Abstract
Rothmund–Thomson syndrome is a rare genodermatosis caused by biallelic mutations of the RECQL4 gene and is characterised by poikiloderma, sparse hair, eyelashes and/or eyebrows, small stature, skeletal and dental abnormalities and cancer predisposition. Mutations predicted to result in the loss of RECQL4 protein have been associated with osteosarcoma risk, but mutation(s)–phenotype correlations are better addressed by combined DNA and RNA analyses. We describe two siblings with a mild phenotype, mainly restricted to the skin, who carry the unreported paternal c.2272C>T alteration in exon 14 and the previously reported maternal exon 15 c.2492_2493delAT, both predicted to result in premature termination codons (p.(Arg758*), p.(His831Argfs*52)). However real-time and transcript analysis showed, in the carrier father and affected daughter, increased levels of a novel RECQL4 physiological alternative transcript with partial in-frame skipping of exon 14, generated by increased usage of a weak cryptic splice site. This alternative transcript is expressed in all controls and tested tissues, its upregulation is specific to the paternal c.2272C>T mutation and depends on the abrogation of the binding motifs for SF2 and SRp55 serine/arginine-rich proteins with bypass of the mutation site located in the skipped exon 14 portion. Moreover, in the proband the increased levels of the alternative transcript, likely encoding a protein isoform with residual activity, may compensate for the dearth of the canonical transcript with the c.2492_2493delAT, accounting for the mild clinical phenotype of the siblings. Our results emphasise the value of RNA analysis to better predict the effects of RECQL4 mutations on the clinical phenotype.
Similar content being viewed by others
Introduction
Rothmund–Thomson syndrome (RTS, MIM#268400) is a clinically and genetically heterogeneous autosomal recessive disorder caused by biallelic mutations of the RECQL4 gene (MIM*603780)1 in up to 66% of patients.2 To date, more than 60 different RECQL4 mutations, only a few recurrent, have been described in RECQL4-related diseases, 45 of which were in patients with RTS, defined as RTS type II.3, 4, 5, 6 RECQL4-negative patients are classified as RTS type I.3 The relationship between RECQL4 genotype and syndromic phenotype depends on mutation type and intragenic position and the specific combination of two different mutations, which makes (with a few exceptions) each case incomparable to others previously described. It has been proposed that in RTS patients, ‘deleterious’ RECQL4 mutations, predicted to lead to truncated proteins, predispose to osteosarcoma2 and skeletal malformations.7 Until functional studies, currently available for a restricted number of RECQL4 mutations,8 unravel possible genotype–phenotype correlations, the analysis of mutant alleles at the transcript level may complement DNA analysis in predicting the effect of mutations on the clinical phenotype, including cancer outcome.
Here, we report two siblings with RTS found to be compound heterozygotes for the previously reported exon 15 mutation, c.2492_2493delAT (p.(His831Argfs*52)),1, 2 and the yet undescribed exon 14 mutation, c.2272C>T (p.(Arg758*)). According to DNA analysis, both variants are predicted to result in premature termination codons. Conversely, cDNA sequence analysis revealed the expression of full-length transcripts carrying the c.2492_2493delAT variant but failed to detect full-length transcripts containing the C>T transition. Moreover, substantial enhancement of levels of a novel RECQL4 physiological alternative in-frame transcript, which skips the 3′ region of exon 14 harbouring the C>T mutation, was observed by RT-PCR and quantified by real-time PCR. This finding provided the key to unravel the correlation between the RECQL4 genotype and the mild phenotype of the two siblings.
Materials and methods
Patients
Two siblings (II-1 and II-2) were referred to our laboratory with a clinical diagnosis of RTS by dermatologist colleagues and were enrolled in the study after their parents provided appropriate informed consent to the genetic tests.
Another RTS patient, whose sample was used for quantitative analysis of RECQL4 transcripts, has been previously reported.9
Cell cultures
EBV-transformed lymphoblastoid cell lines (LCLs) were established from peripheral blood lymphocytes of the elder sibling II-1, the parents (I-1 and I-2) and five healthy controls.
LCLs were cultured in complete RPMI-1640 medium (EuroClone, Milano, Italy) supplemented with 10% fetal bovine serum (Lonza, Walkersville, MD, USA) and 1% penicillin, streptomycin and ampicillin in a 37 °C humidified incubator with 5% CO2.
DNA isolation and mutational analysis
Genomic DNA was extracted from peripheral blood samples from I-1, I-2, II-1 and II-2 according to standard protocols. The whole RECQL4 gene was amplified (primer sequences and PCR conditions are available upon request) and PCR products were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit and an ABI 3730 capillary sequencer (Applied Biosystems, Foster City, CA, USA).
Electropherograms were analysed with ChromasPro software 1.42 (Technelysium Pty Ltd, Tewantin, QLD, Australia) using the wild-type sequence of the RECQL4 gene (GenBank: NG_016430.1) as reference.
RNA isolation, RT-PCR and cDNA analysis
Total RNA was isolated from LCLs of patients, healthy parents, controls, fresh tissues and cell cultures using TRI reagent (Sigma, St Louis, MO, USA) and treated with DNase I (RNase-free, New England Bio-Labs, Inc., Ipswich, MA, USA). cDNA was synthesised from 250 ng of total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with random hexamers. All samples were reverse transcribed in two independent experiments.
RECQL4 cDNA, from exon 11 to exon 15, was amplified and sequenced as described above. Nucleotide sequences were compared with the major RECQL4 transcript reference sequence (GenBank: NM_004260.3).
RECQL4 cDNA, from exon 13 to exon 15, was amplified from 20 tissue RNAs of the Human Total Master Panel II (Clontech, Mountain View, CA, USA), 4 RNAs from fresh tissues (blood, skin, bone marrow and tooth) and 3 RNAs from cell cultures (adult and fetal fibroblasts and keratinocytes).
RECQL4 cDNA from exon 5 to exon 15 and from exon 13 to exon 21 was also amplified. Primer sequences and PCR conditions are available upon request.
NG_016430.1 was used as reference for exon numbering.
Real-time PCR analysis
The relative levels of RECQL4 full-length exon 14 and RECQL4 partially skipped exon 14 mRNAs were determined by real-time RT-PCR using the StepOne Real-Time PCR System (Applied Biosystems). Isoform-specific TaqMan assays (Assay ID Hs00355454_g1 and custom made assay; Applied Biosystems) were purchased from Applied Biosystems and used, following validation, according to the manufacturer’s instructions. GAPDH (Assay ID: 4333764F, Applied Biosystems) was chosen as endogenous normalising gene, according to assay efficiency and stability experiments previously performed. Relative gene expression was determined using the ΔΔCt method.10
Bioinformatic tools
Human Splicing Finder (http://www.umd.be/HSF/) and Alternative Splice Site Predictor (http://wangcomputing.com/assp/index.html) were used to identify alternative donor and acceptor splicing sites. The ESEFinder web interface (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home)11, 12 was consulted for prediction of exon splicing enhancer (ESE) sequences across the mutation site and for score analysis of serine/arginine-rich (SR) proteins.
Results
Clinical report of study subjects
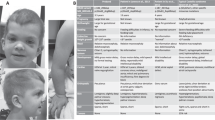
The index family includes two siblings with clinical diagnoses of RTS, born to nonconsanguineous healthy parents. Their clinical signs are listed in Table 1 and representative photographs are provided in Figure 1.
Representative pictures of siblings with RTS. II-1 at the age of (a) 4 and (b) 9 years. Note the poikiloderma on the cheeks and the thin eyebrows. (c) The skin of the hand dorsum shows only minimal atrophy and an inflammatory scar at the site of a previous mosquito bite, but no poikiloderma. Nails are normal. (d) Café-au-lait spots can be seen on the back. (e) Mosquito stings on the legs led to marked inflammatory reactions. (f) The younger brother, II-2, at age 1 year and 7 months: note the erythema on the cheeks and sparse eyebrows and eyelashes.
The elder sibling (II-1) is currently aged 9 years and 3 months. Birth weight was 2.720 kg. Since the age of 6 months, she showed a fixed erythema of the cheeks, sensitive to sun and heat exposure.
By the age of 15 months, the time of clinical RTS diagnosis, the malar erythema had evolved into a true poikiloderma, with a pattern of reticulated hypo-hyperpigmentation, thin desquamation, mild atrophy and telangiectases. The girl also presented thin, sparse scalp hair and sparse eyelashes and eyebrows, whereas teeth and nails were normal. At 4 years, the time of molecular diagnosis, poikiloderma, mostly restricted to the face, was clearly apparent (Figure 1a). Weight and height were moderately under mean values for age. An osteolytic area between the third medial and third distal femoral diaphysis was identified by radiography (data not shown). This lesion became less evident in time and at a recent control it was no more detectable, attesting its benign nature, likely unrelated to RTS. No other signs of RTS, such as hyperkeratotic lesions around elbows and knees, skeletal defects, haematological dysfunction or neoplasia, were identified. During the following years, the parents noticed the progressive appearance of café-au-lait spots on the trunk as well as an improvement of scalp hair density and texture. At 9 years, the clinical follow-up recorded normal scalp hair and eyebrows sparse only in the lateral third (Figure 1b). Nails (Figure 1c) and teeth were normal. The poikiloderma pattern was unaltered and limited to the cheeks, as the extremities only presented slight atrophy in the absence of telangiectases and dyspigmentation (Figure 1c). In addition, ∼15 café-au-lait spots, mainly localised on the trunk, were counted (Figure 1d). She also presented numerous atrophic, hyperpigmented scars on the lower limbs. The parents reported that they were the sequelae of insect (mosquito) bites that, in their daughter, always led to marked inflammatory reactions lasting several weeks that scarred on healing (Figure 1e). Growth parameters were slightly defective. Bone age was 1 year delayed.
The younger male sibling (II-2) is currently 1 year and 7 months old. Birth weight was 3.400 kg (70th percentile) and he is currently 13th percentile for weight and 11th percentile for height. At 3 months, he developed malar erythema that is not yet evolved into poikiloderma. At the current age, atrophic skin areas are apparent on the face, hair is sparse and thin and eyebrows and eyelashes are sparse to absent (Figure 1f). Nails are normal whereas, to date, only four incisors have erupted, the first one at 12 months. A few café-au-lait spots, similar to those observed on his sister, are developing on the abdominal skin. No radiologic investigations have been performed to date.
Molecular characterisation of study subjects
Direct sequencing of the whole RECQL4 gene from genomic DNA of II-1 showed that the patient is a compound heterozygote for a new and a previously described mutation, inherited from the father and mother, respectively. Subsequent investigation confirmed the presence of the same mutations in the newborn brother (II-2) (Figure 2a). The paternally inherited mutation is an exon 14 c.2272C>T transition (NM_004260.3) predicted to result in a stop codon at residue 758 (p.(Arg758*)) in the helicase domain. The second and previously reported mutation,1, 2 c.2492_2493delAT in exon 15, causes a frameshift resulting in a termination codon 51 residues downstream (p.(His831Argfs*52)). These data have been submitted to LOVD database (http://databases.lovd.nl/shared/genes/RECQL4).
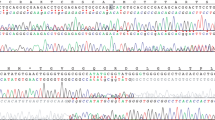
DNA and RNA characterisation of RECQL4 mutations in the index family. (a) Pedigree of the family with two siblings with Rothmund–Thomson syndrome (filled diamond) found to be compound heterozygotes for the paternal c.2272C>T and the maternal c.2492_2493delAT mutations shown by the electropherograms. (b) cDNA analysis of the RECQL4 region spanning exons 11–15 showing the expected 746 bp amplicon as well as a shorter, 548 bp, band observed in all samples but with a higher intensity in the affected daughter II-1 and carrier father I-1. MW, molecular weight; C−, negative control; C1, C2, C3, C4, control samples; I-1, father; I-2, mother; II-1, index case. (c) Diagram based on sequence analysis of the two gel bands in (b) showing the canonical and alternative splicing of IVS14. Two different transcripts originate from the RECQL4 pre-mRNA (right): the full-length exon 14 transcript of 746 bp (upper right) and the partially skipped exon 14 transcript of 548 bp (lower right). The two different SR proteins recognising the ESE motifs in the region of exon 14 including the mutation site (C>T in bold) are framed in purple (SF2/ASF) and yellow (SRp55) on RECQL4 pre-mRNA. The dashed line indicates the exonic cryptic splice site leading to the partially skipped exon 14 transcript. (d) Sequence electropherograms showing the physiological partially skipped exon 14 transcript (r.2266_2463del). (e) The table on the left shows the two SR proteins recognising the ESE motifs in RECQL4 exon 14 close to the c.2272 mutation site (in red) and their scores in the presence of the wild-type and c.2272C>T sequence. The diagram on the right shows ESE binding site(s) of SF2/ASF (purple bars) and SRp55 (yellow bar) proteins on the RECQL4 wild-type and mutant sequences reported on the x axis (C and T red encircled). The C>T mutation abrogates one of the binding sites for SF2/ASF and the binding site for SRp55.
As regards the mutant alleles, their transcripts might be expected to undergo degradation by nonsense-mediated decay (NMD) or, if translated, should produce nonfunctional truncated proteins. The involvement of the helicase domain in both mutant alleles prompted us to perform RT-PCR analysis on a portion of the RECQL4 functional helicase domain, from exon 11 to exon 15.
Two different RECQL4 products were observed in the proband (II-1), her parents and four different controls: a 746-bp fragment, corresponding to the expected amplicon, and a faster migrating product of 548 bp (Figure 2b).
Sequence analysis revealed that the 746-bp band included the entire exon 14 sequence; however, no fragment bearing the c.2272C>T transition was detected in the affected girl and her father (data not shown). Moreover, the electropherogram of the 548-bp amplicon showed a previously undescribed exon 14–15 junction because of an in-frame skipping of the last 198 nucleotides of exon 14, including the c.2272C>T mutation (r.2266_2463del; p.Val756_Gln821del) (GenBank: KJ147518) (Figure 2c and d). In the proband and her mother, sequence analysis of both RECQL4 fragments revealed the presence of transcripts carrying the c.2492_2493delAT mutation (r.2492_2493delAT).
Furthermore, we retrieved the novel transcript lacking 198 nucleotides (nt) of exon 14 (r.2266_2463del) in all the analysed samples (four controls and family members) and all 27 tissues/cells of different embryogenetic origin that have been investigated, including those most affected in RTS, that is, skin, fibroblasts, dental tissue and keratinocytes (Supplementary Figure 1). We verified, with further PCR experiments, that the region of the helicase domain lacking the 198 nt of exon 14 of this novel amplicon is a part of a longer transcript of 3036 nt (from exon 5 to exon 21) (Supplementary Figure 2) and probably includes the first 4 exons and the canonical ATG. We thus consider this novel transcript a physiological isoform resulting from unreported RECQL4 alternative splicing (AS), involving a weak donor splice site predicted by both Human Splicing Finder and Alternative Splice Site Predictor.
Transcripts analysis in the family also highlighted that the intensity of the full-length transcript band was fainter in II-1 compared with the parents and controls, whereas the intensity of the alternative transcript (AltTr) band was stronger in II-1 and I-1 than in the other samples (Figure 2b).
We thus suspected that the c.2272C>T mutation might enhance the physiological AS in the proband and her father and found this hypothesis supported by ESEFinder bioinformatic analysis. Indeed, the C>T substitution is predicted to abolish both Exonic Splicing Enhancer motifs recognised by the SR proteins SF2 and SRp55. Another SF2 predicted binding site is located 12 nt upstream the C>T mutation and is not affected by the transition (Figure 2c and e).
Real-time experiments confirmed increased levels of the RECQL4 AltTr in the C>T mutation carrier and the patient (Figure 3a). I-1 showed levels of the AltTr that were twice those of controls, whereas II-1 displayed markedly higher levels of the AltTr than both the controls and the father (Figure 3a). However, compared with controls and the father, she showed notably decreased levels of the full-length exon 14 mRNA, likely also attributable to instability of transcripts from the maternal c.2492_2493delAT allele (Figure 3b). Indeed, the mother showed reduced levels of the canonical full-length exon 14 transcript but levels of the AltTr were comparable to those observed in controls (Figure 3a and b). The same quantitative analysis of RECQL4 transcripts was performed in another compound heterozygous RTS patient with two different RECQL4 mutations, one affecting the 3′ splice site of exon 13,9 and hence close to the paternally inherited mutation of our siblings. This analysis highlighted decreased levels of both the full-length and the partially skipped exon 14 RECQL4 isoforms compared with controls and family members, suggesting that enhancement of AS leading to a partially skipped exon 14 is a mutation-specific mechanism.
Real-time expression profile of RECQL4 isoforms. (a) RECQL4 partially skipped exon 14 and (b) RECQL4 full-length exon 14 mRNA expression in case II-1, her healthy parents and an RTS patient9 with different RECQL4 mutations compared with five controls (CTRL). Samples were run in triplicate and data are expressed as mean±SD. I-1, father; I-2, mother; II-1, index case; RTS, previously described RTS patient.
Discussion
RNA analysis of LCLs from RTS family members and controls led to the identification of a novel mRNA splice variant of RECQL4 generated by AS and leading to in-frame loss of 198 nt of exon 14. This AltTr is not listed among the different RECQL4 transcripts in the UCSC Genome Browser (http://genome.ucsc.edu/), Entrez NCBI (http://www.ncbi.nlm.nih.gov/gene/) or Ensembl (http://www.ensembl.org/) databases, even if an EST clone from a B-cell library was found to carry the AltTr exon 10 through exon 15 portion. According to our analysis, the AltTr is likely full length (Supplementary Figure 2) and should be ubiquitous, being detected in all 27 tissues/cells tested (Supplementary Figure 1).
This RECQL4 isoform is a fraction of the physiological RECQL4 transcripts (Figure 2b and Supplementary Figure 1), but is markedly increased in the compound heterozygous RTS proband (II-1) and her father (I-1) (Figure 2b). The enhancement of the physiological AltTr appears to be dependent on the paternal c.2272 C>T mutation, as its expression is not affected by other RECQL4 mutations, such as that of the proband’s mother or those carried by a previously characterised RTS individual9 (Figure 3a and b). Indeed, the c.2272 C>T mutation, abolishing the binding sites for SF2 and SRp55, enhances the use of a weak cryptic splice site lying 7 nt upstream to the detriment of the canonical IVS14 donor splice site (Figure 2c). This mechanism could lead to undetectable levels of transcripts including the C>T mutation in the proband and her father.
In the father, the c.2272C>T mutant allele yields increased levels of the AltTr and decreased expression levels of the full-length exon 14 mRNA. The mother shows levels of the AltTr comparable to those of controls and decreased levels of the full-length exon 14 mRNA likely because of her c.2492_2493delAT allele, containing a premature termination codon. In the proband (II-1), the expression of the AltTr is hugely enhanced by both the paternal c.2272C>T mutation and a possible compensatory mechanism resulting in stabilisation of the AltTr to overcome the deficiency of the maternal c.2492_2493delAT derived transcripts.
Upregulation of RECQL4 AS in the proband allows the bypass of the predicted deleterious effect of the C>T mutation via skipping of the exon 14 region containing the mutation site. We therefore speculate that the AltTr probably encodes for a functional protein, even if lacking a portion (66 amino acids) of the helicase domain, and its high expression levels may account for the mild phenotype of the RTS proband (II-1).
This hypothesis is supported by the finding of extremely low levels of both the AltTr and full-length exon 14 transcripts in a nonrelated RTS patient who displayed a much more severe phenotype than our proband. Functional studies of the protein translated by the AltTr could confirm whether the c.2272C>T is a hypomorphic mutation that might reduce, but not eliminate, gene activity. In keeping with this assumption, the overall clinical phenotype of both siblings currently appears quite mild.
Indeed, the patients reported here have clinical signs mainly restricted to the skin and the skin annexes: the skin poikiloderma exclusively affected the face, and scalp hair density and texture showed a spontaneous improvement over time in the elder patient. The presence of multiple café-au-lait spots manifested in early childhood in the siblings is notable (Table 1 and Figure 1). This sign has been reported in a few previous cases of RTS,4, 13, 14 but has not been emphasised. Café-au-lait spots may represent an underreported skin manifestation of RTS that would add RTS to the long list of syndromes that can present with this particular skin phenotype.15 Persistent insect bite reactions leaving scars, as observed in the elder RTS sibling, have not been previously reported in RTS. Although no histopathological study was performed, they may indicate a modest immune dysregulation, as recently reported in an RTS patient developing granulomas.4
To date, no malignancies have developed in the elder sibling, despite the fact that RTS is a syndrome predisposing to cancer, particularly osteosarcoma, that has an estimated prevalence of 30% and a mean age at diagnosis of 11 years.16
Interestingly, from literature revision we noticed that 10 reported patients (9 RTS and 1 RAPADILINO) carry the c.2269C>T mutation (p.Q757*),1, 2, 17, 18 mapping 3 nt upstream the here described c.2272C>T mutation. No transcript analysis has been performed for the c.2269C>T, but according to our ESEFinder analysis (data not shown), this mutation is also predicted to be a splicing instead of a truncating mutation. Out of 10 patients, 4 developed osteosarcoma.18 However, the comparison is hard as the mutation was combined with a different truncating mutation in each of the nine heterozygous patients, whereas enhancement of the AltTr, likely driven by the deficit of the transcripts from the second mutant allele, might not occur in the only homozygous patient. Only a prolonged and careful follow-up of the siblings, now 9 years and 3 months old and 1 year and 7 months old, will allow validation of the suggested correlation between their genotype at the RECQL4 locus and the mild clinical presentation.
Two main conclusions can be drawn from our study. First, the novel C>T mutation carried by the investigated siblings permitted us to unveil a novel physiological alternative transcript of the RECQL4 gene. The role of this AltTr is currently unknown, although its expression in all the tested tissues suggests that it may have a functional role. Apart from the alternative splicing evidenced here, no other information is available on AS of RECQL4 and its contribution to disease. The second conclusion relates to the refinement of molecular diagnostics for patients with RTS and other RECQL4-related diseases. The RECQL4 gene has long been described to have a peculiar splicing pattern because of the short size of 13 of its 20 introns, accounting for improper splicing and deleterious effects of some intronic mutations.2, 19 Molecular characterisation of transcripts has occasionally disclosed the effect of RECQL4 mutations overlooked by DNA analysis.20
The current study shows that the functional effects of mutations identified by DNA analysis may be better interpreted by RNA analysis. Because of the above considerations, the clinical utility gene card for RTS21 recommends that the characterisation of RECQL4 mutations should always include RNA analysis.
References
Kitao S, Shimamoto A, Goto M et al: Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet 1999; 22: 82–84.
Wang LL, Gannavarapu A, Kozinetz CA et al: Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst 2003; 95: 669–674.
Larizza L, Roversi G, Volpi L : Rothmund-Thomson syndrome. Orphanet J Rare Dis 2010; 5: 2.
De Somer L, Wouters C, Morren MA et al: Granulomatous skin lesions complicating Varicella infection in a patient with Rothmund-Thomson syndrome and immune deficiency: case report. Orphanet J Rare Dis 2010; 5: 37.
Simon T, Kohlhase J, Wilhelm C, Kochanek M, De Carolis B, Berthold F : Multiple malignant diseases in a patient with Rothmund-Thomson syndrome with RECQL4 mutations: case report and literature review. Am J Med Genet A 2010; 152A: 1575–1579.
Fradin M, Merklen-Djafri C, Perrigouard C et al: Long-term follow-up and molecular characterization of a patient with a RECQL4 mutation spectrum disorder. Dermatology 2013; 226: 353–357.
Mehollin-Ray AR, Kozinetz CA, Schlesinger AE, Guillerman RP, Wang LL : Radiographic abnormalities in Rothmund-Thomson syndrome and genotype-phenotype correlation with RECQL4 mutation status. AJR Am J Roentgenol 2008; 191: W62–W66.
Croteau DL, Rossi ML, Ross J et al: RAPADILINO RECQL4 mutant protein lacks helicase and ATPase activity. Biochim Biophys Acta 2012; 1822: 1727–1734.
Beghini A, Castorina P, Roversi G, Modiano P, Larizza L : RNA processing defects of the helicase gene RECQL4 in a compound heterozygous Rothmund-Thomson patient. Am J Med Genet A 2003; 120A: 395–399.
Livak KJ, Schmittgen TD : Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25: 402–408.
Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR : An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 2006; 15: 2490–2508.
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR : ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acid Res 2003; 31: 3568–3571.
Sznajer Y, Siitonen HA, Roversi G et al: Atypical Rothmund-Thomson syndrome in a patient with compound heterozygous mutations in RECQL4 gene and phenotypic features in RECQL4 syndromes. Eur J Pediatr 2008; 167: 175–181.
Carlson AM, Thomas KB, Kirmani S, Lindor NM : Chronic tibial nonunion in a Rothmund-Thomson syndrome patient. Am J Med Genet A 2012; 158A: 2250–2253.
Shah KN : The diagnostic and clinical significance of cafe-au-lait macules. Pediatr Clin North Am 2010; 57: 1131–1153.
Wang LL, Levy ML, Lewis RA et al: Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. Am J Med Genet 2001; 102: 11–17.
Cabral RE, Queille S, Bodemer C et al: Identification of new RECQL4 mutations in Caucasian Rothmund-Thomson patients and analysis of sensitivity to a wide range of genotoxic agents. Mutat Res 2008; 643: 41–47.
Siitonen HA, Sotkasiira J, Biervliet M et al: The mutation spectrum in RECQL4 diseases. Eur J Hum Genet 2009; 17: 151–158.
Kitao S, Lindor NM, Shiratori M, Furuichi Y, Shimamoto A : Rothmund-Thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics 1999; 61: 268–276.
Balraj P, Concannon P, Jamal R et al: An unusual mutation in RECQ4 gene leading to Rothmund-Thomson syndrome. Mutat Res 2002; 508: 99–105.
Larizza L, Roversi G, Verloes A : Clinical utility gene card for: Rothmund-Thomson syndrome. Eur J Hum Genet 2013; 21: 792.
Acknowledgements
We thank the patients and their relatives for intensive cooperation in the study and Galliera Genetic Bank (Network of Telethon Genetic Biobank; project GTB07001) for providing us with lymphoblastoid cell lines from affected patient and control cases. We are grateful to the colleagues of the Pediatric Department, San Gerardo Hospital, Fondazione MBBM, University of Milano-Bicocca, Monza, Italy, for providing us the panel of tissue RNAs. This work was partially supported by AIRC Associazione Italiana per la Ricerca sul Cancro (Grant No. 4217 to LL) and Dote di Ricerca FSE, Regione Lombardia funding to EAC (ID project: 16304, 2011-2012).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Colombo, E., Fontana, L., Roversi, G. et al. Novel physiological RECQL4 alternative transcript disclosed by molecular characterisation of Rothmund–Thomson Syndrome sibs with mild phenotype. Eur J Hum Genet 22, 1298–1304 (2014). https://doi.org/10.1038/ejhg.2014.18
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.18
Keywords
This article is cited by
-
Translational biology of osteosarcoma
Nature Reviews Cancer (2014)