient and control cDNA isolated from fibroblast mRNA. Following PCR amplification, the patient cDNA contained an extra product at 463 bp, indicating differential splicing. (b) Visualization of the cDNA region amplified by PCR. Locations of the forward and reverse PCR primers are indicated with arrows. The position in exon 6 of the coding region for the DFG motif is indicated by a black box. The variant is located on the last base of exon 6. The expected full-length PCR product is 531 bp. (c) Zoom in view of the splicing effect of c.464G>C. The wild-type allele G at position 464 of the cDNA (top) results in normal splicing, retaining exon 6, as seen in control cDNA and one allele of the patient cDNA. Sanger sequencing of splice junctions is shown. The product due to alternative splicing caused by variant c.464G>C (bottom) lacks exon 6, including the coding region for the DFG motif, and as a result is only 463 bp. (d) Alternative splicing results in a frameshift and introduction of a premature stop codon. The wild-type allele G (top) results in correct splicing and translation. The splicing variant C (bottom) results in a frameshift due to the loss of exon 6 and introduces a premature stop codon 18 amino acids after the frameshift, presumably resulting in nonsense-mediated decay. (e) Primers flanking the variant c.1226G>A were used for PCR on cDNA from mRNA isolated from patient and control-derived fibroblasts. Analysis of the PCR product on 1% Agarose gel showed that the patient cDNA transcript was 75 bp shorter than the control cDNA transcript (882 bp). (f) Representation of the amplified cDNA region. Location of the forward and reverse PCR primers is indicated by arrows. The expected full-length PCR product is 882 bp. (g) Detailed view of Sanger sequences surrounding the splice junctions caused by the nonsense variant in exon 15. Nonsense-associated alternative splicing causes skipping of exon 15 (75 bp) in the patient. (h) Skipping of mutated exon 15 in the patient results in a continuation in the same reading frame. The nucleotide sequence containing the wild-type allele G results in correct splicing and translation (top). The nonsense variant c.1226G>A removes exon 15 (75 bp) from the transcript, which continues in frame (bottom) resulting in a near-full length transcript.
To assess the effect of nonsense variant c.1226G>A in the same cDNA, we designed primers covering several exons upstream and downstream of exon 15 (Supplementary Methods S7). We selected the allele containing the c.1226G>A variant using a primer in exon 6, which was confirmed by the absence of the c.464G>C variant. Sanger sequencing of this region showed that exon 15 is skipped in frame in nearly all transcripts (Figures 3e–h). The resulting transcripts, r.1192_1266del, are near-full length. The loss of exon 15 removes the first coiled-coil domain of NEK1. The region containing coiled-coil domains 1 and 2 has been shown to interact strongly with ATRX and weakly with MRE11 and 53BP1 that are involved in dsDNA break repair and homologous recombination.19 The coiled-coil domains 1 and 2 are also required for localization to the primary cilium, although the ciliary signal is strongest in transcripts containing coiled-coil domain 2.20
Cilia number is reduced in patient-derived fibroblasts compared with controls
To determine the cellular effects of NEK1 variants in the patients, we examined cilia number and length in primary skin fibroblasts from the elder brother and a healthy control using anti-acetylated α-tubulin and anti-ARL13B antibodies to mark the ciliary axoneme and anti-RPGRIP1L antibody to mark the basal body at the base of the cilium (Figure 4). Patient-derived fibroblasts showed a statistically significant reduction in cilia number compared with control fibroblasts (18.0 vs 48.9%, P<0.0001). In addition, patient-derived fibroblasts showed a marked increase in cytoplasmic acetylated α-tubulin signal compared with control fibroblasts (12.3 vs 3.8 gray units, P=0.002). There was no significant difference in mean cilia length (3.7 vs 3.5 μm, P=0.09). Reconstitution of the cilia phenotype by transfecting fibroblasts with a full-length wild-type NEK1 construct was attempted; however, robust results were not obtainable (data not shown).
Patient-derived fibroblasts show a severe ciliation defect. (a) Immunofluorescence of ciliary axoneme (ARL13B, green), basal body (RPGRIP1L, red) and nucleus (Hoechst, blue) shows a severe reduction in cilia number but not in cilia length in patient- compared with control-derived fibroblasts. Scale bars represent 20 μm. (b) An average of 48.9% of control-derived fibroblasts are ciliated compared with 18.0% of patient-derived fibroblasts (>800 cells analyzed). This difference is statistically significant (**** indicates two-tailed P-value <0.0001, calculated using Mann–Whitney test). (c) Mean cilia length in control-derived fibroblasts was 3.5±0.1 μm and in patient-derived fibroblasts 3.7±0.2 μm (±200 and 50 cilia analyzed for control and patient, respectively). This difference is not statistically significant (two-tailed P-value equals 0.09, calculated using Mann–Whitney test). (d) Immunofluorescence of acetylated α-tubulin (green), pericentrin (red) and Hoechst (blue) shows a marked increase in cytoplasmic acetylated α-tubulin in patient- compared with control-derived fibroblasts. Scale bars represent 20 μm. (e) Comparison of the intensity of cytoplasmic acetylated α-tubulin signal in control- and patient-derived fibroblasts (3.8 vs 12.3 gray units, ** indicates P=0.002, calculated using Mann-Whitney test).
Discussion
In this study, we identified compound heterozygous variants in NEK1 in two brothers with OFD syndrome type II, Mohr syndrome. Homozygous variants in NEK1 are associated with short-rib polydactyly syndrome (SRPS) Majewski type (OMIM 263520),17, 21 which in the past has been considered allelic to OFD (Mohr syndrome).22, 23 Our results confirm that these syndromes may be part of the same phenotypic spectrum.
NEK1 is a known ciliary gene comprising a kinase domain in addition to a basic and coiled-coil domain for localization and protein interaction (Figure 2).20, 24, 25 NEK1 kinase is a member of the NIMA (Never In Mitosis Gene A)-related kinase family and is localized at the cilium’s basal body.24 This kinase family is considered to have a role in cell-cycle regulation and DNA damage repair, with some kinases having an additional role in the formation of the primary cilium.20, 24, 26
Immunocytochemistry in patient-derived fibroblasts demonstrated severely reduced ciliation and an increase in cytoplasmic acetylated α-tubulin signal compared with control-derived fibroblasts. Our findings are consistent with the reduced number of normal cilia and the expression of long, thick microtubular extensions marked by acetylated α-tubulin in fibroblasts of a fetus with SRPS Majewski type resulting from NEK1 variants.17 This increase in cytoplasmic acetylated α-tubulin may be due to the inability of variant NEK1 to phosphorylate VHL, which stabilizes microtubules.27 A reduction in ciliated cells was also noted in embryonic fibroblasts of kat2J mice homozygous for a frameshift variant in the kinase domain.24 Contrary to previous reports, we did not detect a difference in mean cilia length.17 This could be related to using the anti-ARL13B antibody to visualize the ciliary axoneme instead of the anti-acetylated α-tubulin antibody, as increased microtubule staining prevented reliable measurements of cilia.
Decreased ciliation could negatively affect adequate signal transduction during morphogenesis and craniofacial development as cilia are involved in multiple signaling pathways such as Hedgehog and Wingless (Wnt) signaling.8, 28, 29 For example, variants in the NEK1 interaction partner KIF3A, a component of the intraflagellar transport system, lead to cilia defects resulting in an inappropriate increase in canonical Wnt signaling.30 Although homozygous Kif3a null variants are lethal in mice,31 mice with conditional Kif3a deletions in cranial neural crest cells are characterized by impaired skeletogenesis and proliferation defects in the facial midline, similar to the anomalies detected in our patients.32 Additionally, inactivation of OFD1 is also associated with defective ciliation and subsequent disturbed Hedgehog and canonical Wingless signaling, showing overlap in the pathogenic disease mechanism.5, 33, 34, 35 This might therefore explain the considerable overlap in phenotype between patients with OFD type I and our patients.36
In addition to localizing to the basal body of the cilium, NEK1 resides in the cilium, the cytoplasm and the nucleus.24, 37, 38 There is a canonical nuclear localization signal in residues 355–378 within the coiled-coil domain of NEK1, and a cryptic nuclear localization signal within residues 258–321 of the basic domain (Figure 2).37 The coiled-coils 1 and 2 are essential for localization to the primary cilium.20 However, in a study investigating the role of Nek1 in murine IMCD3 cells, a tagged-Nek1 construct was only retained in the ciliary compartment long enough to be visualized if the kinase domain (which normally targets the protein to the cytoplasm) was non-functional, and endogenous mNek1 could not be visualized in the primary cilium.20 Cycling of NEK1 through the ciliary, cytoplasmic and nuclear compartments can partly explain why reconstitution assays yielded no robust results.20 Transient expression of NEK1 at the basal body prevented assessment of transfected cells only; and therefore, the rescue of the cilia phenotype could not be quantified. Moreover, rescue of the cilia phenotype was limited because NEK1 overexpression also inhibits ciliogenesis.20, 24 Similar to other ciliary proteins with a dual role in the nucleus, NEK1 is involved in DNA damage response and cell-cycle control.39, 40 Expression and kinase activity of NEK1 are upregulated in irradiated cells and NEK1-null cells are hypersensitive to DNA damage by ionizing radiation.40, 41 A portion of NEK1 can redistribute to form discrete nuclear foci at sites of DNA damage.40, 41 The mechanism for this DNA damage response has not yet been fully established, but OFD1 has been linked to DNA repair through its interaction with the chromatin-remodeling complex TIP60.42 NEK1-deficient primary renal tubular epithelial (RTE) cells of kat2J mice showed increased chromosome missegregation events and aneuploidy after mitosis.40 The DNA damage response of these ciliary proteins likely also has a role during morphogenesis when progenitor cells are under increased replication stress. Impairment of this response would therefore possibly result in tissue dysplasia.39 Second, the impaired DNA damage response can underlie degenerative instead of developmental phenotypes in ciliopathies, eg, nephronophthisis.39, 43 Renal cysts were present in five out of eight reported lethal NEK1-related SRPS type Majewski patients17, 21 and NEK1-deficient kat and kat2J mice display slowly progressive polycystic kidney disease.44, 45 Although our patients had a normal renal ultrasound at age 20 and 16 years respectively, we cannot exclude that a renal phenotype may develop at an older age.
The phenotype in our patients is mild compared with the previously described lethal cases with NEK1 variants. This could reflect the extreme clinical heterogeneity associated with variants in ciliary genes; for example, in the ciliary gene MKS1 biallelic null variants cause the embryonically lethal Meckel-Gruber syndrome while missense variants cause Joubert syndrome.46 In NEK1, missense, splice-site, nonsense and frameshift variants have been reported in the patients with SRPS (Supplementary Table S8).17, 21, 47
Most variants were identified in the kinase domain of NEK1. In addition, one patient showed a homozygous frameshift variant in the C-terminal domain of NEK1 and in another patient digenic inheritance with DYNC2H1 has a role (Figure 2).17, 21, 47 The splice variant c.464G>C identified in the present study is predicted to result in nonsense-mediated decay of the mRNA transcript due to the premature stop codon and thus no protein translation. The nonsense variant p.(Trp409*) removes the first coiled-coil domain. A possible explanation for the milder phenotype in our patients compared with previously published patients is basal exon skipping of exon 15 that contains the premature stop codon. Basal exon skipping is a mechanism whereby detrimentally mutated exons that begin and end in the same reading frame can be selectively spliced out of the final gene transcript.48 This mechanism has been described in other ciliopathies.48, 49 Nonsense-associated alternative splicing results in a detectable level of near-full length NEK1 transcript that contains the complete kinase domain and is expected to escape nonsense-mediated decay. The resulting protein likely has some residual kinase function. As the N-terminal kinase domain is required for cilia formation,17, 20 the remaining 18% ciliation in patient-derived fibroblasts could suggest that some functioning kinase domain is (partly) preserved. Moreover, variants in modifier genes could have a role in the milder phenotype seen in our patients.
Similar to other ciliopathies, OFD type II is probably a genetically heterogeneous condition. Further studies in a cohort of patients with autosomal recessive types of OFD for variants in NEK1 and genes coding for proteins interacting with NEK1, such as KIF3A, might reveal more insight into the underlying pathogenic mechanisms (NEK1 interacting proteins reviewed in Surpili et al19). The fact that we found compound heterozygous variants in a gene outside homozygous regions illustrates that consanguinity can be misleading and that WES or whole-genome sequencing can provide more information in cases in which homozygosity mapping does not lead to the causative variants.
In this study, we provide the first genetic and functional evidence that variants in NEK1 can be involved in the etiology of OFD type II. Similar to the cellular phenotype seen in SRPS type Majewski, fibroblasts derived from our patient with OFD type II show a severe defect in cilia formation and an increase in cytoplasmic acetylated α-tubulin. Analysis of NEK1 should be considered in patients with autosomal recessive OFD.
References
Gurrieri F, Franco B, Toriello H, Neri G : Oral-facial-digital syndromes: review and diagnostic guidelines. Am J Med Genet A 2007; 143a: 3314–3323.
Toriello HV, Franco B : Oral-facial-digital syndrome type I. In: Pagon RA, Adam MP, Ardinger HH et al (eds): Gene Reviews(R). University of Washington: Seattle, WA, USA, 1993.
Ferrante MI, Giorgio G, Feather SA et al: Identification of the gene for oral-facial-digital type I syndrome. Am J Hum Genet 2001; 68: 569–576.
Macca M, Franco B : The molecular basis of oral-facial-digital syndrome, type 1. Am J Med Genet C Semin Med Genet 2009; 151c: 318–325.
Romio L, Fry AM, Winyard PJ, Malcolm S, Woolf AS, Feather SA : OFD1 is a centrosomal/basal body protein expressed during mesenchymal-epithelial transition in human nephrogenesis. J Am Soc Nephrol 2004; 15: 2556–2568.
Fry AM, Leaper MJ, Bayliss R : The primary cilium: guardian of organ development and homeostasis. Organogenesis 2014; 10: 62–68.
Powles-Glover N : Cilia and ciliopathies: classic examples linking phenotype and genotype-an overview. Reprod Toxicol 2014; 48: 98–105.
Satir P, Pedersen LB, Christensen ST : The primary cilium at a glance. J Cell Sci 2010; 123: 499–503.
Adly N, Alhashem A, Ammari A, Alkuraya FS : Ciliary genes TBC1D32/C6orf170 and SCLT1 are mutated in patients with OFD type IX. Hum Mutat 2014; 35: 36–40.
Lopez E, Thauvin-Robinet C, Reversade B et al: C5orf42 is the major gene responsible for OFD syndrome type VI. Hum Genet 2014; 133: 367–377.
Shamseldin HE, Rajab A, Alhashem et al: Mutations in DDX59 implicate RNA helicase in the pathogenesis of orofaciodigital syndrome. Am J Hum Genet 2013; 93: 555–560.
Thauvin-Robinet C, Lee JS, Lopez E et al: The oral-facial-digital syndrome gene C2CD3 encodes a positive regulator of centriole elongation. Nat Genet 2014; 46: 905–911.
Thomas S, Legendre M, Saunier S et al: TCTN3 mutations cause Mohr-Majewski syndrome. Am J Hum Genet 2012; 91: 372–378.
Valente EM, Logan CV, Mougou-Zerelli S et al: Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nat Genet 2010; 42: 619–625.
Huber C, Cormier-Daire V : Ciliary disorder of the skeleton. Am J Med Genet C Semin Med Genet 2012; 160c: 165–174.
Sun Y, Ruivenkamp CA, Hoffer MJ et al: Next-generation diagnostics: gene panel, exome, or whole genome? Hum Mutat 2015; 36: 648–655.
Thiel C, Kessler K, Giessl et al: NEK1 mutations cause short-rib polydactyly syndrome type majewski. Am J Hum Genet 2011; 88: 106–114.
Ung PM, Schlessinger A : DFGmodel: predicting protein kinase structures in inactive states for structure-based discovery of type-II inhibitors. ACS Chem Biol 2015; 10: 269–278.
Surpili MJ, Delben TM, Kobarg J : Identification of proteins that interact with the central coiled-coil region of the human protein kinase NEK1. Biochemistry 2003; 42: 15369–15376.
White MC, Quarmby LM : The NIMA-family kinase, Nek1 affects the stability of centrosomes and ciliogenesis. BMC Cell Biol 2008; 9: 29.
El Hokayem J, Huber C, Couve et al: NEK1 and DYNC2H1 are both involved in short rib polydactyly Majewski type but not in Beemer Langer cases. J Med Genet 2012; 49: 227–233.
Franceschini P, Guala A, Vardeu MP, Signorile F, Franceschini D, Bolgiani MP : Short rib-dysplasia group (with/without polydactyly): report of a patient suggesting the existence of a continuous spectrum. Am J Med Genet 1995; 59: 359–364.
Silengo MC, Bell GL, Biagioli M, Franceschini P : Oro-facial-digital syndrome II. Transitional type between the Mohr and the Majewski syndromes: report of two new cases. Clin Genet 1987; 31: 331–336.
Shalom O, Shalva N, Altschuler Y, Motro B : The mammalian Nek1 kinase is involved in primary cilium formation. FEBS Lett 2008; 582: 1465–1470.
Meirelles GV, Perez AM, de Souza EE et al: 'Stop Ne(c)king around': How interactomics contributes to functionally characterize Nek family kinases. World J Biol Chem 2014; 5: 141–160.
Fry AM, O'Regan L, Sabir SR, Bayliss R : Cell cycle regulation by the NEK family of protein kinases. J Cell Sci 2012; 125: 4423–4433.
Patil M, Pabla N, Huang S, Dong Z : Nek1 phosphorylates Von Hippel-Lindau tumor suppressor to promote its proteasomal degradation and ciliary destabilization. Cell Cycle 2013; 12: 166–171.
Zaghloul NA, Brugmann SA : The emerging face of primary cilia. Genesis 2011; 49: 231–246.
Veland IR, Awan A, Pedersen LB, Yoder BK, Christensen ST : Primary cilia and signaling pathways in mammalian development, health and disease. Nephron Physiol 2009; 111: 39–53.
Corbit KC, Shyer AE, Dowdle WE et al: Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol 2008; 10: 70–76.
Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS : Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA 1999; 96: 5043–5048.
Liu B, Chen S, Johnson C, Helms JA : A ciliopathy with hydrocephalus, isolated craniosynostosis, hypertelorism, and clefting caused by deletion of Kif3a. Reprod Toxicol 2014; 48: 88–97.
Ferrante MI, Romio L, Castro S et al: Convergent extension movements and ciliary function are mediated by ofd1, a zebrafish orthologue of the human oral-facial-digital type 1 syndrome gene. Hum Mol Genet 2009; 18: 289–303.
Ferrante MI, Zullo A, Barra et al: Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat Genet 2006; 38: 112–117.
Bimonte S, De Angelis A, Quagliata L et al: Ofd1 is required in limb bud patterning and endochondral bone development. Dev Biol 2011; 349: 179–191.
Thauvin-Robinet C, Cossee M, Cormier-Daire V et al: Clinical, molecular, and genotype-phenotype correlation studies from 25 cases of oral-facial-digital syndrome type 1: a French and Belgian collaborative study. J Med Genet 2006; 43: 54–61.
Hilton LK, White MC, Quarmby LM : The NIMA-related kinase NEK1 cycles through the nucleus. Biochem Biophys Res Commun 2009; 389: 52–56.
Mahjoub MR, Trapp ML, Quarmby LM : NIMA-related kinases defective in murine models of polycystic kidney diseases localize to primary cilia and centrosomes. J Am Soc Nephrol 2005; 16: 3485–3489.
Chaki M, Airik R, Ghosh AK et al: Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 2012; 150: 533–548.
Chen Y, Chen CF, Riley DJ, Chen PL. : Nek1 kinase functions in DNA damage response and checkpoint control through a pathway independent of ATM and ATR. Cell Cycle 2011; 10: 655–663.
Polci R, Peng A, Chen PL, Riley DJ, Chen Y : NIMA-related protein kinase 1 is involved early in the ionizing radiation-induced DNA damage response. Cancer Res 2004; 64: 8800–8803.
Giorgio G, Alfieri M, Prattichizzo C, Zullo A, Cairo S, Franco B : Functional characterization of the OFD1 protein reveals a nuclear localization and physical interaction with subunits of a chromatin remodeling complex. Mol Biol Cell 2007; 18: 4397–4404.
Slaats GG, Saldivar JC, Bacal J et al: DNA replication stress underlies renal phenotypes in CEP290-associated Joubert syndrome. J Clin Invest 2015; 125: 3657–3666.
Upadhya P, Birkenmeier EH, Birkenmeier CS, Barker JE : Mutations in a NIMA-related kinase gene, Nek1, cause pleiotropic effects including a progressive polycystic kidney disease in mice. Proc Natl Acad Sci USA 2000; 97: 217–221.
Upadhya P, Churchill G, Birkenmeier EH, Barker JE, Frankel WN : Genetic modifiers of polycystic kidney disease in intersubspecific KAT2J mutants. Genomics 1999; 58: 129–137.
Romani M, Micalizzi A, Kraoua I et al: Mutations in B9D1 and MKS1 cause mild Joubert syndrome: expanding the genetic overlap with the lethal ciliopathy Meckel syndrome. Orphanet J Rare Dis 2014; 9: 72.
Chen CP, Chang TY, Chen CY et al: Short rib-polydactyly syndrome type II (Majewski): prenatal diagnosis, perinatal imaging findings and molecular analysis of the NEK1 gene. Taiwan J Obstet Gynecol 2012; 51: 100–105.
Drivas TG, Wojno AP, Tucker BA, Stone EM, Bennett J : Basal exon skipping and genetic pleiotropy: a predictive model of disease pathogenesis. Sci Transl Med 2015; 7: 291ra297.
Littink KW, Pott JW, Collin RW et al: A novel nonsense mutation in CEP290 induces exon skipping and leads to a relatively mild retinal phenotype. Invest Ophthalmol Vis Sci 2010; 51: 3646–3652.
Acknowledgements
We would like to thank the patients and their parents for their participation in this study. We wish to thank the Utrecht Sequencing Facility, UMC Utrecht, for WES using the Illumina HiSeq 2500 and further data processing with the bioinformatics pipeline. We thank the Cell Microscopy Core at the Department of Cell Biology, Center for Molecular Medicine, UMC Utrecht for use of the Zeiss Confocal laser microscope LSM700 and Corlinda ten Brink for help with the Volocity 6.3 software. We thank Dr R Roepman and Dr H Arts for providing us with the anti-RPGRIP1L antibody and J Hoogeboom (Department of Clinical Genetics, Erasmus Medical Center, Rotterdam, the Netherlands) for providing us with clinical data on patients in the OFD cohort. This work was supported by the Dutch Kidney Foundation (KOUNCIL consortium: CP11.18 to NK and RG).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Monroe, G., Kappen, I., Stokman, M. et al. Compound heterozygous NEK1 variants in two siblings with oral-facial-digital syndrome type II (Mohr syndrome). Eur J Hum Genet 24, 1752–1760 (2016). https://doi.org/10.1038/ejhg.2016.103
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2016.103
This article is cited by
-
Primary cilia proteins: ciliary and extraciliary sites and functions
Cellular and Molecular Life Sciences (2018)
-
NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin
Scientific Reports (2017)