Abstract
Primary ovarian insufficiency (POI), or premature ovarian failure, is defined as the cessation of ovarian function before the age of 40. An insufficient ovarian follicle pool derived from primordial germ cells (PGCs) is an important cause of POI. Although the Nanos gene family is known to be required for PGC development and maintenance in diverse model organisms, the relevance of this information to human biology is not yet clear. In this study, we screened the coding regions of the NANOS1, NANOS2 and NANOS3 genes in 100 Chinese POI patients and identified four variants in the coding regions of these three genes, including one synonymous variant in NANOS3, one missense variant in each of NANOS1 and NANOS2 and one potentially relevant mutation (c.457C>T; p.Arg153Trp, heterozygous) in NANOS3. We demonstrated that the p.Arg153Trp substitution decreases the stability of NANOS3, potentially resulting in a hypomorph. Furthermore, an investigation of the relationship between the number of PGCs and the dosage of NANOS3 in mouse models showed that the population of PGCs is controlled by the level of NANOS3 protein. Taken together, our results provide new insight into the properties of the NANOS3 protein and establish that NANOS3 mutation is one possible cause of POI.
Similar content being viewed by others
Main
Primary ovarian insufficiency (POI) is a heterogeneous disorder that affects ∼1% of women under 40 years of age, including 1 : 10 000 women by age 20 and 1 : 1000 women by age 30.1 The age at which menopause naturally occurs is largely determined by the size of the initial primordial follicle pool. The follicles are derived from primordial germ cells (PGCs), a transient population of germline stem cells that undergo several rounds of cell proliferation2 and subsequently arrest at prophase I. The arrested cells make up the pool of primordial follicles3 that will provide the basis for the future reproductive lifespan of the adult female, although it should be noted that some aspects of this model have recently been questioned.4, 5 PGC behaviour is tightly regulated by intrinsic and microenvironmental factors.
The Nanos genes are evolutionarily conserved across many organisms and have important roles during germ cell development.6, 7, 8 In Drosophila, the single nanos (Nos) is required for development of the abdomen as well as for the germline maintenance.9, 10 Generally, NANOS is recruited by its cofactor pumilio to Nanos-response elements on target mRNAs, where the NANOS protein represses translation.11 In mouse, three homologues exist, with Nanos2 and Nanos3 functioning primarily in male germ cell development and maintaining PGCs viability, respectively.12, 13 However, the function of NANOS homologues in human is not yet known. We speculated that there is a relationship between POI and defects in NANOS genes, as POI is largely dependent on the size of the pool of PGCs.
Here, we found a novel mutation in the NANOS3 gene through screening the coding regions of the NANOS1, NANOS2 and NANOS3 genes in 100 Chinese POI patients. In addition, we investigated the effect of this mutation on protein stability by biochemical assays. Finally, we established the relationship between the dosage of NANOS3 protein and the numbers of PGCs in mouse models. These findings provide new insights into the pathogenic mechanism of POI and have important implications for understanding the properties of NANOS3 and its role in PGC maintenance.
Results and Discussion
Identifying NANOS family mutations in POI patients
We sequenced all coding regions of the NANOS1, NANOS2 and NANOS3 genes in 100 Chinese POI patients. This analysis identified a total of four variants in the coding regions of these three genes, including one synonymous variant in NANOS3 and one missense variant in each of NANOS1, NANOS2 and NANOS3 (Table 1). Among these variants, three were also found in the National Center for Biotechnology Information (NCBI) SNP database (dbSNP) or the 1000 Genomes Project database. In contrast, the variant in NANOS3 (c.457C>T; p.Arg153Trp) was not observed in the two databases14 and was not present in the control group of our study. The affected woman was 23 years old, heterozygotes for the variant, unmarried and the offspring of consanguineous parents. Her mother was 45 years old, premenopausal and had a normal menstrual cycle. She had elevated follicle-stimulating hormone (>124.89 U/l) and luteinising hormone (>43.17 U/l) on several examinations, with the cessation of periods. Her estradiol (E2) level was 70.2 pmol/l.
Characterising the Arg153Trp mutation in NANOS3
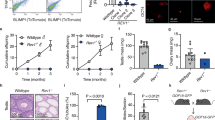
We aligned the amino-acid sequences of NANOS3 homologues in different species to determine the location of the identified mutation, and we found that it was located within a region of the NANOS3 gene that is highly conserved among Homo sapiens, Pan troglodytes, Mus musculus and Rattus norvegicus (Figure 1a). To confirm that the potential mutation (Arg153Trp) is pathological, we analysed the biochemical effect of the mutation on the stability of HA-tagged human NANOS3 (hNANOS3) protein in transiently transfected HEK293 cells using a cycloheximide (CHX)-chase analysis. The hNANOS3 protein has a half-life of 3 h, suggesting that hNANOS3 is normally an unstable protein; however, hNANOS3 with the Arg153Trp mutation has an even shorter half-life of 1.5 h (Figures 1b and c). Using co-transfected GFP as a reporter, we confirmed that the reduction in hNANOS3 protein level was not due to variability in the transfection efficiency (Figure 1b).
Characterisation of the properties of wild-type and mutant hNANOS3 proteins. (a) The alignment of amino-acid sequences of NANOS3 homologues in different species shows that the arginine (grey shadow) is conserved. (b) Turnover of hNANOS3 and hNANOS3 R153W proteins. (c) The levels of hNANOS3 and hNANOS3 R153W proteins were quantified and plotted relative to the corresponding hNANOS3 levels at 0 h. Data represent the means±S.E. of the results from at least three independent experiments. Two stars represent statistical significance, P<0.001. (d) Turnover of hNANOS3 protein with R153 substituted with Ala, Phe, Leu, Met, Pro and Val. (e) Turnover of mNANOS3 and NANOS3 R133W with or without T132 substituted with Val. (f) Turnover of hNANOS3 protein with MG132 (20 mM), chloroquine (100 mM) or NH4Cl (50 mM) treatment. Treatment with MG132 obviously slowed the turnover of both wild-type and mutant NANOS3 proteins. (g) The increased accumulation of hNANOS3 R153W in the insoluble fraction
Because of the non-polar nature of Trp, we substituted six other non-polar amino acids (Ala, Met, Leu, Phe, Val and Pro) at position 153 in the hNANOS3 protein. Five of the six non-polar amino-acid substitutions (excluding Pro) decreased hNANOS3 stability (Figure 1d), indicating that the polar nature of the residue at position 153 is important for protein stability. However, in marked contrast to hNANOS3, in mouse NANOS3 (mNANOS3), the Arg133Trp mutation (Arg 133 is the murine analogue to human Arg 153) has less effect on protein stability (Figure 1e). We noted that in mNANOS3, the amino acid immediately before the arginine is threonine, a polar amino acid (at position 132), whereas in hNANOS3, it is valine, a non-polar amino acid (at position 152). To test whether the non-polar nature at this position can affect mNANOS3 stability, we replaced the polar amino acid Thr with the non-polar amino acid Val at position 132, or we replaced both Thr and Arg at positions 132 and 133 with non-polar Val and Trp. We found that mNANOS3 with T132V degraded rapidly compared with the wild type, and mNANOS3 with both T132V and R133W degraded even more rapidly in CHX-chase analysis (Figure 1e), suggesting that NANOS3 is sensitive to nonpolar amino-acid substitution at the conserved arginine and the preceding amino acid, and that the hydrophilic environment at these positions is crucial for NANOS3 stability.
To identify the degradation pathway responsible for the clearance of hNANOS3 protein, we assessed the effects of proteasome and lysosome inhibition on HA-tagged hNANOS3 and hNANOS3 with the Arg153Trp mutation in transfected HEK293 cells. We found that the levels of both proteins were increased significantly by treatment with the proteasome inhibitor MG132 (Figure 1f, upper panel), whereas these levels were not affected by treatment with the lysosome inhibitors chloroquine (Figure 1f, middle panel) or ammonium chloride (NH4Cl) (Figure 1f, lower panel). These data suggest that the hNANOS3 protein is degraded primarily by the proteasomal machinery.
The ubiquitin–proteasome system is the major pathway that degrades misfolded proteins. The rapid degradation of mutant proteins, generally due to their impaired folding, which is a common pathological mechanism in disease-causing missense mutations.15, 16 Therefore, we examined the potential mechanism of rapid degradation of hNANOS3 with the Arg153Trp mutation. We found no significant difference in the levels of these proteins in the soluble fraction (Figure 1g). However, exposure to MG132 resulted in much higher levels of mutant NANOS3 in the insoluble fraction (Figure 1g). This result suggests that the mutant NANOS3 protein tends to be structurally unstable and form aggregates, which are cleared by the ubiquitin–proteasome system, leading to the rapid decrease in the level of NANOS3 protein after CHX treatment.
Functional consequence of decreased NANOS3 protein
To investigate the possible causal link between NANOS3 dosage and POI, we generated a mouse model with attenuated Nanos3 transcription levels instead of making an R133W knock-in mouse, as mNANOS3 stability was less sensitive to degradation affected by the R133W mutation. A Neo cassette was inserted into the promoter region of the Nanos3 gene (Figure 2a), and quantitative real-time polymerase chain reaction (PCR) and immunofluorescence were used to observe NANOS3 expression levels in the female gonad at E12.5. As shown in Figure 2b, the expression level of Nanos3 is obviously lower in Nanos3Neo/Neo mice than in Nanos3Neo/+ mice. We found that most PGCs express NANOS3 in Nanos3Neo/+ mice (Figures 2c and d). In contrast, Nanos3Neo/Neo mice strikingly display not only reduced numbers of PGCs but also very low levels of NANOS3 protein in OCT4-positive PGCs (Figures 2c and d), suggesting that the Neo cassette influences the expression level of Nanos3 in the female gonad as early as E12.5. Next, gonadal ridges of E12.5 embryos were used to evaluate the correlation between NANOS3 dosage and PGC number using anti-MVH antibody, a marker for germ cells. Female Nanos3Neo/Neo mice exhibited a significant decrease in germ cell number (Figures 3a and b). Consequently, at postnatal day 4, germ cells in Nanos3Neo/Neo ovaries were notably reduced in number compared with Nanos3Neo/+ ovaries (Figures 3c and d). Taken together, our data suggested that the dosage of NANOS3 has a very important role in the maintenance and survival of PGCs.
Neo cassette insertion into the promoter region decreased Nanos3 expression level. (a) A PGK-Neo cassette flanked by two flippase recognition target (FRT) sites and followed by a LoxP site was inserted in the Nanos3 promoter region 1476 bp upstream of the Nanos3 initiator. Another LoxP site was inserted 906 bp downstream of the Nanos3 3′UTR. (b) Quantitative real-time PCR analysis of Nanos3 mRNA from E12.5 female gonad in indicated genotype mice. (c) Immunofluorescence analysis of NANOS3 expression in the E12.5 female gonad. OCT4 is used as a marker of PGCs. NANOS3 is in the cytoplasm of germ cells in Nanos3Neo/+ mice, whereas NANOS3 is hardly detected in Nanos3Neo/Neo mice. Representative PGCs are magnified (insets). (d) Quantification of NANOS3 expression in OCT4-positive cells. Data represent the means±S.E. of the results from three independent sections. Two stars represent statistical significance, P<0.001
The causal link between the dosage of NANOS3 and the number of PGCs. (a) Immunofluorescence analysis of MVH expression in the gonadal ridges at E12.5 from mice of the indicated genotype. (b) Quantification of germ cell number in gonadal ridges at E12.5 from mice of the indicated genotype. (c) Immunofluorescence analysis of MVH expression in the ovaries at postnatal day 4 from mice of the indicated genotype. (d) Quantification of ovaries at postnatal day 4 from mice of the indicated genotype. Two stars represent statistical significance, P<0.001
We realise that the PGC reduction in Nanos3Neo/Neo mice may not directly reflect the result effected by the Arg153Trp mutation, but there is a clear correlation between NANOS3 dosage and PGC survival. We considered the nature of the Arg133Trp mutation and the neighbouring amino acid to determine whether it was advisable to generate a knock-in mouse model. We observed a strong polar region preceding the arginine in mNANOS3, which enhanced its stability; this direct experimental evidence suggested that the pathogenic nature of this point mutation might not be conserved across species. Furthermore, heterozygous knockout mice showed almost normal pools of PGCs, whereas homozygous mice exhibited a complete loss of PGCs, suggesting that the expression of Nanos3 from a single Nanos3 allele is sufficient to maintain the number of PGCs (data not shown). To gauge the relative contribution of the decreased NANOS3 level to PGC number, we made every effort to generate Nanos3Neo/Neo mice. As expected, these mice showed a marked reduction in the pool of PGCs during development. The identified human NANOS3 mutation accelerates hNANOS3 degradation by altering polar amino acids to a non-polar nature, consistent with the observation that a decreased dosage of NANOS3 protein reduces the pool of PGCs, which leads to decreased germ cell number in the mouse model. Although only one patient was found to carry this mutation in this study, it is challenging to study the genetics of PGC formation and related human diseases that cause infertility due to the reduced chance of germline transmission of the transferred genes. The successful elucidation of the molecular mechanism of the Arg153Trp mutation in NANOS3 and its absence from unaffected controls argue that the Arg153Trp mutation was the causative mutation. Thus, our results provide insights that NANOS3 haplo-insufficiency caused by the Arg153Trp mutation disrupts the process of human germ cell development. This and other studies have addressed the physiological importance of Nanos3 in PGC development for most species; however, accurate information from a primate model may help to elucidate the specific mechanistic effect of Nanos3 on human fertility.
Materials and Methods
Study population
A total of 100 Chinese POI patients and 200 healthy controls were recruited from the First Affiliated Hospital, Anhui Medical University, China. The ages of these patients ranged from 15 to 39 years. The diagnostic criteria were menopause occurring before age 40, at least two serum FSH concentrations of >40 IU/l and an exclusion of chromosomal abnormalities. Patients with associated endocrinopathies or autoimmune disorders, infections or iatrogenic agents, such as chemotherapy or radiotherapy, were also excluded. Controls were individuals of proven fertility, with normal menstrual cycles and ovary morphology, without a history of subfertility treatment. All human-related studies were approved by the Anhui Medical University ethics board and informed consent was obtained from responsible persons on behalf of all the study participants.
Mouse models
Animal studies were performed in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International-accredited SPF animal facility, and all animal protocols were approved by the Animal Care and Use Committee of the Model Animal Research Center, the host for the National Resource Center for Mutant Mice in China, Nanjing University.
Mutation screen
Genomic DNA was extracted from the peripheral blood leukocytes using standard methods. The coding regions of the NANOS1, NANOS2 and NANOS3 genes were amplified by PCR using specific primer sets (Table 2).
PCR products were sequenced on an ABI 3730XL automated sequencer (Applied Biosystems, Foster City, CA, USA) for mutation analysis.
Plasmids, antibodies and reagents
Full-length human NANOS3 WT, R153W, R153A, R153F, R153L, R153M, R153P and R153V and mouse Nanos3 WT, R133W, T132V, T132V/R133W cDNA coding sequences were subcloned into the pCGN-HA vector to generate N-terminal HA-tagged NANOS3.
The antibodies used in this study include anti-ACTIN (5A7, Abmart, Shanghai, China), anti-GFP (sc-8334, Santa Cruz, Santa Cruz, CA, USA), anti-HA (HA7, Sigma, St. Louis, MO, USA), anti-OCT3/4 (sc-5279, Santa Cruz), anti-NANOS3 (ab70001, Abcam, Cambridge, MA, USA) and anti-MVH (ab13840, Abcam).
The reagents used for protein stability assays include CHX (C7698, Sigma), MG132 (C2211, Sigma), chloroquine (C6628, Sigma) and ammonium chloride (0612, AMRESCO, Solon, OH, USA).
Protein stability assay
HEK293T cells were transfected with HA-tagged NANOS3 as indicated. At 24 h post transfection, the cells were treated with the protein biosynthesis inhibitor CHX at 50 μg/ml and incubated for an indicated time, then lysed. Total cell lysates were analysed by western blotting.
Treatment of cells with proteasome and lysosome inhibitors
HEK293T cells were transfected with HA-tagged hNANOS3 WT or R153W. At 24 h post transfection, the cells were treated with the protein biosynthesis inhibitor CHX at 50 μg/ml plus the proteasome inhibitor MG132 at 20 mM, the lysosome inhibitor NH4Cl at 50 mM or the lysosome inhibitor chloroquine at 100 mM.
Preparation of soluble and insoluble fraction of cell lysates
Cells were lysed in RIPA buffer. After incubating on ice for 15 min, the lysates were cleared by centrifugation at 13 000 × g for 10 min and the supernatant was collected as the soluble fraction. The pellets were washed two times with RIPA buffer, and then they were suspended in RIPA buffer and sonicated three times. The samples were then centrifuged at 13 000 × g for 10 min to obtain the insoluble fraction.
Generation of conditional nanos3 knockout mice
The Nanos3 targeting construct was generated according to our standard protocol.17 One LoxP site was inserted 906 bp downstream of Nanos3 3′UTR, and a neomycin resistance cassette was introduced 1476 bp upstream of Nanos3 5′UTR. The neomycin resistance coding sequences were under the control of the PGK promoter. This cassette was flanked by FRT sites and followed by one LoxP site. The targeting construct was electroporated into strain 129 ES cells, and targeted clones were identified by Southern blotting. Germline chimaeras were generated by morula aggregation and succeed in germline transmission. The resulting chimaeric males were mated to C57BL/6J mice. Heterozygous mice were continuously backcrossed to C57BL/6J mice for at least five generations before being used for experiments.
Immunohistochemistry
E12.5 embryos and ovaries were fixed in 4% PFA, embedded in paraffin and sectioned (6 μm). Tissue sections were rehydrated, incubated in Coplin jars filled with 1 mM EDTA (pH8.0) in a 95–99 °C water bath for 30 min, cooled at room temperature (RT) for 60 min and then blocked in 10% normal goat serum for 60 min. The sections were incubated overnight at 4 °C with anti-MVH (1 : 200), anti-OCT3/4 (1 : 50) and anti-NANOS3 (1 : 400). The following day, the sections were washed in PBS and incubated for 1.5 h at RT with secondary antibodies. The sections were then counterstained with DAPI and analysed by confocal laser microscopy.
Statistical analysis
All experiments were repeated at least three times. Data were subjected to statistical analyses by ANOVA. A P-value of less than 0.05 was considered statistically significant.
Abbreviations
- POI:
-
primary ovarian insufficiency
- PGC:
-
primordial germ cell
- NCBI:
-
National Center for Biotechnology Information
- SNP:
-
single-nucleotide polymorphism
- E2:
-
estradiol
- CHX:
-
cycloheximide
References
Coulam CB, Adamson SC, Annegers JF . Incidence of premature ovarian failure. Obstet Gynecol 1986; 67: 604–606.
Pepling ME, Spradling AC . Female mouse germ cells form synchronously dividing cysts. Development 1998; 125: 3323–3328.
Saitou M, Barton SC, Surani MA . A molecular programme for the specification of germ cell fate in mice. Nature 2002; 418: 293–300.
Zou K, Yuan Z, Yang Z, Luo H, Sun K, Zhou L et al. Production of offspring from a germline stem cell line derived from neonatal ovaries. Nat Cell Biol 2009; 11: 631–636.
Johnson J, Canning J, Kaneko T, Pru JK, Tilly JL . Germline stem cells and follicular renewal in the postnatal mammalian ovary. Nature 2004; 428: 145–150.
Mosquera L, Forristall C, Zhou Y, King MLA . mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development 1993; 117: 377–386.
Koprunner M, Thisse C, Thisse B, Raz E . A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev 2001; 15: 2877–2885.
Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S et al. Conserved role of nanos proteins in germ cell development. Science 2003; 301: 1239–1241.
Sano H, Mukai M, Kobayashi S . Maternal Nanos and Pumilio regulate zygotic vasa expression autonomously in the germ-line progenitors of Drosophila melanogaster embryos. Dev Growth Differ 2001; 43: 545–552.
Kobayashi S, Yamada M, Asaoka M, Kitamura T . Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 1996; 380: 708–711.
Bergsten SE, Gavis ER . Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development 1999; 126: 659–669.
Suzuki A, Saga Y . Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes Dev 2008; 22: 430–435.
Suzuki H, Tsuda M, Kiso M, Saga Y . Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev Biol 2008; 318: 133–142.
Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA et al. A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061–1073.
Kwok JB, Taddei K, Hallupp M, Fisher C, Brooks WS, Broe GA et al. Two novel (M233T and R278T) presenilin-1 mutations in early-onset Alzheimer’s disease pedigrees and preliminary evidence for association of presenilin-1 mutations with a novel phenotype. Neuroreport 1997; 8: 1537–1542.
Corydon TJ, Bross P, Jensen TG, Corydon MJ, Lund TB, Jensen UB et al. Rapid degradation of short-chain acyl-CoA dehydrogenase variants with temperature-sensitive folding defects occurs after import into mitochondria. J Biol Chem 1998; 273: 13065–13071.
Shi G, Xing L, Liu Z, Qu Z, Wu X, Dong Z et al. Dual roles of FBXL3 in the mammalian circadian feedback loops are important for period determination and robustness of the clock. Proc Natl Acad Sci USA 2013; 110: 4750–4755.
Acknowledgements
We thank the other members in Xu lab for the helpful discussion and technical support. This work was supported by the National Basic Research Program of China Grant 2010CB945100 (to YX), 2012CB944704 (to Y C), Jiangsu Science Foundation BK2010011 and National Science Foundation 31171343 (to YX).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Wu, X., Wang, B., Dong, Z. et al. A NANOS3 mutation linked to protein degradation causes premature ovarian insufficiency . Cell Death Dis 4, e825 (2013). https://doi.org/10.1038/cddis.2013.368
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.368
Keywords
This article is cited by
-
Premature ovarian insufficiency: a review on the role of tobacco smoke, its clinical harm, and treatment
Journal of Ovarian Research (2024)
-
Modeling primary ovarian insufficiency-associated loci in C. elegans identifies novel pathogenic allele of MSH5
Journal of Assisted Reproduction and Genetics (2022)
-
Gene Alterations and Expression Spectrum of NANOS3 in Nonobstructive Azoospermia
Reproductive Sciences (2022)
-
Onco-fertility and personalized testing for potential for loss of ovarian reserve in patients undergoing chemotherapy: proposed next steps for development of genetic testing to predict changes in ovarian reserve
Fertility Research and Practice (2021)
-
Premature ovarian insufficiency: pathogenesis and therapeutic potential of mesenchymal stem cell
Journal of Molecular Medicine (2021)