Abstract
The transcription factor signal transducer and activator of transcription 3 (STAT3) is frequently activated in human cancers. Interestingly, STAT3 also maintains the pluripotency and self-renewal of murine embryonic stem cells, and several tissue stem cell types. To investigate whether STAT3 also maintains the small-intestine crypt stem cell, we conditionally inactivated a Floxed Stat3 allele (Stat3fl) in murine small-intestine crypt stem cells. Following Cre recombinase expression, apoptosis increased in Stat3fl/− experimental crypts relative to Stat3wt/− controls before declining. Control Stat3wt/− mice carrying a Flox-STOP LacZ reporter transgene stably expressed LacZ after Cre induction. In contrast, Stat3fl/− intestine LacZ expression initially increased modestly, before declining to background levels. Quantitative PCRs revealed a similar transient in recombined Stat3fl allele levels. Long-term bromodeoxyuridine labelling directly demonstrated that functional STAT3 is required for +4 to +6 region label-retaining small-intestine stem cell survival. Rapid clearance of recombined Stat3fl/− cells involves apoptosis potentially induced by elevated c-Myc in non-recombined cells and involves elevated p53 expression and caspase 3 activation. Intriguingly, Stat3fl/− intestine recombination triggered dramatically upregulated polycomb transcriptional repressor Bmi1 – potentially accelerating recombined crypt repopulation. In summary, STAT3 activity is absolutely required for small-intestine crypt stem cell survival at both the +4 to +6 label-retaining and crypt base columnar cell locations.
Similar content being viewed by others
Main
The signal transducer and activator of transcription (STAT) family of transcription factors are activated in response to extracellular cytokine and growth factor ligands. Classically, following phosphorylation at a single-tyrosine residue by JAK family tyrosine kinases, STAT monomers dimerise via reciprocal phosphotyrosine-Src homology 2 (SH2) domain interactions, acquiring nuclear localisation activity and high-affinity DNA-binding activity to activate gene expression.1, 2 Inappropriate activation of STAT family members, especially STAT3, occurs in numerous human cancers and cancer cell lines.3, 4 STAT3 activation is also critical for maintaining pluripotency and preventing differentiation of murine embryonic stem (mES) cells. Thus, inducible expression of a dominant negative interfering STAT3F mutant in mES cells abolished their self-renewal and promoted differentiation.5 STAT3 function is also important in several tissue stem cell types – for example, porcine skin stem cells express STAT3 at high levels.6 Similarly, transduction of murine haematopoietic stem cells with a dominant negative STAT3-expressing retrovirus caused a large and permanent reduction in their in vivo lympho-myeloid reconstituting ability.7
Here, we examine the requirement of murine small-intestine crypt stem cells for functional STAT3. The small-intestine epithelium undergoes constant renewal – a process dependent upon stem cells located near the crypt base.8 The crypt stem cells generate a population of rapidly proliferating cells – the transit-amplifying cells – which undergo a limited number of divisions while migrating up the crypt. On reaching the crypt-villus junction, these transit-amplifying cells exit the cell cycle and terminally differentiate, yielding mature enterocytes, goblet cells and enteroendocrine cells, which migrate towards and eventually shed from, the villus tip. The crypt base, in contrast, comprises two other cell types – Paneth cells and slowly proliferating crypt base columnar (CBC) cells. The precise identity of the small-intestine crypt stem cell has been controversial for many years, with one long-accepted candidate being the radiation-hypersensitive, long-term label-retaining cell located at the +4 to +6 positions from the crypt base.9 Another long-time crypt stem cell candidate, the CBC cell interleaved between crypt base Paneth cells10 has attracted renewed interest following studies demonstrating that Lgr5-expressing CBC cells can regenerate all the cell types in the small-intestine crypt-villus unit,11 and indeed, can generate crypt-like structures in vitro with self-renewal activity.12
Given the importance of aberrantly activated STAT3 in many cancers, the resurgent interest in the cancer stem cell concept as a therapeutic target, and the importance of STAT3 function in mES and several tissue stem cell types, we investigated STAT3's role in the murine small-intestine crypt. We used an inducible cytochrome P450 Cyp1A1 promoter-Cre recombinase (AH-Cre) transgene13 and a Stat3 allele carrying LoxP sites flanking the exon encoding STAT3's tyrosine 705 residue (Stat3fl)14 to conditionally inactivate STAT3 in murine small-intestine crypts in response to treatment with the xenobiotic β-naphthoflavone. The inducible AH-Cre transgene allows Cre expression in both the CBC crypt stem cells and in the +4 to +6 position long-term label-retaining crypt stem cells.15 The second Stat3 allele in both control and experimental mice was a Null(−) allele whose exons encoding the SH2 domain and the tyrosine 705 residue have been removed – yielding a STAT3 protein which cannot undergo dimerisation.16
In this study, we provide evidence that STAT3 function is necessary for the survival of both the murine small-intestine crypt +4 to +6 position long-term label-retaining stem cell and the slowly proliferating CBC stem cell.
Results
Use of a Flox-STOP LacZ transgene reveals the transient appearance and subsequent loss of STAT3-null crypts
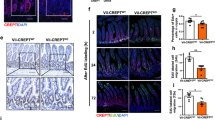
The Flox-STOP LacZ reporter transgene was used as a surrogate marker of recombined small-intestine crypts. By 1 day after the induction of Cre expression, the control Stat3wt/− small intestine expressed LacZ activity at medium level, whereas the experimental Stat3fl/− intestine expressed low levels of LacZ activity. By 2 days post induction, the control Stat3wt/− intestine showed homogeneous high-level LacZ activity, whereas the experimental Stat3fl/− intestine showed only a modest rise in LacZ activity. By 3 days post induction, the control intestine retained high-level LacZ expression as expected. In contrast, the Stat3fl/− intestine showed essentially no macroscopically observable LacZ activity (Figure 1a). Cross-sections of day 3 experimental and control small intestines demonstrated LacZ staining consistent with Cre-recombined Flox-STOP LacZ cells migrating out of the crypt (Figure 1b). Stripping away the smooth muscle layer allowed direct examination of the small-intestine crypt bases. By 3 days post induction, virtually 100% of Stat3wt/− crypt bases had strong LacZ staining, reflecting the continued viability of the Stat3wt/− crypt stem cell(s). In contrast, in the Stat3fl/− experimental intestine, virtually none of the crypt bases retained any LacZ staining activity (Figure 1c).
Use of a Flox-STOP LacZ transgene as a surrogate marker of Stat3fl recombination to visualise the transient appearance and disappearance of STAT3-null crypts. (a) LacZ enzyme activity in Stat3fl/− and Stat3wt/− small intestine at 1, 2 and 3 days after β-naphthoflavone injection/induction of Cre expression. (b) Cross-section of small intestine showing LacZ enzyme activity in Stat3fl/− and Stat3wt/− mice at 3 days after β-naphthoflavone injection/induction of Cre expression. (c) Crypt bases in Stat3fl/− and Stat3wt/− small intestine at 3 days following β-naphthoflavone injection/induction of Cre expression viewed at high power ( × 90 magnification) after removal of the smooth muscle layer. LacZ enzyme activity panels are representative of three Stat3fl/− versus three Stat3wt/− mice per time point. (d) Anti-STAT3 immunoreactivity (with the NEB/CST #9132 primary antibody recognising an epitope close to the STAT3 tyrosine 705 residue) in Stat3fl/− versus Stat3wt/− small intestine at 1.7 days after induction of Cre expression. Immunohistochemistry panels are representative of three Stat3fl/− versus three Stat3wt/− mice, the black bars represent 50 μm. (e) Anti-STAT3 phosphotyrosine 705 immunoreactivity (using the NEB/CST #9131 primary antibody) in un-induced Stat3wt/− control small intestine. The panel is representative of three Stat3wt/− mice, the black bar represents 50 μm. (f) Schematic of the small-intestine crypt base showing the thin CBC/Lgr5+ slowly proliferating stem cells interleaved by Paneth cells with the over-lying +4 to +6 region containing long-term, label-retaining, quiescent stem cells
To confirm that the Stat3fl/− small intestine was functionally STAT3 null following Cre induction, anti-STAT3 immunohistochemistry was performed using the New England Biolabs/Cell Signaling Technology #9132 (NEB/CST#9132; NEB/CST, Ipswich, MA, USA) antibody (recognising an epitope around the STAT3 tyrosine 705 residue) – this revealed a loss of STAT3 immunoreactivity from the Stat3fl/− small-intestine crypts by 1.7 days post induction relative to the Stat3wt/− control (Figure 1d). To confirm that STAT3 is constitutively activated in the crypt epithelium, immunohistochemistry was performed using the NEB/CST #9131 antibody, which recognises tyrosine 705 phosphorylated STAT3. This revealed frequent, strong, nuclear phosphotyrosine 705 immunoreactivity in un-induced Stat3wt/− control crypt epithelial cells (Figure 1e). The small-intestine crypt CBC and +4 to +6 position stem cell locations where AH-Cre transgene expression is induced following β-naphthoflavone treatment are illustrated (Figure 1f).
Individual crypt cell position immunohistochemical analysis of Stat3fl/− versus Stat3wt/− small-intestine epithelium reveals a wave of functionally STAT3-null cells moving up the crypt
Immunohistochemistry using the NEB/CST #9132 antibody (recognising an epitope around the STAT3 tyrosine 705 residue) was performed on Stat3fl/− and Stat3wt/− small intestine from 1.7 , 2 and 3 days after Cre induction. Individual crypt cell positions were assessed for STAT3 staining intensity in 56 half-crypts. To avoid cells with a low level of expression of intact STAT3 confounding the signal from cells which no longer possessed a functional Stat3fl allele, only crypt cells showing absolutely no traces of anti-STAT3 #9132 immunoreactivity (nuclear or cytoplasmic) were counted as negative. By 1.7 days after Cre induction, there was a clearly elevated frequency of STAT3-negative cells in the bottom quarter of the Stat3fl/− crypts relative to the Stat3wt/− crypts (Figure 2a). By 2 days, the elevated STAT3-negative cell frequency was apparent over most of the length of the Stat3fl/− crypts relative to the Stat3wt/− crypts (Figure 2b), while by 3 days, the elevated STAT3-negative cell distribution was confined to the upper half of the Stat3fl/− crypts relative to the Stat3wt/− crypts (Figure 2c) – consistent with a wave of functionally STAT3-null cells migrating upwards from the base of the Cre-recombined Stat3fl/− crypts.
Use of anti-STAT3 immunohistochemistry to trace migration of functionally STAT3-null small-intestine crypt epithelial cells after induction of Cre expression. (a) Average frequency of cells lacking any anti-STAT3 immunoreactivity for Stat3fl/− (dashed line/triangles) and Stat3wt/− (solid line/squares) crypts at individual crypt cell positions at 1.7 days after β-naphthoflavone injection/induction of Cre recombinase expression. (b) Average frequency of cells lacking any anti-STAT3 immunoreactivity for Stat3fl/− (dashed line/triangles) and Stat3wt/− (solid line/squares) crypts at individual crypt cell positions at 2 days after β-naphthoflavone injection/induction of Cre recombinase expression. (c) Average frequency of cells lacking any anti-STAT3 immunoreactivity for Stat3fl/− (dashed line/triangles) and Stat3wt/− (solid line/squares) crypts at individual crypt cell positions at 3 days after β-naphthoflavone injection/induction of Cre recombinase expression. The error bars indicate the values of the standard error of the mean (S.E.M.), N=3 Stat3fl/− versus 3 Stat3wt/− mice per time point
A surge in apoptotic index in Stat3fl/− small-intestine crypt epithelium at 2 days post induction of Cre recombinase expression
Following Cre induction, mice were harvested at various time points and apoptotic and mitotic indices counted from 56 half-crypts in the first 8 cm of (methacarn-fixed, Mayer's haematoxylin and eosin (H+E)-stained) small intestine. The apoptotic indices in both Stat3fl/− and Stat3wt/− small-intestine crypts increased modestly by 1.7 days after Cre induction. By 2 days after Cre induction, the Stat3wt/− control crypt average apoptotic index had essentially returned to baseline values (1.443%), whereas the Stat3fl/− crypt average apoptotic index had increased dramatically (3.803%) and statistically significantly (P=0.0404, N=3 Stat3fl/− versus 3 Stat3wt/−) versus the control (Figure 3a). A statistically significant increase in apoptotic index in Stat3fl/− crypts versus Stat3wt/− controls was also observed at 3 days (2.43 versus 1.513%, P=0.0404, N=3 Stat3fl/− versus 3 Stat3wt/−), while by 4 days after induction, the Stat3fl/− apoptotic index had returned to the baseline values of the Stat3wt/− control crypts.
Changes in an average apoptotic and mitotic indices and average crypt length in Stat3fl/− and wt/− small-intestine crypts in response to induction of Cre recombinase expression. (a) Apoptotic indices (average percentage of crypt cells undergoing apoptosis) in Stat3fl/− (dashed line/triangles) and Stat3wt/− (solid line/squares) crypts between 1 and 4 days following β-naphthoflavone-driven induction of Cre recombinase expression. (b) Mitotic indices (average percentage of crypt cells undergoing mitosis) in Stat3fl/− (dashed line/triangles) and Stat3wt/− (solid line/squares) crypts between 1 and 4 days following induction of Cre expression. (c) Average length of small-intestine crypts in Stat3fl/− (dashed line/triangles) and Stat3wt/− (solid line/squares) mice between 1 and 4 days following induction of Cre expression. The error bars indicate S.E.M. values, N=3 Stat3fl/− versus 3 Stat3wt/− mice per time point
However, examination of the Stat3fl/− small-intestine crypt mitotic indices and average crypt cell number showed no statistically significant differences compared with Stat3wt/− control crypts (Figures 3b and c). Similarly, there were no obvious changes in crypt-villus phenotype in Stat3fl/− versus Stat3wt/− small intestine following two cycles of Cre induction (at 4 and 8 weeks after birth), followed by aging mice for 1–2 years (see Supplementary Figure S1).
When the overall apoptotic indices were broken down into apoptotic frequencies at individual crypt cell positions, the apoptotic frequency had clearly increased at positions +5 and +6 up from the crypt base in Stat3fl/− versus Stat3wt/− control crypts by 1.7 days post induction (Figure 4a). By 2 days post induction, there was a general increase in apoptotic frequency throughout the lower half of the Stat3fl/− versus Stat3wt/− crypts (Figure 4b), while by 3 days, the Stat3fl/− crypt positions showing elevated apoptotic frequencies relative to control crypts had shifted to the upper half of the crypt (Figure 4c) – again consistent with the proliferation, migration and apoptosis of functionally STAT3-null crypt epithelial cells.
Changes in individual small-intestine crypt cell apoptotic indices in Stat3fl/− and Stat3wt/− crypts in response to induction of Cre recombinase expression. (a) Apoptotic indices for Stat3fl/− (dashed line/triangles) and Stat3wt/− (solid line/squares) crypts at individual crypt cell positions at 1.7 days after β-naphthoflavone injection/induction of Cre recombinase expression. (b) Apoptotic indices for Stat3fl/− (dashed line/triangles) and Stat3wt/− (solid line/squares) crypts at individual crypt cell positions at 2 days after β-naphthoflavone injection/induction of Cre recombinase expression. (c) Apoptotic indices for Stat3fl/− (dashed line/triangles) and Stat3wt/− (solid line/squares) crypts at individual crypt cell positions at 3 days after β-naphthoflavone injection/induction of Cre recombinase expression. The error bars indicate S.E.M. values, N=3 Stat3fl/− versus 3 Stat3wt/− mice per time point
Activation of caspase 3 correlates with loss of functional STAT3 in the crypt
To confirm the apoptotic index measurements, activated (cleaved) caspase 3 immunohistochemistry was performed on the small intestine. By day 2, there was an elevated frequency of activated caspase 3-positive cells in the Stat3fl/− small-intestine crypt versus the Stat3wt/− control (Figure 5a). When activated caspase 3-positive cell frequencies were assessed at individual crypt cell positions, there was a clear elevation in the Stat3fl/− crypt relative to the Stat3wt/− in the central third of the crypt (Figure 5b). Further, the overall frequency of activated caspase 3-positive cells by 2 days after Cre induction was significantly higher (4.207 versus 2.37%, P=0.0404, N=3 Stat3fl/− versus 3 Stat3wt/−) in the Stat3fl/− versus Stat3wt/− crypts (Figure 5c). These activated caspase 3 frequencies were in reasonable agreement with the H+E-derived apoptotic indices presented earlier (Figure 3a), a representative H+E-stained apoptotic body is illustrated (Figure 5d).
Correlation of elevated apoptotic indices in Stat3fl/− crypts following induction of Cre recombinase expression with elevated activated (proteolytically cleaved) caspase 3 immunoreactivity. (a) Anti-activated caspase 3 immunoreactivity in Stat3wt/− and Stat3fl/− small-intestine crypts at 2 days after Cre induction, the black bars represent 50 μm. (b) Frequency of activated caspase 3 immunoreactive cells at individual crypt cell positions in Stat3fl/− (dashed line/triangles) versus Stat3wt/− (solid line/squares) small intestine at 2 days after Cre induction. The error bars indicate S.E.M. values, N=3 Stat3fl/− versus 3 Stat3wt/− mice per time point. (c) Overall frequency of activated caspase 3 immunoreactive cells in Stat3wt/− (solid bar) versus Stat3fl/− (open bar) small-intestine crypts at 2 days after Cre induction, error bars indicate S.E.M. values, N=3 Stat3fl/− versus 3 Stat3wt/− mice. (d) Representative apoptotic body (arrowed) in Stat3fl/− small-intestine crypt at 2 days after induction of Cre expression, the black bar represents 20 μm
Loss of bromodeoxyuridine-labelled long-term label-retaining cells at positions +4 to +6 in the Stat3fl/− crypt following Cre induction
Long-lived label-retaining cells in small-intestine crypts were identified using a bromodeoxyuridine (BrdU)-labelling approach, allowing investigation of the requirement for functional STAT3 for long-term label-retaining crypt stem cell survival.
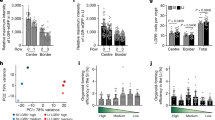
Following anti-BrdU immunohistochemistry, all well-formed crypts were counted, and those containing BrdU-positive nuclei identified. Subsequently, any BrdU-positive cells showing any trace of eosin-staining granules and a clear cytoplasm – that is any remaining labelled Paneth cells – were excluded. The remaining BrdU-positive cells were then assigned to the crypt base region (crypt positions +1 to +3 from the base), the crypt +4 to +6 region (Figure 6a), or to higher positions within the crypt. The highest frequency of long-term label-retaining cells was clearly located in the +4 to +6 region up from the Stat3wt/− crypt base (Figure 6b) – as expected from previous studies of crypt stem cells/label-retaining cells.9 Interestingly, the only region showing any significant difference in the frequency of BrdU-positive nuclei between the Stat3fl/− and Stat3wt/− genotypes was the +4 to +6 region – with the frequency of BrdU-positive nuclei being much lower in the Stat3fl/− crypts compared with Stat3wt/− crypts (0.62 versus 1.85%, P=0.0404, N=3 Stat3fl/− versus 3 Stat3wt/−). Hence, functional STAT3 is required for crypt +4 to +6 region long-term label-retaining stem cell survival.
Long-term BrdU labelling of Stat3fl/− and Stat3wt/− small-intestine crypts demonstrating the loss of BrdU-labelled +4 to +6 crypt cells in response to STAT3 inactivation. Stat3fl/− and Stat3wt/− mice were given BrdU in their drinking water for 3 weeks, followed by an 8-week water-only chase phase. This was followed by β-naphthoflavone injection and induction of Cre recombinase expression with a 7 day delay before harvesting. Small-intestine samples were then analysed by anti-BrdU immunohistochemistry and all BrdU-positive, non-Paneth, nuclei identified. The BrdU-positive cells were then divided up into three groups – crypt base positions +1 to +3 (base), the +4 to +6 positions (+4 to +6) and crypt positions from +7 to the crypt-villus junction (higher). (a) Anti-BrdU immunohistochemistry of a (Stat3wt/−) crypt demonstrating a representative BrdU-positive nucleus (arrowed) in the +4 to +6 crypt position range, the black bar represents 20 μm. (b) Frequency of crypts retaining a BrdU-positive nucleus at the crypt base, +4 to +6 and higher positions, Stat3wt/− crypts are represented by hatched bars, whereas Stat3fl/− crypts are represented by open bars. The error bars indicate S.E.M. values, N=3 Stat3fl/− versus 3 Stat3wt/− mice
Transient appearance of the recombined Stat3fl allele
Given the transient appearance of LacZ activity around 2 days after Cre induction and the peak in Stat3fl/− crypt apoptotic activity by 2 days after induction, it seemed likely that STAT3 inactivation resulted in the elimination of Cre-recombined Stat3fl/− crypt stem cells. To confirm this, a quantitative polymerase chain reaction (Q-PCR) was performed on small-intestine DNA using a primer pair, which would amplify only the recombined Stat3fl allele. A control Q-PCR using primers specific for a distant region of all three Stat3 allele variants ensured that all reactions had equal amounts and integrity of template DNA.
With the control primer pair, 400 ng of genomic DNA yielded low and essentially constant threshold cycle (Ct) values of ∼21 cycles for both the Stat3fl/− and Stat3wt/− small intestines before induction and over the time course of 1–4 days following Cre induction (Figure 7a). In contrast, the Ct values for the Q-PCR reaction detecting the recombined Stat3fl allele were initially very high in both the Stat3fl/− and Stat3wt/− intestine DNA at ∼34 cycles. Following Cre induction, the Ct values for the recombined Stat3fl allele reaction remained very high and fairly constant in the Stat3wt/− control intestine DNA at about 35 cycles throughout the time course. In contrast, in the Stat3fl/− experimental intestine, the Ct values for the recombined Stat3fl Q-PCR decreases rapidly, reaching a minimum around 2 days after induction, after which the Ct values began increasing again towards the recombined Stat3fl allele Ct values seen in the Stat3wt/− control intestine Q-PCR (Figure 7a) – behaviour consistent with the transient appearance and disappearance of the recombined Stat3fl allele in the Stat3fl/− small intestine.
Q-PCR demonstrating the transient appearance and disappearance of the recombined Stat3fl allele. (a) Ct values (cycles) were plotted versus time after induction of Cre expression for the recombined Stat3fl allele Q-PCR reaction using Stat3fl/− small-intestine genomic DNA (solid line, solid squares) and for the recombined Stat3fl allele Q-PCR reaction using Stat3wt/− small-intestine genomic DNA (dashed line, solid triangles). Ct values were also plotted versus time after induction of Cre expression for the control Stat3 (wt/fl/− alleles) Q-PCR reaction using Stat3fl/− small-intestine genomic DNA (solid line, open squares) and for the control Q-PCR reaction using Stat3wt/− small-intestine genomic DNA (dashed line, open triangles). The error bars indicate S.E.M. values, N=3 Stat3fl/− versus 3 Stat3wt/− mice per time point. (b) Q-PCR Ct values for the recombined Stat3fl allele reaction versus time after induction of Cre expression were then converted into relative, linear-scaled, values for the level of the recombined Stat3fl allele. This involved subtracting the average control Stat3 Q-PCR Ct value from the average recombined Stat3fl Q-PCR Ct value to yield a difference value (difference). The un-induced (day 0) time point difference value from Stat3wt/− intestine genomic DNA was then normalised to the Stat3fl/− intestine genomic DNA difference value to give a normalised difference value (normalised difference). Then the Stat3fl/− difference values were subtracted from the Stat3wt/− normalised difference values to yield the difference of these difference values (difference of difference). Finally, the difference of difference values were raised to 2x to yield a relative measure of the recombined Stat3fl allele, which could be plotted directly on a linear scale (no error bars are provided). N=3 Stat3fl/− versus 3 Stat3wt/− mice per time point
Relating the recombined Stat3fl and control Stat3 Q-PCR Ct values from Stat3fl/− and Stat3wt/− intestine DNA, normalising to un-induced (day 0) values and raising the output value to 2x allowed a relative recombined Stat3fl allele level to be plotted linearly. This approach revealed a dramatic rise and fall in the relative level of the recombined Stat3fl allele in the Stat3fl/− versus the Stat3wt/− intestine, peaking at around 2 days after Cre induction (Figure 7b).
Elevated c-Myc and p53 expression implicated in the apoptotic removal of Cre-recombined Stat3fl/− crypt cells
We also demonstrate statistically significantly (P=0.0404, N=3 Stat3fl/− versus 3 Stat3wt/−) elevated c-Myc expression (1.283 versus 0.816 positive nuclei per crypt) in Stat3fl/− relative to Stat3wt/− crypts at 0.5 days after Cre induction (Figures 8a and b). STAT3-driven c-Myc expression17 in non-recombined Stat3fl/− crypt cells could trigger apoptosis in Cre-recombined Stat3fl/− cells through a cellular competition mechanism,18 accelerating crypt repopulation with stem cells retaining a functional Stat3fl allele. The apoptotic mechanism triggered in the recombined Stat3fl/− cells may be p53 mediated as the frequency of p53-positive crypt nuclei is significantly elevated in Stat3fl/− crypts relative to Stat3wt/− crypts at 2 days (2.169 versus 0.492%, P=0.0259, N=4 Stat3fl/− versus 3 Stat3wt/−) and 3 days (2.204 versus 0.335%, P=0.0259 or P=0.0249 adjusted for ties, N=4 Stat3fl/− versus 3 Stat3wt/−) after Cre induction (Figure 8c).
Upregulation of c-Myc expression in Stat3fl/− relative to Stat3wt/− crypts at day 0.5 after induction of Cre expression as a potential mechanism for non-recombined Stat3fl/− cells to trigger p53-mediated apoptosis in Cre-recombined Stat3fl/− cells. (a) Anti-c-Myc immunohistochemistry in Stat3fl/− versus Stat3wt/− crypts at day 0.5 after induction of Cre expression. Immunohistochemistry panels are representative of three Stat3fl/− versus three Stat3wt/− mice, the black bars represent 20 μm. (b) Statistically significant increase in numbers of c-Myc-positive nuclei in Stat3fl/− (open bar) versus Stat3wt/− (hatched bar) crypts at day 0.5 after induction of Cre expression (P=0.0404), the error bars indicate S.E.M. values, N=3 Stat3fl/− versus 3 Stat3wt/− mice. (c) Quantitation of anti-p53 immunohistochemistry demonstrating a statistically significant increase in the frequency of p53-positive cell nuclei in Stat3fl/− (open bars) relative to Stat3wt/− (hatched bars) crypts at 2 days (P=0.0259, N=4 Stat3fl/− versus 3 Stat3wt/− mice) and 3 days (P=0.0259 or 0.0249 adjusted for ties), N=4 Stat3fl/− versus 3 Stat3wt/− mice) after induction of Cre expression
Caspase 3 activation in STAT3-depleted Cre-recombined Stat3fl/− crypt epithelial cells
To directly demonstrate that STAT3 inactivation in a crypt epithelial cell can lead to the apoptosis of that cell, we performed confocal fluorescent immunohistochemistry on serial small-intestine tissue sections using an anti-activated caspase 3 antibody combined with a 4′,6-diamidino-2-phenylindole (DAPI) nuclear counterstain versus, on alternating slides, the NEB/CST #9132 anti-STAT3 antibody, again combined with a DAPI counterstain. This demonstrated the presence in day 2 Cre-recombined Stat3fl/− crypts of epithelial cells, which showed essentially no detectable STAT3 but which displayed clear activated caspase 3 immunoreactivity (Figure 9).
Confocal fluorescence immunohistochemistry demonstrating activation of caspase 3 cleavage in a Stat3fl/− crypt at 2 days after induction of Cre expression and its correlation with loss of STAT3 immunoreactivity in corresponding cells identified (arrowed) on the subsequent serial tissue section. The white scale bars represent 20 μm
Elevated Bmi1 in the recombined Stat3fl/− crypt as a possible mechanism to accelerate crypt repopulation
It has been suggested that the polycomb transcriptional repressor family member Bmi1 marks the small-intestine crypt +4 to +6 region stem cell, with expression at the +4 and +5 positions up from the crypt base.19 Hence, we performed anti-Bmi1 immunohistochemistry on Stat3fl/− and Stat3wt/− small intestine. In contrast to the expected obvious Bmi1 immunoreactivity at the +4 and +5 positions in Stat3wt/− crypts, there was a generally very low frequency of Bmi1-positive nuclei, which were usually found in the lower half of the crypt. In contrast, there was a very obvious significant increase in the frequency of strongly Bmi1-positive nuclei throughout the crypt in Stat3fl/− relative to Stat3wt/− crypts at 2 days (10.85 versus 3.39%) and 3 days (28.81 versus 1.02%) after Cre induction (P=0.0404, N=3 Stat3fl/− versus 3 Stat3wt/−; Figures 10a–c), with Stat3fl/− Bmi1 immunoreactivity reverting to control values by 4 days after Cre induction.
Quantitation of a transient peak in Bmi1 expression in Stat3fl/− crypts following β-naphthoflavone injection and induction of Cre expression. (a) Stat3fl/− and Stat3wt/− small intestines were subjected to anti-Bmi1 immunohistochemistry. Strongly anti-Bmi1 immunoreactive crypt nuclei were counted and the average percentage of strongly positive nuclei in Stat3wt/− (hatched bars) and Stat3fl/− (open bars) crypts charted. The error bars indicate S.E.M. values, N=3 Stat3fl/− versus 3 Stat3wt/− mice per time point. (b) Anti-Bmi1 immunohistochemistry from a Stat3wt/− small intestine at 3 days after induction of Cre expression. The immunohistochemistry panel is representative of three Stat3wt/− mice, the black bar represents 20 μm. (c) Anti-Bmi1 immunohistochemistry from a Stat3fl/− small intestine at 3 days after induction of Cre expression. The immunohistochemistry panel is representative of three Stat3fl/− mice, the black bar represents 20 μm
It might be difficult to reconcile massively increased Bmi1 expression with a role as a robust marker of the crypt +4 to +6 region stem cell. Others have suggested that elevated Bmi1 expression (at least in murine ES cells) is rather a marker of differentiation and associated with exit from the self-renewing/pluripotent state.20 Elevated Bmi1 expression on exiting the self-renewing state in murine ES cells might also occur in recombined Stat3fl/− crypt stem cells, hastening crypt repopulation with stem cells retaining a functional Stat3fl allele.
Discussion
This study demonstrates the requirement for functional STAT3 in the crypt stem cells of the murine small intestine, with recombined Stat3fl/− crypt stem cells being rapidly eliminated and the intestinal epithelium being repopulated with cells retaining functional STAT3.
The AH-Cre transgene allows Cre expression in both small-intestine crypt stem cell compartments – the +4 to +6 position long-term label-retaining master stem cells, and the proliferating, non-label-retaining, Lgr5+ CBC stem cells driving tissue homeostasis. The +5 to +6 location of the initial burst of apoptosis seen in the Stat3fl/− crypt relative to the Stat3wt/− crypt at day 1.7 following induction of Cre expression corresponds well with the location of the +4 to +6 label-retaining quiescent master stem cell. Whereas by day 2 following Cre induction, the elevated apoptotic index in the Stat3fl/− crypt is also clear at lower positions in the crypt. This might reflect two separate events – an initial recombination-driven apoptotic pulse by day 1.7 in the Stat3fl/− +4 to +6 master stem cells, followed at around day 2 by a wave of functionally STAT3-null cells generated from the recombined Stat3fl/− Lgr5+ CBC cells moving up the crypt and undergoing apoptosis. The rapid removal of Cre-recombined Stat3fl/− cells might involve STAT3-driven c-Myc expression17 and cellular competition18 whereby non-recombined Stat3fl/− cells could trigger p53-dependent apoptosis in Cre-recombined Stat3fl/− cells. The upregulated c-Myc expression in Stat3fl/− versus Stat3wt/− crypts at day 0.5 and the upregulated p53 expression in Stat3fl/− crypts at days 2 and 3 following Cre induction could support such a mechanism. Further, confocal fluorescence microscopy experiments suggested that the loss of functional STAT3 in recombined Stat3fl/− crypts sensitises cells to caspase 3-driven apoptosis.
The failure to observe obvious anti-Bmi1 immunoreactivity at the +4 and +5 positions in the Stat3wt/− crypt base (in the expectation of reduced immunoreactivity following Stat3fl/− crypt recombination) was unexpected. Instead, there was a massive upregulation of strong anti-Bmi1 immunoreactivity in the Stat3fl/− crypt at 2–3 days following Cre induction.
One explanation for the upregulated crypt Bmi1 expression following STAT3 inactivation might lie in a recent study of a murine ES cell chromatin remodelling complex, esBAF, critical for maintaining ES cell pluripotency and self-renewal.20 Chromatin immunoprecipitation-sequencing demonstrated a highly statistically significant 70% co-occupation of murine ES cell target genes by both STAT3 and the esBAF ATPase component Brg.20
Further, knocking-down Brg expression upregulated the expression of several polycomb group (PcG) genes – including Bmi1, Cbx7 and Ring1a – which normally contribute to the polycomb repressive complex 1. These genes also contained a statistically significant region bound by Brg, with the suggestion that esBAF/Brg normally antagonises PcG function in murine ES cells by binding to and repressing PcG gene transcription. In a striking parallel with Brg knockdown in murine ES cells, STAT3 inactivation in the small-intestine crypt also upregulated Bmi1 expression, implying that the frequent co-occupation of murine ES cell target genes by STAT3 and esBAF/Brg is functionally important – with a potentially analogous interaction in the crypt stem cell(s).
In contrast, several PcG subunit genes were not upregulated following Brg knockdown, these included Suz12, Eed and Ezh2, encoding Polycomb repressive complex 2 components.20 Interestingly, the Eed gene is transcriptionally controlled by STAT3 (and Oct4) in murine ES cells, with STAT3/Oct4 (via Eed) reversibly silencing differentiation-associated genes such as Tbx5, GATA6 and Cdx2.21 Further, Eed and Bmi1 can function antagonistically – in haematopoietic cell proliferation, Eed demonstrated an anti-proliferative function, negatively regulating lymphoid and myeloid progenitor cell numbers in the bone marrow,22 whereas Bmi1 positively regulated primitive bone marrow progenitor cell proliferation.22
Thus, one model for STAT3 function in the murine small-intestine crypt stem cell(s) could involve an interaction between STAT3 and a stem cell chromatin remodelling complex to bind to the Bmi1 gene, suppressing Bmi1 expression and restraining crypt stem cell proliferation. In addition, STAT3 could drive Eed gene expression, again downregulating crypt stem cell proliferation. STAT3 inactivation would impair both of these potential mechanisms to restrain crypt stem cell proliferation. The STAT3-null crypt stem cells would then exit the pluripotent/self-renewing state, proliferate, and be removed through apoptosis, accelerating the repopulation of the recombined Stat3fl/− small intestine.
Materials and Methods
Generation of mouse strains
Mice carrying the Floxed(fl) and Null(−) Stat3 alleles were the kind gift of Professor S Akira,14, 16 whereas mice carrying the AH-Cre transgene were the kind gift of Dr. DJ Winton.13 Mice used in this study were outbred, segregating for the C57BL6/J 129/Ola and C3H genomes, with littermates being used as controls. Murine genomic DNA was isolated using a commercial DNA purification kit (5 Prime GmbH, Hamburg, Germany) and genotyped by PCR using primers which are specific for the Stat3wt versus fl alleles, for the Stat3− allele and the Cre and LacZ transgenes. The following oligodeoxynucleotide primer pairs were used for PCR genotyping: 5′-CCTGAAGACCAAGTTCATCTGTGTGAC-3′ and 5′-CACACAAGCCATCAAACTCTGGTCTCC-3′ yields an ∼320-bp PCR product for the Stat3fl allele versus an ∼230-bp PCR product for the Stat3wt allele. For the Stat3 Null(−) allele PCR reaction, primers 5′-AGCAGCTGACAACGCTGGCTGAGAAGCT-3′ and 5′-ATCGCCTTCTATCGCCTTCTTGACGA-3′ yield an ∼1-kbp PCR product. For the combined PCR reaction for Cre and LacZ transgenes, the 5′-TGACCGTACACCAAAATTTG-3′ and 5′-ATTGCCCCTGTTTCACTATC-3′ primer pair yields an ∼1-kbp PCR product for the Cre transgene and the 5′-CTGGCGTTACCCAACTTAAT-3′ and 5′-ATAACTGCCGTCACTCCAAC-3′ primer pair yields an ∼500-bp PCR product for the LacZ transgene. All mice used in this study were male, between 3 and 6 months old (with the exception of Supplementary Figure S1), with three or more mice per time point for both genotypes, and were cared for in accordance with UK Home Office guidelines.
Induction of recombination and harvesting experimental and control mice
To induce Cre recombinase expression, male mice were injected intraperitoneally with a 10 mg/ml β-naphthoflavone (Sigma, St. Louis, MO, USA) solution in corn oil (Sigma) to give a final β-naphthoflavone dose of 100 mg per kg body weight. Three intraperitoneal injections were administered in 1 day with injections spaced 2 h apart for the 1, 1.7 and 2 day time points and 3–4 h apart for the 3 and 4 day time points, all mice were harvested in the afternoon. After harvesting, the first 8 cm of small intestine and the final remaining bulk part of the small intestine were transferred into methacarn fixative (4 : 2 : 1 volumes methanol : chloroform : glacial acetic acid) overnight before transfer to neutral-buffered formalin (Sigma) and tissue processing. The intervening section of small intestine was divided into samples for overnight fixation in cold formalin and paraffin embedding for immunohistochemistry (quick fixed), and for genomic DNA extraction.
Long-term BrdU label-retention experiment
For long-term BrdU label-retention experiments, mice were given BrdU (Sigma) dissolved in their drinking water at 3 mg/ml for 3 weeks, followed by an 8 week chase period with water alone. Mice were then given three 100 mg/kg β-naphthoflavone (in corn oil) intraperitoneal injections 4 h apart to induce Cre recombinase expression and harvested 7 days later. Following harvesting, small-intestine samples were quick fixed and then subjected to anti-BrdU immunohistochemistry, brief haematoxylin counterstaining and extended eosin staining. All well-formed crypts were counted and any nuclei showing BrdU immunoreactivity were identified, with any BrdU-positive cells showing histochemical or morphological features of Paneth cells being discounted.
LacZ staining of small intestine
For LacZ staining, small intestines were harvested, flushed through with ice-cold phosphate-buffered saline (PBS; Invitrogen, Carlsbad, CA, USA) and then flushed through with ice-cold X-gal fixative solution (0.5 volumes formalin, 0.5 volumes PBS, 0.004 volumes 25% glutaraldehyde (wt/vol) (Sigma) solution). Sections of small intestine were then cut open, pinned flat and lumen-uppermost on a paraffin wax-filled Petri dish and fixed for 1 h in X-gal fixative, then washed with PBS and incubated with demucifying solution (10% (vol/vol) glycerol, 10 mM Tris-HCl pH8.2, 20% (vol/vol) ethanol, 92 mM NaCl, 22 mM dithiothreitol) for 1 h. Subsequently, the intestine was washed with PBS and any adhering mucus removed. The dish was then incubated overnight in the dark with gentle agitation with X-gal staining solution (0.004 volumes of 5% (wt/vol) X-gal in dimethylformamide (Promega, Madison, WI, USA) in 10 mM MgCl2, 29 mM potassium ferricyanide, 30 mM potassium ferrocyanide in PBS). Finally, the X-gal staining solution was discarded and the intestine washed with PBS and formalin.
Measurements of apoptotic and mitotic indices in murine small intestine
For measurements of apoptotic and mitotic indices in the crypts of the small intestine, methacarn-fixed small-intestine slides were stained with H+E (RA Lamb, Thermofisher Scientific, Waltham, MA, USA). Apoptotic and mitotic cells were identified by their characteristic morphologies with each individual cell position within 56 half-crypts being assessed and counted to yield apoptotic and mitotic indices at each individual position within the crypt. These individual crypt position frequencies were then summed to yield overall crypt apoptotic and mitotic indices.
Immunohistochemistry
STAT3 immunohistochemistry used a primary antibody (NEB/CST #9132) recognising an epitope close to the STAT3 tyrosine 705 residue. Antigen retrieval used heating slides in 1 × citrate buffer (LabVision, Thermofisher Scientific). The primary rabbit anti-STAT3 antibody NEB/CST #9132 was diluted 1 : 100 in 5% normal goat serum (NGS (vol/vol); Dako, Glostrup, Denmark) in 1 × Tris-buffered saline (TBS; Sigma). The secondary antibody step used anti-rabbit horseradish peroxidase (HRP)-labelled polymer (EnVision+, Dako). HRP activity was then visualised with a 3,3′-diaminobenzidine (DAB) chromogenic substrate system (EnVision+, Dako), DAB staining used a 10 min development time.
STAT3 phosphotyrosine 705 immunohistochemistry used the NEB/CST #9131 primary antibody. Antigen retrieval used heating slides in 1 × EDTA buffer (LabVision). The primary rabbit anti-phosphotyrosine 705 STAT3 antibody (NEB/CST #9131) was diluted 1 : 100 in 5% NGS (vol/vol) in 1 × TBS, 0.1% (vol/vol) Tween-20 (Sigma; TBS/T). The secondary antibody step used anti-rabbit HRP-labelled polymer (EnVision+, Dako), DAB (Dako) staining used a 10 min development time.
Activated (cleaved) caspase 3 immunohistochemistry used antigen retrieval with slides heated in 1 × EDTA buffer (LabVision), the rabbit monoclonal anti-activated caspase 3 primary antibody (NEB/CST #9664) was diluted 1 : 200 in 5% NGS (Dako) (vol/vol) in 1 × TBS/T (Sigma). The secondary antibody step used an anti-rabbit HRP-labelled polymer (EnVision+, Dako). DAB (Dako) staining used a 15 min development time.
BrdU immunohistochemistry/eosin staining for the long-term label-retention experiment used antigen retrieval by heating slides in 1 × citrate buffer (LabVision). The mouse anti-BrdU primary antibody (Becton Dickinson #347580, San Diego, CA, USA) was diluted 1 : 250 in 10% NGS (vol/vol) in 1 × TBS/T. The secondary antibody step used an anti-mouse HRP-labelled polymer (EnVision+, Dako). DAB (Dako) staining used a 40 min development time. Following counterstaining in haematoxylin for 20 s and washing, slides were stained in eosin for 8 min to identify Paneth cells, then briefly washed, dehydrated and mounted.
c-Myc immunohistochemistry used antigen retrieval with slides heated in 1 × citrate buffer (LabVision). The rabbit anti-c-Myc primary antibody (sc-788; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was diluted 1 : 100 in TBS/T, with an anti-rabbit HRP-labelled polymer (EnVision+, Dako) secondary antibody step and a 30 min DAB (Dako) staining development time.
Bmi1 immunohistochemistry used antigen retrieval with slides heated in 1 × EDTA buffer (LabVision). The rabbit anti-Bmi1 primary antibody (NEB/CST #2830) was diluted 1 : 100 in 10% NGS (vol/vol) in TBS/T. The secondary antibody step used anti-rabbit HRP-labelled polymer (EnVision+, Dako). DAB (Dako) staining used a 15 min development time.
Quantitative polymerase chain reaction assay for recombined Stat3fl allele
Q-PCR assays to determine the level of the recombined Stat3fl allele were carried out on genomic DNA prepared from small-intestine tissue samples using a QIAamp DNA Mini kit (Qiagen, Hilden, Germany) and performed using a StepOne Plus real-time PCR system (Applied Biosystems, Carlsbad, CA, USA). For each genotype/time point, Q-PCR reactions were performed on small-intestine genomic DNA from three independent mice, with each 20 μl reaction containing 400 ng of template DNA, an oligodeoxynucleotide primer pair at a concentration of 1.25 μM each and 1 × SYBR Green and 1 × ROX passive reference dye (Dynamo, Finnzymes, Espoo, Finland). To detect the recombined Stat3fl allele, a primer pair was designed with a coding sense primer hybridising close to the start of intron 21–22 (5′-TTCTCCAGTCAGTGGGTTCCA-3′) and a non-coding sense primer hybridising beyond the HindIII restriction site in intron 22–23 (5′-AGGGCCAGGTTGACAAGACAC-3′) – which yielded a single Q-PCR product of ∼250 bp in the presence of the recombined Stat3fl allele, but did not yield any product with the non-recombined Stat3fl allele, nor with the Stat3wt or Stat3− alleles. As controls, Q-PCR reactions were performed using a primer pair, which hybridised with a distant region of all three Stat3 alleles. A coding sense primer hybridising near the 5′-end of Stat3 exon 5 (5′-GGCCAGGCCAACCACCCAAC-3′) and a non-coding sense primer hybridising near the 3′-end of intron 5–6 (5′-AACACAGGTTACCTTTGGGA-3′) yielded a single Q-PCR product of ∼230 bp with all three Stat3 alleles. To amplify the recombined Stat3fl product, the following Q-PCR programme was used: 95 °C for 10 min, 95 °C for 15 s, 65 °C for 1 min, repeated for 40 cycles, and finally 95 °C for 15 s. To amplify the control total Stat3 product, the following programme was used: 95 °C for 10 min, 95 °C for 15 s, 60 °C for 1 min, repeated for 40 cycles.
Confocal fluorescence immunohistochemical analysis of murine small intestine
Confocal fluorescence microscopy was performed using a Leica (Heidelberg, Germany) TCS SP2 AOBS spectral confocal microscope using a × 63 oil-immersion objective and analysed using proprietary Leica confocal software. Serial small-intestine tissue section slides were stained for either STAT3 or activated caspase 3. Anti-STAT3 fluorescence immunohistochemistry used antigen retrieval by heating slides in 1 × citrate buffer (LabVision). The anti-STAT3 NEB/CST #9132 rabbit primary antibody was used at a 1 : 100 dilution in 5% (vol/vol) NGS in TBS/T. The secondary antibody step used an Alexa 488-conjugated goat anti-rabbit reagent (Invitrogen) at a 1 : 400 dilution in TBS/T, with coverslips eventually mounted using Vectashield Hard-Set aqueous mounting medium containing DAPI (Vector, Burlingame, CA, USA). Anti-activated caspase 3 fluorescence immunohistochemistry used antigen retrieval by heating slides in 1 × EDTA buffer (LabVision). The anti-activated caspase 3 NEB/CST #9661 rabbit primary antibody was used at a 1 : 100 dilution in 5% (vol/vol) NGS in TBS/T. The secondary antibody step used an Alexa 594-conjugated goat anti-rabbit reagent (Invitrogen) at a 1 : 400 dilution in TBS/T, with coverslips mounted using DAPI-containing Vectashield Hard-Set mounting medium (Vector).
Statistical analysis
Tests for statistical significance used the non-parametric Mann–Whitney U-test with a 95% confidence interval.
Abbreviations
- AH-Cre:
-
Cyp1A1 promoter-Cre recombinase
- BrdU:
-
bromodeoxyuridine
- CBC:
-
crypt base columnar cell
- Ct:
-
threshold cycle
- DAPI:
-
4′,6-diamidino-2-phenylindole
- Fl:
-
floxed
- H+E:
-
haematoxylin plus eosin
- HRP:
-
horseradish peroxidase
- mES:
-
murine embryonic stem cell
- NGS:
-
normal goat serum
- PcG:
-
polycomb group
- Q-PCR:
-
quantitative polymerase chain reaction
- SH2:
-
src homology 2
- STAT:
-
signal transducer and activator of transcription
- TBS:
-
Tris-buffered saline
- WT:
-
wild type
- –:
-
null
References
Schindler C, Darnell JE . Transcriptional responses to polypeptide ligands – the JAK-STAT pathway. Ann Rev Biochem 1995; 64: 621–651.
Zhong Z, Wen ZL, Darnell JE . STAT3 – a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 1994; 264: 95–98.
Yu H, Jove R . The stats of cancer – new molecular targets come of age. Nat Rev Cancer 2004; 4: 97–105.
Haura EB, Turkson J, Jove R . Mechanisms of disease: insights into the emerging role of signal transducers and activators of transcription in cancer. Nat Clin Pract Oncol 2005; 2: 315–324.
Niwa H, Burdon T, Chambers I, Smith A . Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 1998; 12: 2048–2060.
Dyce PW, Zhu H, Craig J, Li JL . Stem cells with multilineage potential derived from porcine skin. Biochem Biophys Res Comm 2004; 316: 651–658.
Oh IH, Eaves CJ . Overexpression of a dominant negative form of STAT3 selectively impairs hematopoietic stem cell activity. Oncogene 2002; 21: 4778–4787.
Barker N, van de Wetering M, Clevers H . The intestinal stem cell. Genes Dev 2008; 22: 1856–1864.
Potten CS, Booth C, Pritchard DM . The intestinal epithelial stem cell: the mucosal governor. Int J Exp Path 1997; 78: 219–243.
Cheng H, Leblond CP . Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine V. Unitarian theory of the origin of the four epithelial cell types. Am J Anat 1974; 141: 537–561.
Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007; 449: 1003–1007.
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009; 459: 262–265.
Ireland H, Kemp R, Houghton C, Howard L, Clarke AR, Sansom OJ et al. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta-catenin. Gastroenterology 2004; 126: 1236–1246.
Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S . Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol 1998; 161: 4652–4660.
Ireland H, Houghton C, Howard L, Winton DJ . Cellular inheritance of a Cre-activated reporter gene to determine Paneth cell longevity in the murine small intestine. Dev Dynam 2005; 233: 1332–1336.
Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA 1997; 94: 3801–3804.
Bowman T, Broome MA, Sinibaldi D, Wharton W, Pledger WJ, Sedivy JM et al. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci USA 2001; 98: 7319–7324.
Moreno E, Basler K . dMyc Transforms cells into super-competitors. Cell 2004; 117: 117–129.
Sangiorgi E, Capecchi MR . Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 2008; 40: 915–920.
Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR . An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA 2009; 106: 5187–5191.
Ura H, Usuda M, Kinoshita K, Sun C, Mori K, Akagi T et al. STAT3 and Oct-3/4 control histone modification through induction of Eed in embryonic stem cells. J Biol Chem 2008; 283: 9713–9723.
Lessard J, Schumacher A, Thorsteinsdottir U, van Lohuizen M, Magnuson T, Sauvageau G . Functional antagonism of the Polycomb-group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev 1999; 13: 2691–2703.
Acknowledgements
This work was supported by a Cancer Research UK programme grant, we also gratefully thank M Bishop for providing mouse genotyping services, A Hayes for performing confocal fluorescence immunohistochemistry, C Oliver for preliminary data and thank L Parry for providing two cycles of Cre induction small-intestine slides.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Matthews, J., Sansom, O. & Clarke, A. Absolute requirement for STAT3 function in small-intestine crypt stem cell survival. Cell Death Differ 18, 1934–1943 (2011). https://doi.org/10.1038/cdd.2011.77
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2011.77
Keywords
This article is cited by
-
STAT3 and HIF1α cooperatively mediate the transcriptional and physiological responses to hypoxia
Cell Death Discovery (2023)
-
TIMP-2 regulates proliferation, invasion and STAT3-mediated cancer stem cell-dependent chemoresistance in ovarian cancer cells
BMC Cancer (2020)
-
Brf1 loss and not overexpression disrupts tissues homeostasis in the intestine, liver and pancreas
Cell Death & Differentiation (2019)
-
RETRACTED ARTICLE:Deficiency in intestinal epithelial Reg4 ameliorates intestinal inflammation and alters the colonic bacterial composition
Mucosal Immunology (2019)
-
Potential role of CBX7 in regulating pluripotency of adult human pluripotent-like olfactory stem cells in stroke model
Cell Death & Disease (2018)