Abstract
Noonan syndrome (NS) is a developmental disorder characterized by short stature, facial dysmorphisms and congenital heart defects. To date, all mutations known to cause NS are dominant, activating mutations in signal transducers of the RAS/mitogen-activated protein kinase (MAPK) pathway. In 25% of cases, however, the genetic cause of NS remains elusive, suggesting that factors other than those involved in the canonical RAS/MAPK pathway may also have a role. Here, we used family-based whole exome sequencing of a case–parent trio and identified a de novo mutation, p.(Arg802His), in A2ML1, which encodes the secreted protease inhibitor α-2-macroglobulin (A2M)-like-1. Subsequent resequencing of A2ML1 in 155 cases with a clinical diagnosis of NS led to the identification of additional mutations in two families, p.(Arg802Leu) and p.(Arg592Leu). Functional characterization of these human A2ML1 mutations in zebrafish showed NS-like developmental defects, including a broad head, blunted face and cardiac malformations. Using the crystal structure of A2M, which is highly homologous to A2ML1, we identified the intramolecular interaction partner of p.Arg802. Mutation of this residue, p.Glu906, induced similar developmental defects in zebrafish, strengthening our conclusion that mutations in A2ML1 cause a disorder clinically related to NS. This is the first report of the involvement of an extracellular factor in a disorder clinically related to RASopathies, providing potential new leads for better understanding of the molecular basis of this family of developmental diseases.
Similar content being viewed by others
Introduction
Noonan syndrome (NS) is an autosomal dominant, clinically variable condition, with an estimated prevalence of 1 in 1000–2500.1 NS patients are characterized by facial dysmorphism, a wide spectrum of cardiac disease, reduced postnatal growth, variable cognitive deficits, and ectodermal and skeletal defects.2, 3, 4 Congenital heart defects are observed in a large proportion of NS patients, in particular pulmonary stenosis (66%), and hypertrophic cardiomyopathy (14%).5 Other relatively frequent clinical features of NS patients include webbed neck, cryptorchidism, bleeding tendency and hydrops fetalis. NS is genetically heterogeneous and mutations in PTPN11, SOS1, KRAS, NRAS, RAF1, BRAF, SHOC2, CBL and RIT1 account for approximately 75% of affected individuals.4, 6 To date, all mutations causing NS result in enhanced activation of signal transducers belonging to the RAS/mitogen-activated protein kinase (MAPK) pathway.7
To identify new genetic causes of NS, we used a family-based whole-exome-sequencing approach in a case–parent trio with sporadic disease to detect de novo changes in the proband. Genetic testing had previously excluded mutations in known NS genes. We identified a de novo mutation in α-2-macroglobulin (A2M)-like-1 (A2ML1), which encodes the secreted protease inhibitor A2ML1. A2ML1 was resequenced in a cohort of cases with a clinical diagnosis of NS. Moreover, to provide more evidence for the involvement of the gene in this disorder resembling NS, protein modeling was performed and a2ml1 mutants were functionally characterized in zebrafish.
Materials and methods
Patients
The individual (case 1) selected for exome sequencing had a clinical diagnosis of NS based on the criteria defined by van der Burgt et al.3 For Sanger sequencing of the A2ML1 gene, we obtained a total of 295 DNA samples from unrelated individuals with NS or a clinically related phenotype. All patients, including patient 1, had been tested negative for mutations in previously identified disease genes (PTPN11, KRAS, SOS1, NRAS, SHOC2, CBL, RAF1, BRAF, MAP2K1, MAP2K2, HRAS and RIT1). These samples were collected from three genetic centers (35 patients selected by IPATIMUP, Porto, Portugal, 120 patients from the Department of Human Genetics, RUNMC, Nijmegen, the Netherlands and 140 patients from the Department of Pediatrics, Università Cattolica del Sacro Cuore (UCSC) and Department of Hematology, Oncology and Molecular Medicine, Istituto Superiore di Sanità (ISS), Rome, Italy). The clinical diagnosis for NS or suggestive of a related trait was made on the basis of standardized clinical criteria assessed by experienced clinical geneticists.
Genomic DNA from whole blood was extracted using standard protocols. This study was approved by the Review Boards of all participating institutions. Informed consent to participate in the study was obtained for all individuals as well as permission to publish photographs of individuals shown in Figure 1.
Photographs and pedigrees of families diagnosed with Noonan-like syndrome with likely pathogenic mutations in A2ML1. (a) Family 1 – case 1. De novo mutation p.(Arg802His); (b) Family 2 – cases 2, 3 and 4. Familial p.(Arg802Leu). The sister of case 2 is reported upon heteroanamnesis to have the same clinical phenotype. However, neither DNA nor detailed clinical information was available for this study. (c) Family 3 – cases 5, 6, 7 and 8. Familial p.(Arg592Leu). Clinical details of all cases are presented in Table 3. Note, the absence of genotypes for (un)affected family members indicates that DNA samples of these individuals were not available for testing. Black solid squares/circles represent clinically affected individuals, whereas as open squares/circles represent healthy individuals.
Exome sequencing for de novo variants
Exome sequencing was performed as described before.8 In brief, exome enrichment was performed using a SOLiD optimized SureSelect Human Exome Kit (version 1, 37 Mb; Agilent Technologies Inc., Santa Clara, CA, USA), subsequently followed by SOLiDv3 PLUS sequencing. Read mapping and variant calling for the patient–parent trio was performed as described before.8 All candidate de novo mutations present in the individual with NS were independently validated using Sanger sequencing.
Mutation analysis
For 120 individuals with NS from the Department of Human Genetics, Nijmegen, the Netherlands and 35 individuals with NS from IPATIMUP, Porto, Portugal, all coding exons and flanking intronic sequences of A2ML1 were PCR amplified and analyzed by Sanger sequencing or the Ion PGM system, respectively, using standardized protocols. All variants and protein codons are referred to according to NM_144670.3 using HGVS nomenclature. Mutations identified were checked for de novo occurrence whenever parental DNAs were available. For the 140 individuals with NS or a clinically related phenotype selected from the ISS and UCSC, Rome, Italy, exons 15, 16, 19, 24, 25, 26, 29, 31 and 32 and flanking intronic sequences were considered, based on the identification of possible pathogenic mutations in the first cohort. For all variants detected, an in silico-based method was used to assess the effect of the mutation (Alamut software, version 2.1; http://www.interactive-biosoftware.com/) in addition to an assessment of variant pathogenicity according to guidelines by the CMGS and VKGL, for the British and Dutch Molecular Genetic Societies, respectively.9
Protein modeling of mutations identified in A2ML1
Pathogenic mutations were modeled for their effect on protein function. Because the three-dimensional structure of A2ML1 (Uniprot entry A8K2U0) is unknown, protein modeling was performed using PDB entry 4ACQ, which represents the crystal structure of A2M. Overall, A2M and A2ML1 share 40% sequence identity. Mutations used for modeling occur within a sequence stretch that is highly conserved (Supplementary Figure 1). Graphical representations for mutations modeled on 4ACQ were generated and analyzed through project HOPE and YASARA.10, 11 Residue numbers in the paper are based on residues in A2ML1 (not 4ACQ). The residues in A2ML1 and their counterparts in 4ACQ and zebrafish (see section below) are listed in Table 1.
Cell culture, transfection and immunoblotting
COS7 cells and 293T cells were maintained using standard protocols and transfected using polyethyleneimine (PEI) (Sigma-Aldrich, St Louis, MO, USA). Cells were lysed directly in 2 × SDS sample buffer (125 mM Tris-HCl (pH 6.8), 20% glycerol, 4% SDS, 2% β-mercaptoethanol and 0.04% bromophenol blue) and boiled. Lysates were separated on a 10% SDS-polyacrylamide gel and blotted using mouse anti-phospho-extracellular signal-regulated kinase (ERK), rabbit anti-ERK (Cell Signalling Technologies, Boston, MA, USA) and rabbit anti-GFP (Torrey Pines Biolabs, Secaucus, NJ, USA). Enhanced chemiluminescence was used to detect signal from HRP-conjugated secondary antibodies (BD Bioscience, San Jose, CA, USA).
Zebrafish injections and in situ hybridization
Zebrafish were kept and embryos were raised under standard conditions. Zebrafish a2ml1 (GenBank: BC125959.1) was cloned into pCS2+. Mutants were derived by PCR and verified by sequencing. The gene encoding EGFP was fused in frame to the 3′ end of (mutant) a2ml1, which allows monitoring of the expression of (mutant) A2ml1 protein in zebrafish embryos. Cytomegalovirus (CMV)-driven expression vectors for (mutant) A2ml1, CMV:a2ml1-egfp, were injected into zebrafish embryos at the one-cell stage. Synthetic RNA encoding mutant Shp2-D61G was injected at the one-cell stage as a control.12 Morphological phenotypes were assessed at 4 days postfertilization (dpf). Embryos were anesthesized at 4 dpf with MS-222 (Sigma-Aldrich), fixed in 4% PFA and the cartilage was stained with Alcian Blue. The width of the ceratohyal and the distance to the tip of Meckel’s cartilage were determined using the Image J software (http://rsb.info.nih.gov/ij/) and the ratio was determined as a direct measure for craniofacial defects. Averages were determined and a Student’s t-test was carried out to determine whether the differences between the different conditions were statistically significant. To investigate cardiac defects, embryos were fixed at 55 hpf and in situ hybridizations were carried out essentially as described13 using probes specific for myl7.14
Results
Exome sequencing in an NS case–parent trio
Exome sequencing in case 1 (Figure 1a) and her parents revealed four potential de novo mutations, two of which were validated by Sanger sequencing and confirmed to be of de novo origin (Supplementary Table 1). The first mutation is predicted to lead to p.(Arg129Pro) in OR12D3 (NM_030959.2:c.386G>C), encoding an olfactory receptor. Based on the function of the gene product (odor perception) combined with the low evolutionary conservation at base pair level (PhyloP 0.55), we do not consider this variant to be relevant for the phenotype of the patient, but assume that this de novo mutation reflects the human per-generation background mutation rate.15 The second de novo mutation was detected in A2ML1 (NM_144670.3:c.2405G>A) and is predicted to lead to p.(Arg802His). This de novo mutation occurred at a highly conserved nucleotide (PhyloP 3.41) and affects a residue within the A2M domain (Figure 2a). It is noteworthy that the c.2405G>A variant in A2ML1 is also reported in 4/12 194 alleles in the NHLBI Exome Sequencing Project (ESP) (http://evs.gs.washington.edu/EVS/). Further analysis of the ESP database indicated that several frequently reported causal mutations in known NS genes, including PTPN11, are described in this database at similar frequency (Table 2).
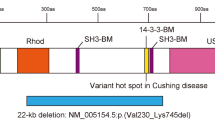
A2ML1 domain structure and modeling of the intramolecular interactions involving p.Arg802. (a) Schematic representation of A2ML1 protein domain structure and mutations in individuals with Noonan syndrome-like disorder. Numbers correspond to amino-acid positions of A2ML1. MG, macroglobulin-like domain; BRD, bait region domain; CUB, complement C1r/C1s, Uegf, Bmp1 domain; TED, thiol-ester domain; RBD, receptor binding domain. (b) Crystal structure of A2M (pdb code 4ACQ). Overall structure with p.Arg598 and p.Arg804 highlighted, corresponding to p.Arg592 and p.Arg802 in A2ML1, respectively. (c) Closeup of A2M-R804, corresponding to A2ML1-R802 (in green), which interacts with A2M-E908 (A2ML1-E906) (in blue) through hydrogen bonds (yellow dashed lines) and electrostatic interactions. A2M-E941 and A2M-E942 (in blue) may also interact with A2M-R802, but are not conserved in A2ML1 (p.P939 and p.D940, respectively).
Mutation analysis of the A2ML1 gene in an NS cohort
Next, 155 individuals with a clinical diagnosis of NS were analyzed for mutations in A2ML1 by direct Sanger sequencing and Ion PGM system. This screen led to the identification of two additional likely causal missense mutations in two unrelated individuals (Figures 1b and c and Table 2). The mutation identified in case 2, c.2405G>T, affects the same residue as identified in family 1, but is predicted to result in a different substitution, p.(Arg802Leu). Segregation analysis showed that the mutation is transmitted to his son (case 3), who is also diagnosed with the same disorder resembling NS. His mother (case 4), who does not show typical features of NS, also carries the mutation (Figure 1b). In the proband of family 3 (case 5), a c.1775G>T mutation (p.(Arg592Leu)) was detected. Familial segregation studies revealed the same mutation in her daughter (case 6) and her father (case 7), who were both clinically diagnosed with a disorder resembling NS (Figure 1c). Strikingly, the patient’s first pregnancy resulted in intrauterine fetal death at 29 weeks of gestation because of hydrops fetalis (case 8; Figure 1c). Pathological examination of the fetus showed facial features suggestive for NS. Molecular analysis of fetal DNA indicates maternal inheritance of the A2ML1 mutation p.(Arg592Leu). The clinical features of all individuals with an A2ML1 mutation are summarized in Table 3. A full description of the clinical data is provided in the supplement.
Screening of selected exons of A2ML1 in an additional cohort of 140 individuals with a clinical phenotype fitting or suggestive for NS did not reveal any pathogenic mutations. All variants detected are shown in Supplementary Table 2.
Protein modeling of mutations detected in A2ML1
All three A2ML1 mutations, p.(Arg592Leu), p.(Arg802His) and p.(Arg802Leu), affect highly conserved residues among orthologs (Supplementary Figure 2). For protein modeling of the mutation locations and their predicted effect, the crystal structure of A2M (pdb 4ACQ) was used (Figure 2 and Table 1). A2ML1 is highly homologous to A2M and it is highly likely that protein folding of the two related proteins is similar (Supplementary Figure 1). Arginine 592, corresponding to A2M p.Arg598, is located at the edge of the bait region domain and is buried in a large cavity on the back face of the A2ML1 monomer,16 whereas arginine 802, corresponding to A2M p.Arg804, is located within macroglobulin-like domain 7 and is positioned more at the surface of the protein (Figure 2b). Mutations of either residue likely lead to conformational changes in A2ML1 by loss of hydrogen bonds or charge interactions. In more detail, within the intramolecular binding network, disease-associated mutations of p.Arg802 were predicted to disrupt the interaction with p.Glu906 (Figure 2c).
Functional consequences of A2ML1 mutations
A2ml1 is relative highly conserved in zebrafish (38% identity) and we investigated the functional consequences of expression of mutant A2ml1 on zebrafish development. A2ml1 is predominantly expressed in the liver during embryonic development and morpholino-mediated knockdown of a2ml1 has previously been shown to induce defective liver development.17 We introduced human A2ML1 mutations (p.R802H, p.R802L and p.S592L, human numbering for clarity, see Table 1) in zebrafish A2ml1, C-terminally fused with GFP, allowing to monitor (mutant) A2ml1 expression in tissue culture cells and during development (Supplementary Figures 3 and 4).
Expression of most mutant NS-associated genes in tissue culture cells augments activation of the RAS/MAPK pathway, measurable by excessive ERK/MAPK phosphorylation in cycling cells. To investigate the effect of mutant A2ml1 on the RAS/MAPK pathway, we expressed mutant A2ml1-gfp fusion proteins in HEK-293T and COS7 cells to determine the levels of ERK/MAPK phosphorylation. Expression of A2ml1 mutants in normally growing HEK-293T cells or in serum-stimulated COS7 cells did not significantly enhance ERK/MAPK phosphorylation (Supplementary Figure 3). These results suggest either that (mutant) A2ml1 does not modulate ERK/MAPK signaling or that HEK-293T cells and COS7 cells are not responsive to (mutant) A2ml1.
The effect of (mutant) A2ml1 on embryonic development was investigated by microinjection of CMV-promoter-driven expression vectors for wild-type or mutant A2ml1 in zebrafish embryos at the one-cell stage. Exogenous A2ml1-GFP was detected throughout the zebrafish embryo, despite mosaicism of the transgene, which is consistent with A2ml1-GFP being a secreted factor (Supplementary Figure 4). Expression of mutant A2ml1 resulted in developmental defects in zebrafish embryos that involved the heart and craniofacial structures, whereas expression of GFP alone or wild-type A2ml1 did not affect zebrafish development (Figure 3). The morphological defects elicited by mutant A2ml1 resembled those induced by NS-associated Shp2-D61G and N-Ras-I24N.12, 18 Of note, A2ml1-mutation-induced morphological defects appeared later in development (apparent from 3 dpf onwards) and were less severe than those observed for Shp2-D61G-induced defects (from 6 hpf onwards).
Mutant A2ml1 causes a developmental disorder resembling NS. (a) Expression of mutant A2ml1 results in developmental defects in zebrafish embryos at 4 dpf. The craniofacial defects were highlighted by cartilage staining using Alcian Blue. The heads are broader and the faces blunted; ‘n’ refers to the number of embryos examined for x/y ratio. (b) To quantify the craniofacial defects, the ratio of the width of the ceratohyal and the distance to Meckel’s cartilage (x and y, respectively in bottom left panel of (a)) was determined. The averages are plotted, error bars indicate standard error of the mean. Student’s t-test to compare ratios with non-injected control (NIC) indicate that wild-type A2ml1 is not significantly different, but mutant A2ml1 and mutant Shp2 are significantly enhanced compared with NIC as indicated. Note that human numbering of residues was used for A2ml1 mutants. (c) In situ hybridization of 55 hpf embryos using a heart-specific probe (myl7) that stains cardiomyocytes. Cardiac looping was assessed as normal looping (loop) or impaired looping (no loop) as indicated schematically. The number of no loop hearts in embryos expressing mutant A2ml1 or mutant Shp2/total number of embryos is indicated in the bottom right corner of each panel.
Alcian Blue staining of cartilaginous structures in 4 dpf zebrafish embryos revealed craniofacial defects that are characteristic of NS. Morphometric analysis showed statistically significant broadening of the head and blunting of the faces in embryos expressing mutant A2ml1 (Figure 3b). Also, in situ hybridization using the heart-specific probe myl7 (formerly known as cmlc2) indicated that cardiac looping was impaired in mutant a2ml1-injected embryos in a similar manner as in Shp2-D61G-expressing embryos (Figure 3c).
Protein modeling suggested that the p.(Arg802His) and p.(Arg802Leu) mutations would lead to a loss of the interaction between p.Arg802 and p.Glu906. Therefore, p.Glu906 was mutated to leucine and this mutant was expressed in zebrafish embryos to study the functional defect. Expression of mutant A2ml1-E906L induced developmental defects to a similar extent as NS-associated A2ml1-R802L mutants (Figure 3b), further supporting a causal role of A2ML1 mutations involving substitution of p.Arg802.
Discussion
Analysis of a case–parent trio with a clinical suspicion of NS revealed a de novo mutation in A2ML1 (c.2405G>A; p.(Arg802His)). A2ML1 encodes the protease inhibitor A2ML1, which is a member of the α-macroglobulin superfamily of proteins that contains both complement components and protease inhibitors. These proteins display a unique trap mechanism of inhibition, by which the A2M inhibitor undergoes a major conformational change upon its cleavage by a protease, thereby trapping the protease and blocking it from subsequent substrate binding.19 Moreover, A2ML1 binds to the lipoprotein receptor-related protein 1 (LRP1) receptor,20 an upstream activator of the MAPK/ ERK cascade.21 Additionally, LRP1 directly interacts with CBL.22 Taken together, these reports suggest that A2ML1 may act upstream of signaling pathways known to be involved in NS.
Two additional missense mutations in families with a disorder clinically resembling NS were identified by Sanger sequencing of the entire coding region of the gene (c.2405G>T; p.(Arg802Leu) and c.1775G>T; (p.(Arg592Leu)). The frequency of A2ML1 mutations in the cohort tested in this study is therefore approximately 1%. However, as mutations in the major NS genes had already been excluded, the contribution of A2ML1 mutations to the total population clinically diagnosed with NS is expected to be <0.5%. The fact that two of the three A2ML1 mutations have been reported in the EVS database with a frequency comparable to that in the NS patients could either mean that the A2ML1 mutations are not pathogenic or that the phenotypic features of people with an A2ML1 mutation is milder and they are unrecognized in the general population (making A2ML1 a relatively high-frequency gene in mild NS). The fact that case 4 does not show phenotypic features of NS would fit both of these theories, as cases with non-penetrance, which albeit very rare, have previously been described in NS.23 Evidence for the pathogenicity of the A2ML1 mutations therefore had to be provided by functional studies.
Whereas most NS-associated mutations have an effect on RAS/MAPK signaling, we did not detect RAS/MAPK activation in response to expression of A2ML1 mutations in HEK293T cells or COS7 cells. Cell-type-specific MAPK activation by NS-associated genes is not unprecedented; NS-associated mutant SHOC2 enhances stimulus-induced MAPK activation in Neuro2A cells, but not in Cos-1 or 293T cells.24 Hence, our results do not exclude involvement of A2ML1 in NS, but either suggest that A2ML1 does not modulate ERK/MAPK signaling or that COS7 and HEK-293T cells are irresponsive to A2ML1. Future studies of A2ML1 variants in cell systems expressing LRP1 receptor might shed more light on the role of A2ML1 in ERK/MAPK signaling.
Expression of mutant A2ml1 in zebrafish embryos resulted in developmental defects that were characterized by craniofacial and cardiac defects, resembling those induced by the expression of mutant Shp2. The A2ml1-mutation-induced morphological defects appear, however, later in development (apparent from 3 dpf onwards) and are less severe than those observed for Shp2-D61G-induced defects (from 6 hpf onwards). In line with this notion, the affected individuals in families 2 and 3, who transmitted the A2ML1 mutation to their offspring, do not fulfill all classical van der Burgt criteria for NS,3 but present with a less severe phenotype suggestive of the disorder. Of note, phenotypic variability is well known for NS, and more explicitly, not all patients with NS have a cardiac malformation.5, 25 Also, the affected individuals in the two families with an A2ML1 mutation do not have a heart defect, while the same mutations in zebrafish cause cardiac malformation. Zebrafish mutants of the most recently described NS gene, RIT1, show incomplete looping of the heart and hypoplastic heart chambers, whereas the heart phenotype was variable or even absent in patients with RIT1 mutations.6
Finally, we hypothesized that a conformational change in A2ML1 is the underlying mechanism leading to the developmental defects resembling NS. This conformational change may lead to destabilization of A2ML1, or it may interfere with its trap mechanism of inhibition. As both intragenic deletions of A2ML1 (reported in the Database of Genomic Variants (http://projects.tcag.ca/variation/)) and frameshift mutations are detected in healthy controls (Supplementary Table 2), haploinsufficiency is highly unlikely as the underlying disease mechanism. Given that the A2M family of proteins acts in multimeric complexes, it is expected that conformational changes might have a dominant-negative effect or lead to gain of function of the complex. As mutation of p.Arg802 likely disrupts the interaction with p.Glu906, we reasoned that mutation of p.Glu906 should have the same effect on A2ML1 function as mutation of p.Arg802. Indeed, expression of mutant A2ml1-E906L induced developmental defects to a similar extent as NS-associated A2ml1-R802L mutants, providing additional support that A2ML1 is involved in NS.
In summary, our results provide evidence that mutations in A2ML1 are a cause of Noonan-like syndrome, with a variable phenotype ranging from severe (resulting in intrauterine fetal death) to very mild (or even non-penetrance). Although mutations in this gene did not lead to detectable enhanced activation of the RAS/MAPK pathway in HEK293T cells or COS7 cells, expression of A2ml1 mutants in zebrafish embryos induced developmental defects that are comparable to mutations of other NS genes. Thus, we identified a causal role of an extracellular factor in NS for the first time and our results pave the way for further exploration of the function of A2ML1, its binding partners/receptors and other relevant extracellular cascades in the pathogenicity of NS.
Accession codes
References
Mendez HM, Opitz JM : Noonan syndrome: a review. Am J Med Genet 1985; 21: 493–506.
Allanson JE : Noonan syndrome. J Med Genet 1987; 24: 9–13.
van der Burgt I : Noonan syndrome. Orphanet J Rare Dis 2007; 2: 4.
Roberts AE, Allanson JE, Tartaglia M, Gelb BD : Noonan syndrome. Lancet 2013; 381: 333–342.
Colquitt JL, Noonan JA : Cardiac findings in Noonan syndrome on long-term follow-up. Congen Heart Dis 2013; 9: 144–150.
Aoki Y, Niihori T, Banjo T et al: Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am J Hum Genet 2013; 93: 173–180.
Tartaglia M, Gelb BD, Zenker M : Noonan syndrome and clinically related disorders. Best Pract Res Clin Endocrinol Metab 2011; 25: 161–179.
de Ligt J, Willemsen MH, van Bon BW et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929.
Bell JB, D., Sistermans E, Ramsden SC Practice guidelines for the Interpretation and Reporting of Unclassified Variants (UVs) in Clinical Molecular Genetics, 2007 Available at: http://www.cmgs.org/bpgs/pdfs%20current%20bpgs/UV%20 GUIDELINES%20ratified.pdf (last accessed 2 June 2014).
Krieger E, Vriend G : Models@Home: distributed computing in bioinformatics using a screensaver based approach. Bioinformatics 2002; 18: 315–318.
Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G : Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinform 2010; 11: 548.
Jopling C, van Geemen D, den Hertog J : Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet 2007; 3: e225.
Thisse C, Thisse B : High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 2008; 3: 59–69.
Yelon D, Horne SA, Stainier DY : Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol 1999; 214: 23–37.
Roach JC, Glusman G, Smit AF et al: Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 2010; 328: 636–639.
Marrero A, Duquerroy S, Trapani S et al: The crystal structure of human alpha2-macroglobulin reveals a unique molecular cage. Angew Chem 2012; 51: 3340–3344.
Hong SK, Dawid IB : Alpha2 macroglobulin-like is essential for liver development in zebrafish. PLoS One 2008; 3: e3736.
Runtuwene V, van Eekelen M, Overvoorde J et al: Noonan syndrome gain-of-function mutations in NRAS cause zebrafish gastrulation defects. Dis Models Mech 2011; 4: 393–399.
Galliano MF, Toulza E, Gallinaro H et al: A novel protease inhibitor of the alpha2-macroglobulin family expressed in the human epidermis. J Biol Chem 2006; 281: 5780–5789.
Galliano MF, Toulza E, Jonca N, Gonias SL, Serre G, Guerrin M : Binding of alpha2ML1 to the low density lipoprotein receptor-related protein 1 (LRP1) reveals a new role for LRP1 in the human epidermis. PLoS One 2008; 3: e2729.
Geetha N, Mihaly J, Stockenhuber A et al: Signal integration and coincidence detection in the mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) cascade: concomitant activation of receptor tyrosine kinases and of LRP-1 leads to sustained ERK phosphorylation via down-regulation of dual specificity phosphatases (DUSP1 and -6). J Biol Chem 2011; 286: 25663–25674.
Takayama Y, May P, Anderson RG, Herz J : Low density lipoprotein receptor-related protein 1 (LRP1) controls endocytosis and c-CBL-mediated ubiquitination of the platelet-derived growth factor receptor beta (PDGFR beta). J Biol Chem 2005; 280: 18504–18510.
Tartaglia M, Kalidas K, Shaw A et al: PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype–phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet 2002; 70: 1555–1563.
Cordeddu V, Di Schiavi E, Pennacchio LA et al: Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat Genet 2009; 41: 1022–1026.
Zenker M, Voss E, Reis A : Mild variable Noonan syndrome in a family with a novel PTPN11 mutation. Eur J Med Genet 2007; 50: 43–47.
Oishi K, Gaengel K, Krishnamoorthy S et al: Transgenic Drosophila models of Noonan syndrome causing PTPN11 gain-of-function mutations. Hum Mol Genet 2006; 15: 543–553.
Martinelli S, De Luca A, Stellacci E et al: Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. Am J Hum Genet 2010; 87: 250–257.
Acknowledgements
We thank Hanka Venselaar for bioinformatics support in protein modeling and Martina Ruiterkamp-Versteeg, Petra de Vries, Suzanne Keijzers-Vloet and Martine van Zweeden for technical assistance. This work was funded, in part, by a grant from the Research Council for Earth and Life Sciences (ALW 819.02.021) with financial aid from the Netherlands Organization for Scientific Research (NWO) (to JdH), Telethon-Italy (GGP13107 to MT), Fundação para a Ciência e Tecnologia (PTDC/BIM-MEC/0650/2012 to JLC) and PPS5 Consórcio DoIT (ADI – Agência de Inovação to JLC). IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Education and Science and is partially supported by FCT, the Portuguese Foundation for Science and Technology.
Deposition of genetic data
The data obtained in this study are submitted to LOVD, an online gene-centered collection and display of DNA variations (http://databases.lovd.nl/shared/genes).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Vissers, L., Bonetti, M., Paardekooper Overman, J. et al. Heterozygous germline mutations in A2ML1 are associated with a disorder clinically related to Noonan syndrome. Eur J Hum Genet 23, 317–324 (2015). https://doi.org/10.1038/ejhg.2014.115
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.115
This article is cited by
-
Molecular and clinical profile of patients referred as Noonan or Noonan-like syndrome in Greece: a cohort of 86 patients
European Journal of Pediatrics (2022)
-
Integrated in silico MS-based phosphoproteomics and network enrichment analysis of RASopathy proteins
Orphanet Journal of Rare Diseases (2021)
-
The clinical significance of A2ML1 variants in Noonan syndrome has to be reconsidered
European Journal of Human Genetics (2021)
-
LZTR1 facilitates polyubiquitination and degradation of RAS-GTPases
Cell Death & Differentiation (2020)
-
Delineation of LZTR1 mutation-positive patients with Noonan syndrome and identification of LZTR1 binding to RAF1–PPP1CB complexes
Human Genetics (2019)