Abstract
The general transcription factor IID (TFIID) initiates RNA polymerase II-mediated eukaryotic transcription by nucleating pre-initiation complex formation at the core promoter of protein-encoding genes. TAF1, the largest integral subunit of TFIID, contains an evolutionarily conserved yet poorly characterized central core domain, whose specific mutation disrupts cell proliferation in the temperature-sensitive mutant hamster cell line ts13. Although the impaired TAF1 function in the ts13 mutant has been associated with defective transcriptional regulation of cell cycle genes, the mechanism by which TAF1 mediates transcription as part of TFIID remains unclear. Here, we present the crystal structure of the human TAF1 central core domain in complex with another conserved TFIID subunit, TAF7, which biochemically solubilizes TAF1. The TAF1-TAF7 complex displays an inter-digitated compact architecture, featuring an unexpected TAF1 winged helix (WH) domain mounted on top of a heterodimeric triple barrel. The single TAF1 residue altered in the ts13 mutant is buried at the junction of these two structural domains. We show that the TAF1 WH domain has intrinsic DNA-binding activity, which depends on characteristic residues that are commonly used by WH fold proteins for interacting with DNA. Importantly, mutations of these residues not only compromise DNA binding by TAF1, but also abrogate its ability to rescue the ts13 mutant phenotype. Together, our results resolve the structural organization of the TAF1-TAF7 module in TFIID and unveil a critical promoter-binding function of TAF1 in transcription regulation.
Similar content being viewed by others
Introduction
Transcription of protein-encoding genes is vital for the survival of all living organisms. Its precise regulation is critical for normal cell proliferation, differentiation, and development. In eukaryotes, this transcriptional process is orchestrated by RNA polymerase II (RNAPII) and a set of general transcription factors known as TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH1,2. These factors are sequentially recruited to the core promoters of RNAPII-dependent genes and assemble into a transcriptionally competent pre-initiation complex (PIC). By directly recognizing the core promoter region of target genes, TFIID is responsible for initiating and nucleating functional PIC assembly at the first step of transcription3,4.
TFIID is a multi-subunit protein complex, consisting of the TATA-binding protein (TBP) and 12-14 TBP-associated factors (TAFs)5,6,7. TAF1 is the largest subunit of TFIID, which interacts with TBP through an N-terminal TBP-binding sequence. TAF1 has also been reported to possess various biochemical activities including protein phosphorylation, histone acetylation, and acetylated histone tail recognition activities8. In human TAF1, these activities have been mapped to the two terminal kinase domains, a central domain of unknown function (DUF3591 domain), and two tandem bromodomains, respectively (Figure 1A)8,9,10,11,12. Due to its large size and solubility issues, structural information for human TAF1 is limited to the approximate cryoEM mapping of the subunit within TFIID and a high resolution crystal structure of its tandem bromodomains5,12,13. The structural basis of all the other biochemical functions of TAF1 remains elusive.
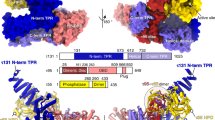
Overall structure of the TAF1-TAF7 complex. (A) Domain organization of TAF1 and the TAF1 and TAF7 fragments crystallized by the in situ proteolysis method. Different structural domains of the crystallized TAF1 and TAF7 fragments are labeled and colored. Regions outlined by dotted lines represent the proteolytically removed segments. (B) Surface representations of the TAF1-TAF7 complex with structural domains colored with the same scheme as shown in A. TAF1 and TAF7 are separately shown on the right from the same angle. (C, D) Ribbon diagram of the TAF1-TAF7 complex structure in two orthogonal views. The TAF1 and TAF7 protein domains are colored with the same scheme as in A. The TAF1 G716 residue mutated in the ts13 hamster cell line is colored in red and indicated by a letter “G”. Dashed lines represent loop regions that are not visible in the crystal structure.
Among the multiple TAF1 domains, the DUF3591 domain is highly conserved from yeast to human. Its functional importance is underscored by a single missense mutation (G716D) within the domain, which causes the temperature-sensitive mutant hamster cell line ts13 to undergo a G1 cell cycle arrest14,15,16. We have also previously demonstrated that a deletion mutation (Δ848-850) within this TAF1 core domain renders the protein unable to complement the ts13 defects in both cell cycle progression and cyclin D1 transcription17. In the present study, we successfully purified the DUF3591 domain of human TAF1 by co-expression with another TFIID subunit, TAF7, which functions as a dissociable regulator of TAF118,19. We herein report the crystal structure of this heterodimeric complex at 2.3 Å. In conjunction with functional analyses and structural comparison with the recently reported yeast TAF1-TAF7 complex20, our results advance the structural understanding of the TAF1-TAF7 module of human TFIID and reveal a critical DNA binding activity within the TAF1 DUF3591 domain.
Results
Overall TAF1-TAF7 Structure
The DUF3591 domain of TAF1 has been linked to the transcriptional regulation of a subset of essential genes, such as G1 cyclins and major histocompatibility class I genes21,22, but has yet to be stably isolated in a functional form in solution. We obtained a soluble form of human TAF1 DUF3591 domain by co-expressing it with full-length human TAF7. The TAF1-TAF7 complex was crystallized using an in situ proteolysis approach. Its structure was determined by single-wavelength anomalous diffraction (SAD) with a platinum-derivatized crystal (Supplementary information, Table S1). The final model contains the majority of the purified TAF1 fragment (amino acids 609-1109) and the N-terminal fragment of TAF7 (amino acids 11-154) with two TAF1 loops and one TAF7 loop missing, none of which is conserved across eukaryotic species (Supplementary information, Figures S1 and S2).
The TAF1-TAF7 crystal structure reveals several unpredicted structural domains, which assemble into a compact architecture with the overall shape of a pyramid (Figure 1). The apex of the pyramid is formed by a winged helix (WH) domain, which is embedded in the middle of the TAF1 DUF3591 fragment. A wide triple barrel constructed by both proteins shapes the base of the architecture. Additional α-helices and loop regions of TAF1 constitute the middle portion of the pyramid, holding the top WH domain and the bottom triple barrel together. At the primary sequence level, the TAF1 DUF3591 domain can be considered an α-helical polypeptide interrupted by the triple barrel-forming sequence and WH domain (Figure 1A). The TAF7 N-terminal fragment contributes mainly to the construction of the triple barrel core (Figure 1B). Its C-terminal region, however, assumes an independent coiled structure, interacting with both the triple barrel and TAF1 C-terminal α-helical regions (Figure 1). With the majority of its polypeptide located at one corner of the pyramid base, TAF7 inserts a long β-hairpin through the center of the complex and reaches the solvent on the other side (Figure 1D). Together, TAF1 and TAF7 bury a total surface area of ∼3900 Å2, consistent with the formation of a stable complex in solution.
The triple barrel interface
The triple barrel core formed between TAF1 and TAF7 constitutes the major extensive interface of this compact heterodimeric complex. It contains 15 parallel and anti-parallel β-strands, which interweave into three continuous β-barrels (Figure 2A). The distal two β-barrels are approximately parallel to each other, giving rise to a third orthogonal barrel in the middle (Figure 2B). The triple barrel fold was first discovered in the crystal structure of the RAP74-RAP30 complex of TFIIF, which acts downstream of TFIID in PIC assembly (Figure 2C)23. It was later found in several other protein complexes, mediating protein hetero-dimerization24,25,26. The TAF1-TAF7 triple barrel can be superimposed on TFIIF with a root-mean-square deviation (RMSD) of 2.8 Å out of 158 aligned Cα atoms, suggesting that parts of the two complexes might share a common evolutionary origin. By convention, we name the two distal β-barrels “TAF1 barrel” and “TAF7 barrel” based on their predominant polypeptide composition.
The TAF1-TAF7 heterodimeric triple barrel. (A) Topology diagram of the TAF1-TAF7 triple barrel in the context of the entire crystallized complex of TAF1 and TAF7. Dashed lines represent loop regions not visible in the crystal structure. The Gly-rich motif of TAF1 and the Arg-rich motif of TAF7 are highlighted. (B) Ribbon diagram of the TAF1-TAF7 triple barrel core. The pseudo two-fold symmetry axis relating the two distal barrels is indicated by the black oval. The residue G716, which is mutated in the ts13 hamster cell line, is colored in red and indicated. (C) Ribbon diagram of the RAP74-RAP30 triple barrel.
Similar to TFIIF, the TAF1 and TAF7 barrels are formed by domain swapping and can be related by a pseudo two-fold symmetry axis (Figure 2B). Distinct from TFIIF, however, the TAF1-TAF7 triple barrel has an asymmetric number of β-strands from the two proteins in the distal barrels – while TAF1 contributes three β-strands to the eight-stranded TAF7 barrel, TAF7 only inserts a two-stranded β-hairpin into the seven β-stranded TAF1 barrel (Figure 2A and 2B). The TAF7 β-hairpin further distinguishes itself from other β-strands of all known triple barrel structures by burying two conserved salt bridges in the TAF1 barrel (Supplementary information, Figure S3). Overall, the asymmetry of the triple barrel core is dictated by the specific interactions between TAF1 and TAF7, while its unique structural properties implicate possible additional functions beyond simple scaffolding.
The G716 residue, which is mutated in the ts13 mutant, is strictly conserved among all TAF1 orthologs. This residue maps to the N-terminal end of the strand β3 and lies at the top of the TAF1 barrel. It is completely buried from the solvent by two α helices in the α-helical domain, which in turn closely pack against the WH domain (Figure 2A and 2B). In this position, the G716D mutation will likely perturb the local structure of the TAF1 barrel and affect the topological configuration between the triple barrel and the WH domain at non-permissive temperature.
Interfaces outside the triple barrel
In addition to the formation of the triple barrel, a large portion of the TAF1 fragment and the C-terminal segment of the TAF7 polypeptide adopt a mixture of α helical and loop structures, which closely wrap around the triple barrel core and hold the TAF1 WH domain in place (Figure 1B and 1C). The very N-terminal portion of the TAF1 fragment forms two short helices flanking a long loop, clamping on top of the TAF1 barrel (Supplementary information, Figure S3). The C-terminal region of the TAF1 DUF3591 domain, meanwhile, folds into five α-helices of variable lengths and covers both the top and the outer wall of the same barrel. Furthermore, the C-terminal segment of the TAF7 fragment adopts a highly coiled structure, plugging one end of the middle barrel and closely interacting with the TAF1 C-terminal helical region (Figure 1C). Overall, the TAF7 barrel in the complex structure is largely exposed to the solvent, whereas the TAF1 barrel is mostly concealed.
Although the crystal structure suggests a central role for the triple barrel in stabilizing TAF1-TAF7 interactions, previous studies have shown that the TAF7 N-terminal sequence involved in triple barrel formation is entirely dispensable for TAF1-TAF7 complex assembly18. Instead, association with TAF1 hinges on a 19-amino acid long central region of TAF718, which contains a highly conserved Arg-rich motif (Figure 3A). In the crystal, this region directly interacts with a highly conserved Gly-rich motif in the C-terminal region of the TAF1 DUF3591 domain (Figure 3A). The TAF1 Gly-rich motif is located in the middle of a long loop, which coils up and nestles between two TAF1 C-terminal helices. The TAF7 Arg-rich motif forms a one-turn helix and packs directly against the TAF1 Gly-rich loop (Figure 3B). Together, they form a second critical interface outside the triple barrel core. Remarkably, this TAF7 structural element is further embraced by the extreme C-terminal end of the TAF1 DUF3591 fragment, which harbors at least two phosphorylated Ser/Thr residues as suggested by their extra side chain electron densities (Supplementary information, Figure S4).
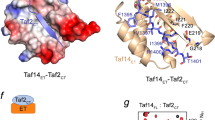
TAF1-TAF7 interactions between conserved loop motifs and C-terminal regions. (A) Schematic diagram of the crystallized TAF1 DUF3591 domain and TAF7 NTD with their conserved Gly-rich and Arg-rich sequence motif, respectively. The interacting TAF1 Gly-rich motif and TAF7 Arg-rich motif are colored pale green and bright yellow, and labeled “G” and “R”, respectively. Black bar above TAF7 represents the 19-amino acid sequence, which has been documented as a critical TAF1-binding region18. (B) A close-up view of the interactions between the conserved TAF1 Gly-rich motif and TAF7 Arg-rich motif, which are colored in the same scheme as shown in A. Dashed lines indicate intermolecular hydrogen bonds and salt bridges. The triple barrel in the complex is shown by surface representation. (C) Schematic diagram of the interactions between the TAF1 RAPiD domain and the TAF7 CTD. (D) Size exclusion chromatography elution profiles of the isolated TAF1-RAPiD domain (brown), the TAF7 CTD (green), and a mixture of the two (purple). Similar to the full-length protein, the TAF7 CTD migrates as a larger species on SDS-PAGE.
The RAPiD domain of TAF1, which is absent from the structure, has been proposed to interact with the C-terminal domain of TAF7 including the Arg-rich motif (Figure 3C)18. Using recombinant proteins, we assessed the ability of the TAF1 RAPiD domain to associate with the TAF7 C-terminal portion beyond the Arg-rich motif. Size exclusion chromatography analysis showed that the two polypeptides were able to form a complex in the absence of the TAF1 DUF3591 domain and the TAF7 N-terminal region (Figure 3D), indicating that TAF1-TAF7 interaction indeed involves yet another independent interface not observed in our current structure.
The TAF1 winged helix domain
The WH fold consists of three α-helices packing against three β-strands (Figure 4A) and is frequently found in basal transcription factors of RNAPII, subunits of RNA Polymerase I and III, and many other transcriptional regulators27,28. These proteins use their WH domains to either recognize DNA or mediate protein-protein interactions. The structure of the TAF1-TAF7 complex unveils a WH domain within the TAF1 DUF3591 fragment, establishing for the first time its existence in TFIID. Our previous mutational studies have shown that removal of three amino acids in one specific (Δ848-850) but not other regions of the DUF3591 domain of TAF1 deprived it of the ability to complement the ts13 defect in cell proliferation and cyclin D1 transcription17. Intriguingly, this deletion mutation maps to the central region of the WH domain, indicating a vital role for the WH fold in supporting TAF1's function (Supplementary information, Figure S2).
The winged helix (WH) domain of TAF1. (A) Superposition of the WH domains of TAF1, the transcription factor E2F4, and the yeast HAT ESA1 protein. (B) Electrostatic potential surface of the TAF1 WH domain viewed from the same angle as in A and 180° away. The surface colors are clamped between red (−83.5kTe−1) and blue (+83.5kTe−1). (C) A close-up view of the α8 helix of TAF1 with three solvent-exposed basic residues. (D) Sequence alignment of α8 helix in the TAF1 WH domain. The three basic residues mutated in our studies are highlighted in cyan.
Out of a structural homology search using the TAF1 WH domain as a query, the DNA-binding E2F4 transcription factor stood out as a top hit with an RMSD of 2.1 Å out of 59 aligned Cα atoms (Figure 4A). Moreover, the 75-residue TAF1 WH domain contains 22 positively charged residues, several of which are strictly conserved among TAF1 orthologs (Supplementary information, Figure S2). With a predicted isoelectric point of 11.3, the TAF1 WH domain features several highly basic surface areas (Figure 4B). One of these areas is presented by the third α-helix of the TAF1 WH domain, α8, which corresponds to a structural element commonly used by DNA-binding WH proteins, such as E2F4, to recognize the major groove of DNA28,29. Collectively, these features of the TAF1 WH domain suggest a possible function in DNA binding, which could contribute to the interactions of TAF1 with core promoter DNA elements in the context of TFIID or its subcomplexes30,31. Because purified full-length TAF1 alone does not bind selectively to specific DNA sequences31, we assume that its WH domain might support promoter recognition by making sequence-independent DNA contacts.
TAF1 WH domain binds promoter DNA
To investigate the DNA-binding activity of TAF1 WH domain, we performed electrophoretic mobility shift assays (EMSA) using TAF1 DUF3591 domain in complex with TAF7 and individually purified TAF7 (TAF1 DUF3591 domain alone is insoluble). Considering that TAF1 has been previously shown to directly and selectively affect the transcription rates of G1 cyclins such as cyclin D1, we carried out the binding assays with 32P-labeled double-stranded DNA containing human cyclin D1 core promoter sequence (positions −22 to +29, CD1). Incubation of TAF1-TAF7 complex with the radiolabeled CD1 probe produced a protein-DNA complex showing reduced mobility compared to free unbound CD1 DNA, with a Kd of 4.1 μM ± 0.8 μM (Figure 5A). By contrast, no mobility-shifted complex was detected with TAF7 alone (Figure 5A). These results indicate that the TAF1 component of the complex is responsible for DNA binding. In follow-up competition assays, we determined that formation of DNA-protein complexes was specific for double-stranded DNA (Figure 5B). The binding affinity of human TAF1 WH domain for DNA is weaker than its closest structural homolog E2F4. This difference in affinity could be attributed to the fact E2F4 binds DNA as a heterodimer, when in complex with its WH-containing partner protein DP2.
DNA binding and ts13 cell complementation activities of WT and mutant TAF1 proteins. (A) TAF1 displays DNA-binding activity. EMSA was carried out with increasing concentrations of the indicated purified proteins. Binding reactions were resolved on native 5% polyacrylamide and subjected to autoradiography. The positions of protein-bound and -unbound 32P-labeled cyclin D1 (CD1) core promoter fragments are shown. (B) TAF1 binds to double-stranded DNA. TAF1-TAF7 (22.5 pmoles) was incubated with radiolabeled CD1 probes in the presence of the indicated molar excess amount of unlabeled double-stranded or single-stranded CD1. Reaction products were analyzed and detected as described in A. (C) Minimal sequence-specificity for TAF1 DNA binding. Increasing concentrations of TAF1-TAF7 complex was subjected to EMSA using different radiolabeled DNA probes. IMD: initiator and downstream region of super core promoter, CD1: cyclin D1 core promoter, Rds2: random double-stranded DNA sequence. Migrations of protein-bound and -unbound DNA fragments are indicated. (D) Quantitation of DNA binding to the DNA probes described in C. Regions containing the bound and unbound radiolabeled DNA probes were excised from the dried gel and quantified by liquid scintillation. The percentage of total counts that the shifted complex represented was calculated for each sample. The graphed results are the average from several independent experiments, IMD: n = 3; CD1: n = 4; Rds2: n = 3. Error bars represent SEM. (E) WH domain mutations abolish DNA-binding activity of TAF1. TAF1-TAF7 complexes assembled in insect cells with WT-TAF1 or TAF1-WH mutant (WH3A) were analyzed by EMSA. Protein-bound and -unbound 32P-labeled CD1 fragments were detected by autoradiography. (F) TAF1 mutations compromise growth complementation efficiency in ts13 cells. Mutant ts13 cells were transfected with the indicated CS2+ control or TAF1 expression plasmid at 33.5 °C and subsequently shifted to 39.5 °C. After 48-72 h at the elevated temperature, the number of viable cells was counted and expressed as a percent relative to cells transfected with WT-TAF1, given a value of 100%. The bar graph depicts the average from 4 independent complementation experiments. Error bars, SEM. CS2+, empty vector; WT, WT-TAF1; Δ848-850, deletion of residues 848-850; WH3A: R864/K865/K868 all changed to alanine; DPT-AAA: D976/P977/T978 all changed to alanine.
To test the sequence preference of TAF1-TAF7 DNA-binding activity, we used additional DNA probes of similar lengths to CD1 including a randomized DNA sequence (Rds2) and the initiator and downstream region (positions −6 to +38, IMD) of the super core promoter (SCP). Intriguingly, yet consistent with our assumption, TAF1-TAF7 complex bound to all three DNA probes with a slight preference for the IMD promoter fragment (Figures 5C and 5D). The SCP was artificially engineered for maximal transcriptional activity by combining the optimal core promoter elements found in eukaryotes32. Accordingly, TFIID bound with higher affinity to the SCP compared to other natural core promoters, which is mirrored by our DNA binding results for the TAF1-TAF7 complex.
Superposition analysis of the TAF1 WH domain with transcription factor E2F4 shows that the third α-helix of the TAF1 WH domain (α8) resembles the DNA-binding element of E2F4 (Figure 4A). The α8 helix contains several highly conserved positively charged residues (Arg or Lys) in a row, forming a solvent-exposed basic surface that might be responsible for DNA-binding within the WH domain (Figure 4C and 4D). In agreement with our prediction, mutation of the conserved positively charged residues R864/K865/K868 to alanine (WH3A) completely eliminated DNA-binding to CD1 (Figure 5E), without disrupting the assembly and biochemical behavior of the TAF1-TAF7 complex (data not shown). These data unveil a promoter-binding function for the TAF1-TAF7 complex, which is dictated by the TAF1 WH domain and requires the conserved basic residues of α8, a feature of canonical WH-DNA interactions. Intriguingly, a recent survey of uterine serous carcinomas by whole exome sequencing revealed the prevalence of missense mutations at R864, one of the three positively charged residues on α8 helix of the WH domain, further implicating the importance of the WH domain in TAF1 function33.
Complementation of ts13 proliferation defect by TAF1 mutants
To determine the biological importance of the TAF1 WH domain and its DNA-binding activity, we next took advantage of the temperature-sensitive ts13 mutant hamster cell line, which shows cell cycle arrest at the nonpermissive temperature of 39.5 °C due to a TAF1 missense mutation. Because expression of wild-type TAF1 is sufficient to restore cell proliferation to ts13 cells at 39.5 °C, the complementation assay allowed us to investigate the ability of various TAF1 mutants to overcome the ts13 proliferation defect. As shown in Figure 5F, expression of WT TAF1 reproducibly resulted in viable proliferation of ts13 cells at 39.5 °C, whereas the CS2+ vector control and the Δ848-850 mutant, which harbors an internal deletion in TAF1 WH domain, completely failed to rescue the defects of the cell line. Remarkably, alanine mutation of R864/K865/K868 in the WH3A mutant severely compromised the ability of TAF1 to rescue ts13 cell proliferation, echoing its effects on the DNA-binding function of the TAF1 WH domain. No visible change in ts13 cell growth was observed at the permissive temperature of 33.5 °C in the presence of WT-TAF1 or WH3A mutant (data not shown). Together, these data strongly indicate that the normal function of TAF1 in transcription regulation depends on the WH domain of the conserved TAF1 DUF3591 core and its DNA-binding activity. Moreover, the effect of G716D mutation in the ts13 mutant further suggests that the intact structural coupling between the WH domain and the triple barrel might also be crucial for TAF1 function.
Outside the WH domain and the triple barrel, the interaction between the Gly-rich loop in TAF1 and the Arg-rich region of TAF7 represents a prominent interface within the complex (Figure 3). To test if disruption of this protein interface has any consequences on TFIID function, we mutated amino acids D976/P977/T978 in the TAF1 Gly-rich loop and examined the ability of the resulting DPT-AAA TAF1 mutant to restore cell proliferation to ts13 cells at 39.5 °C. Interestingly, expression of DPT-AAA was sufficient for ts13 cells to survive and proliferate at the elevated temperature, although the rescue efficiency was twofold lower than WT-TAF1 (Figure 5F). This result suggests that the interface between the two conserved TAF1 and TAF7 loops also supports the primary function of TAF1 in regulating gene transcription.
Discussion
Despite its important role in TFIID, structure-function studies of TAF1 have been held back by its massive yet complicated nature and limited validation of its many proposed activities. Our structure offers the first glimpse of the human TAF1 central DUF3591 domain, which represents the most conserved region of the protein. The crystal structure of the TAF1 DUF3591 domain in complex with TAF7 reveals an intricately organized multi-domain architecture, which is characterized by a heterodimeric triple barrel and a multivalent binding mode between the two proteins. Such a feature makes it possible for TAF7 to act as a dynamic regulator of TAF1 as suggested by recent studies34,35,36. It might also be essential for the large-degree of topological reorganization seen with TFIID upon promoter recognition13.
The human TAF1-TAF7 complex shares overall structural similarity with the recently reported yeast complex20. Several critical structural elements revealed by both studies, such as the triple barrel and WH domain, have been previously characterized as key features of general transcription factors. These structural features validate the physiological authenticity of our structure and suggest that it represents a functional interaction mode of TAF1 and TAF7. Because the entire TFIID complex likely adopts multiple states during the transcription initiation process, the conformation of the TAF1-TAF7 subcomplex captured in our structure may reflect a specific stage of transcription initiation when TAF1 and TAF7 are mutually soluble.
The TAF1 DUF3591 domain has been previously reported to possess HAT activity. Surprisingly, the common structural core shared by many diverse HATs for binding the acetyl-CoA cofactor is absent in the TAF1-TAF7 structure37. Although structural homology can be found between the WH domains of TAF1 and the ESA1 HAT (Figure 4A and Supplementary information, Figure S5), the overall fold of the TAF1 DUF3591 domain argues against its identity as a HAT. Of note, our structure cannot conclusively rule out the HAT function of the TAF1 DUF3591 domain, because TAF7 binding has been previously documented to inhibit the HAT activity of TAF134,35,36. It remains possible that TAF7 binding remodels TAF1 into an enzymatically inactive form that might differ significantly from other known HATs. Isolation and structure determination of a soluble TAF1 DUF3591 domain free of TAF7 will be needed to help resolve this issue.
Regardless of the mystery of TAF1 as a HAT, our structural analysis unravels a previously unrecognized WH sub-domain within the TAF1 DUF3591 domain, which we have shown to possess double-stranded DNA-binding activity. Although promoter DNA binding for TAF1 has been previously implicated with the cross-species TAF1-TAF2 subcomplex of TFIID31, our structure-based studies reveal for the first time the structural basis of this activity. Importantly, we also establish a strong functional correlation between promoter DNA binding and the normal function of TAF1 in transcription and cell cycle regulation. Based on our results, we hypothesize that TAF1, in the context of TFIID, likely relies on its promoter DNA binding activity to achieve and sustain close contacts with the core promoters of genes, including those required for cell cycle progression. Once TAF1 is recruited to the promoter region through either specific promoter recognition by TBP or the assistance of other TFIID subunits, the WH domain of TAF1 provides additional DNA-binding capacity with low sequence preference to allow for the maintenance and subsequent reorganization of the interaction of TFIID with core promoters upon arrival of specific regulatory events. The end result is that TAF1 may become appropriately armed to mediate the transcription of select genes via its proposed enzymatic activity.
Alternatively, the dynamic interaction of the peripheral TFIID subunit TAF7 with TAF1 might differentially modulate the DNA binding activity of TAF1 at the promoters of different genes. The modest promoter DNA binding affinity of the TAF1-TAF7 module may be necessary to allow the remodeling of TFIID during PIC assembly and promote potential interactions between the TAF1 WH domain and other transcription regulatory factors. Interestingly, in the yeast TAF1-TAF7 structure, the TAF1 WH domain is oriented slightly differently from the human ortholog, suggesting potential functional flexibility for the WH domain within TFIID in addition to promoter DNA binding activity.
By combining structural and functional analyses, our studies have established the role of TAF1 in promoter DNA binding and furnished the missing structural framework for delineating the function of the TAF1-TAF7 module within TFIID and for understanding the structural ramifications of TAF1 mutations found in human cancers33.
Materials and Methods
Protein expression and purification
Human TAF1 (amino acids 600-1236) was co-expressed with the human TAF7 full-length protein as a glutathione S-transferase (GST) fusion protein in High Five (Invitrogen) insect cells. The TAF1-TAF7 complex was purified by glutathione affinity chromatography using lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1.0 mM DTT) supplemented with protease inhibitors. The protein complex was further purified by anion exchange and size exclusion chromatography (GE Healthcare) after off-column cleavage by the tobacco etch virus (TEV) protease. The eluted complex was concentrated by ultrafiltration to 12 mg/ml in buffer containing 25 mM Hepes, pH 7.5, 150 mM NaCl, 1.0 mM DTT. Trypsin (0.1% w/w) was added to the complex sample to trim off flexible regions during crystallization screening. The protein samples used for EMSA assays were purified the same way as described above except the final size exclusion was omitted.
Crystallization and data collection
The crystals of human TAF1-TAF7 complex were obtained at 4 °C by the hanging-drop vapor diffusion method, using 1.2 μl protein complex sample mixed with an equal volume of reservoir solution containing 0.2 M potassium sodium tartrate tetrahydrate, pH 7.4, 20% w/v PEG3350. After being briefly equilibrated in the reservoir solution, the crystals were transferred to a cryoprotectant solution containing 25% w/v PEG3350, 0.2 M potassium sodium tartrate tetrahydrate, pH 7.4 and 10% glycerol, and then flash-frozen in liquid nitrogen. The human TAF1-TAF7 complex heavy atom derivative crystals were prepared by soaking the native crystals in a buffer containing 0.2 M potassium sodium tartrate tetrahydrate, pH 7.4, 20% w/v PEG3350 supplemented with 1.0 mM K2Pt(NO2)4 for 12 h, followed by a back soaking in the reservoir solution for an additional hour. The crystals were subsequently harvested the same way as the native ones. All data sets were collected at the BL8.2.1 and BL8.2.2 beamlines at the Advanced Light Source of the Lawrence Berkeley National Laboratory. Native crystals (P212121, a = 83.2 Å, b = 94.5 Å, c = 101.8 Å, α = β = γ = 90°) diffracted to 2.3 Å resolution, and heavy atom derivative crystals (P212121, a = 83.1 Å, b = 94.5 Å, c = 102.2 Å, α = β = γ = 90°) diffracted to 2.8 Å resolution. X-ray diffraction data statistics are shown in Supplementary information, Table S1.
Structure determination
The human TAF1-TAF7 complex structure was determined by SAD phasing of a platinum protein derivative. Reflection data were indexed, integrated and scaled with the HKL2000 package38. Experimentally phased map with a well-defined solvent boundary and readily interpretable electron density for protein was calculated with PHENIX39. An initial model was established using PHENIX and refined against the native data set. The final model was manually built with the program COOT40. The complex structure was refined to an Rfactor = 18.6% and an Rfree = 23.4%. Crystallographic data statistics are shown in Supplementary information, Table S1. 99.81% of all residues of human TAF1-TAF7 complex are in the favored and allowed region of the Ramachandran plot. All structural figures were prepared using PyMOL (http://www.pymol.org/).
Gel filtration analysis
The TAF1-RAPiD (amino acids 1110-1236) and TAF7-CTD (amino acids 154-349) domains were expressed in E. coli in a modified pET-28a (Novagen) vector with a 6× His tag and a Protein GB1 tag, both cleavable by the TEV protease. Both proteins were purified by Ni-NTA affinity columns followed by overnight on-column TEV cleavage, and further by anion-exchange and size exclusion chromatography (GE Healthcare). To form complex, the two polypeptides were mixed in a 1:1 molar ratio, and incubated at 4 °C for one hour. Purified TAF1-RAPiD, TAF7-CTD and the formed complex were analyzed by gel filtration. The eluted complex fractions in the shifted peak were analyzed by 15% (w/v) SDS-PAGE gel and visualized by Coomassie-blue staining.
EMSA
TAF1-TAF7 complex (4-180 pmoles) purified from insect cells was incubated with 4 ng of 32P 5′-end labeled DNA probes in 10 mM Hepes pH 7.9, 5 mM MgCl2, 100 mM NaCl, 10% glycerol, 20 mM tetrasodium pyrophosphate, 0.2 mM dI:dC for 1 hour at 25 °C. For gel electrophoresis, 6x loading buffer (20% Ficoll, 0.025% bromophenol blue) was added and binding reactions were loaded onto nondenaturing 5% polyacrylamide (37.5:1 acrylamide:bis) that was pre-run in 0.5× TBE at 100 V for 30 min. Samples were resolved for 1.5 h at 100 V and shifted complexes detected by autoradiography. Competition assays using 22.5 pmoles of purified TAF1-TAF7 complex were performed as described above with the addition of either unlabeled CD1 core promoter double-stranded (20, 40, 80, 200 ng) or sense single-stranded (10, 20, 40, 100 ng) DNA to the binding reaction. DNA binding was quantified by excising the protein bound and unbound DNA bands from the dried gels. The amount of radioactivity in each band was measured by liquid scintillation (Tri-Carb B2810 TR, PerkinElmer). The percent DNA bound was calculated and plotted using Prism (GraphPad Software). The sequences of DNA probes using for EMSA are provided in Supplementary information, Table S2.
ts13 cell complementation assays
Mammalian TAF1 expression plasmids contain N-terminal HA-tagged TAF1 coding sequence inserted downstream of the CMV promoter in CS2+ vector. Point mutations in TAF1 were introduced by site-directed mutagenesis using the primers shown in Supplementary information, Table S2 and confirmed by DNA sequencing. ts13 cells were grown at 33.5 °C in Dulbecco's modified Eagles medium (Gibco) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and penicillin/streptomycin. For complementation assays, cells were seeded into 60 mm dishes, grown overnight to 70%-80% confluency, and transfected with 2.5 μg of CS2+ or TAF1 expression plasmids using FuGene HD transfection reagent (2:1 FuGene:DNA) according to the manufacturer's protocol (Roche). Transfected cells were maintained an additional 18-24 h at 33.5 C, after which half the cells were shifted to nonpermissive temperature of 39.5 °C. The number of viable cells was determined after 36-48 h at 39.5 °C and the percentage relative to WT-TAF1 expression (given the value of 100%) was calculated.
Accession Code
Coordinates and structure factors have been deposited in the Protein Data Bank with accession code 4RGW.
Accession codes
References
Thomas MC, Chiang CM . The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 2006; 41:105–178.
Grünberg S, Hahn S . Structural insights into transcription initiation by RNA polymerase II. Trends Biochem Sci 2013; 38:603–611.
Buratowski S, Hahn S, Guarente L, Sharp PA . Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 1989; 56:549–561.
Van Dyke MW, Roeder RG, Sawadogo M . Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science 1988; 241:1335–1338.
Papai G, Weil PA, Schultz P . New insights into the function of transcription factor TFIID from recent structural studies. Curr Opin Genet Dev 2011; 21:219–224.
Pugh BF, Tjian R . Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev 1991; 5:1935–1945.
Tanese N, Pugh BF, Tjian R . Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev 1991; 5:2212–2224.
Wassarman DA, Sauer F . TAF(II)250: a transcription toolbox. J Cell Sci 2001; 114:2895–2902.
Mizzen CA, Yang XJ, Kokubo T, et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 1996; 87:1261–1270.
Dikstein R, Ruppert S, Tjian R . TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell 1996; 84:781–790.
Ruppert S, Tjian R . Human TAFII250 interacts with RAP74: implications for RNA polymerase II initiation. Genes Dev 1995; 9:2747–2755.
Jacobson RH, Ladurner AG, King DS, Tjian R . Structure and function of a human TAFII250 double bromodomain module. Science 2000; 288:1422–1425.
Cianfrocco MA, Kassavetis GA, Grob P, et al. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 2013; 152:120–131.
Hisatake K, Hasegawa S, Takada R, Nakatani Y, Horikoshi M, Roeder RG . The p250 subunit of native TATA box-binding factor TFIID is the cell-cycle regulatory protein CCG1. Nature 1993; 362:179–181.
Ruppert S, Wang EH, Tjian R . Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature 1993; 362:175–179.
Hayashida T, Sekiguchi T, Noguchi E, Sunamoto H, Ohba T, Nishimoto T . The CCG1/TAFII250 gene is mutated in thermosensitive G1 mutants of the BHK21 cell line derived from golden hamster. Gene 1994; 141:267–270.
Hilton TL, Li Y, Dunphy EL, Wang EH . TAF1 histone acetyltransferase activity in Sp1 activation of the cyclin D1 promoter. Mol Cell Biol 2005; 25:4321–4332.
Chiang CM, Roeder RG . Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 1995; 267:531–536.
Lavigne AC, Mengus G, May M, et al. Multiple interactions between hTAFII55 and other TFIID subunits. Requirements for the formation of stable ternary complexes between hTAFII55 and the TATA-binding protein. J Biol Chem 1996; 271:19774–19780.
Bhattacharya S, Lou X, Hwang P, et al. Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D. Proc Natl Acad Sci USA 2014; 111:9103–9108.
Weissman JD, Brown JA, Howcroft TK, et al. HIV-1 tat binds TAFII250 and represses TAFII250-dependent transcription of major histocompatibility class I genes. Proc Natl Acad Sci USA 1998; 95:11601–11606.
Dunphy EL, Johnson T, Auerbach SS, Wang EH . Requirement for TAF(II)250 acetyltransferase activity in cell cycle progression. Mol Cell Biol 2000; 20:1134–1139.
Gaiser F, Tan S, Richmond TJ . Novel dimerization fold of RAP30/RAP74 in human TFIIF at 1.7 Å resolution. J Mol Biol 2000; 302:1119–1127.
Shaban NM, Harvey S, Perrino FW, Hollis T . The structure of the mammalian RNase H2 complex provides insight into RNA.NA hybrid processing to prevent immune dysfunction. J Biol Chem 2010; 285:3617–3624.
Geiger SR, Lorenzen K, Schreieck A, et al. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell 2010; 39:583–594.
Taylor NM, Baudin F, von Scheven G, Müller CW . RNA polymerase III-specific general transcription factor IIIC contains a heterodimer resembling TFIIF Rap30/Rap74. Nucleic Acids Res 2013; 41:9183–9196.
Teichmann M, Dumay-Odelot H, Fribourg S . Structural and functional aspects of winged-helix domains at the core of transcription initiation complexes. Transcription 2012; 3:2–7.
Gajiwala KS, Burley SK . Winged helix proteins. Curr Opin Struct Biol 2000; 10:110–116.
Zheng N, Fraenkel E, Pabo CO, Pavletich NP . Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev 1999; 13:666–674.
Lee DH, Gershenzon N, Gupta M, Ioshikhes IP, Reinberg D, Lewis BA . Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol Cell Biol 2005; 25:9674–9686.
Chalkley GE, Verrijzer CP . DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J 1999; 18:4835–4845.
Juven-Gershon T, Cheng S, Kadonaga JT . Rational design of a super core promoter that enhances gene expression. Nat Methods 2006; 3:917–922.
Zhao S, Choi M, Overton JD, et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci USA 2013; 110:2916–2921.
Kloet SL, Whiting JL, Gafken P, Ranish J, Wang EH . Phosphorylation-dependent regulation of cyclin D1 and cyclin A gene transcription by TFIID subunits TAF1 and TAF7. Mol Cell Biol 2012; 32:3358–3369.
Gegonne A, Weissman JD, Singer DS . TAFII55 binding to TAFII250 inhibits its acetyltransferase activity. Proc Natl Acad Sci USA 2001; 98:12432–12437.
Devaiah BN, Lu H, Gegonne A, et al. Novel functions for TAF7, a regulator of TAF1-independent transcription. J Biol Chem 2010; 285:38772–38780.
Marmorstein R, Trievel RC . Histone modifying enzymes: structures, mechanisms, and specificities. Biochim Biophys Acta 2009; 1789:58–68.
Otwinowski Z, Minor W, eds. Processing of X-ray Diffraction Data Collected in Oscillation Mode. New York: Academic Press 1997.
Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 2002; 58:1948–1954.
CCP4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 1994; D50:760–763.
Acknowledgements
We thank the beam line staff of the Advanced Light Source at the University of California at Berkeley for help with data collection, undergraduates E Larson and M Kufeld for assistance with the in vivo complementation experiments, and members of the Zheng and Wang laboratories for discussion. This work is supported by the Howard Hughes Medical Institute (NZ) and the University of Washington Royalty Research Fund (EHW).
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Sequence alignment of representative eukaryotic TAF7 orthologs annotated with secondary structures. (PDF 160 kb)
Supplementary information, Figure S2
Sequence alignment of DUF3591 domains of representative eukaryotic TAF1 orthologs annotated with secondary structure labeled. (PDF 346 kb)
Supplementary information, Figure S3
Two salt bridges formed among three TAF7 β-hairpin residues are buried in the TAF1 barrel. (PDF 634 kb)
Supplementary information, Figure S4
A close-up view of the C-terminal residues in the TAF1 DUF3591 fragment together with positive Fo-Fc electron density contoured at 3.0 σ (red) and 2Fo-Fc electron density contoured at 1.0 σ (blue) calculated before the phosphate groups were built. (PDF 1170 kb)
Supplementary information, Figure S5
Structure comparison between the yeast HAT enzyme ESA1 and TAF1 in complex with TAF7. (PDF 1719 kb)
Supplementary information, Table S1
Summary of crystallographic information (PDF 114 kb)
Supplementary information, Table S2
Nucleic acid sequences of the probes used in EMSA analysis and the primers for the site mutagenesis (PDF 44 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Curran, E., Hinds, T. et al. Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module. Cell Res 24, 1433–1444 (2014). https://doi.org/10.1038/cr.2014.148
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2014.148
Keywords
This article is cited by
-
Hierarchical TAF1-dependent co-translational assembly of the basal transcription factor TFIID
Nature Structural & Molecular Biology (2023)
-
Nonlinear feature selection using sparsity-promoted centroid-encoder
Neural Computing and Applications (2023)
-
TAF1 plays a critical role in AML1-ETO driven leukemogenesis
Nature Communications (2019)
-
TAF1, associated with intellectual disability in humans, is essential for embryogenesis and regulates neurodevelopmental processes in zebrafish
Scientific Reports (2019)
-
Identification of the gene encoding the TATA box-binding protein-associated factor 1 (TAF1) and its putative role in the heat shock response in the protozoan parasite Entamoeba histolytica
Parasitology Research (2019)