Abstract
Death-associated protein kinase (DAPK) is a key player in several modes of neuronal death/injury and has been implicated in the late-onset Alzheimer's disease (AD). DAPK promotes cell death partly through its effect on regulating actin cytoskeletons. In this study, we report that DAPK inhibits microtubule (MT) assembly by activating MARK/PAR-1 family kinases MARK1/2, which destabilize MT by phosphorylating tau and related MAP2/4. DAPK death domain, but not catalytic activity, is responsible for this activation by binding to MARK1/2 spacer region, thereby disrupting an intramolecular interaction that inhibits MARK1/2. Accordingly, DAPK−/− mice brain displays a reduction of tau phosphorylation and DAPK enhances the effect of MARK2 on regulating polarized neurite outgrowth. Using a well-characterized Drosophila model of tauopathy, we show that DAPK exerts an effect in part through MARK Drosophila ortholog PAR-1 to induce rough eye and loss of photoreceptor neurons. Furthermore, DAPK enhances tau toxicity through a PAR-1 phosphorylation-dependent mechanism. Together, our study reveals a novel mechanism of MARK activation, uncovers DAPK functions in modulating MT assembly and neuronal differentiation, and provides a molecular link of DAPK to tau phosphorylation, an event associated with AD pathology.
Similar content being viewed by others
Main
Death-associated protein kinase (DAPK), a death domain (DD)-containing serine/threonine kinase, is a crucial mediator of apoptosis induced by diverse stimuli.1 DAPK also participates in autophagic death2 and caspase-independent necrotic death3 under certain cellular settings. This broad involvement in cell death is attributed to the ability of DAPK to activate multiple death-promoting molecules and pathways.1 Furthermore, the actin cytoskeleton-localized DAPK controls cell morphological changes associated with cell death. This is mainly mediated by its ability to phosphorylate myosin light chain II, thereby inducing apoptotic membrane blebbing.4, 5 Interaction of DAPK with the microtubule (MT)-binding protein MAP1B has also been implicated in the induction of membrane blebbing.6 However, the effect of DAPK on MT dynamics has not been explored.
DAPK is highly expressed in brain, which implies that the activity of DAPK is tightly regulated in the basal state in normal condition to prevent premature neuronal death. However, during a number of stressed conditions, DAPK expression or activity is elevated and DAPK has a critical role in many modes of neuronal injury and death.1, 7, 8 A further connection of DAPK to neuronal diseases comes from a large-scale genetic study, which identifies the association of two single nucleotide polymorphisms (SNPs) in DAPK with late-onset Alzheimer's disease (LOAD).9 However, the causal relationship between DAPK and LOAD has not been established.
Hyperphosphorylation of tau is implicated in the pathogenesis of neurodegenerative disorders including Alzheimer's disease (AD).10 The microtubule-affinity regulating kinase (MARK) was identified by searching for pathological phosphorylation sites of tau.11 Elevated tau phosphorylation at MARK-directed sites is found in the brains of AD patients and AD transgenic mouse models,12, 13 and MARK co-localizes with paired helical filaments in AD brains.14 Additional support for the involvement of MARK in neurodegeneration comes from the Drosophila system, in which PAR-1 (MARK fly ortholog) induces eye degeneration through an enhanced phosphorylation of tau at KXGS motifs.15 These motifs are within the MT-binding domain of tau and their phosphorylation results in tau detachment from MT, thereby inducing MT destabilization. MARK also phosphorylates tau-related MAP2/4 at the same motifs, thereby regulating MT dynamics in both neuronal and non-neuronal cells.16
The MARK/PAR-1 family kinases are crucial for the establishment and maintenance of cell polarity.16 The four mammalian members of MARK (MARK1/2/3/4) have a conserved domain organization, including catalytic, UBA, spacer, and KA1 domains.16 MARK kinase activity is regulated by multiple mechanisms. Phosphorylation of T208 in the activation loop by MARKK/TAO-1 or LKB1 activates MARK.17, 18 Conversely, phosphorylation of S212 in the activation loop by GSK3β19 or T595 in the spacer region by aPKC20, 21 inactivates MARK2. PAK5 inhibits MARK2 through an interaction between their kinase domains.22 Furthermore, an intramolecular interaction between N- and C-terminal segments of yeast MARK homolog Kin2 is suggested to generate an inactive conformation.23 However, it is unknown whether such autoinhibition occurs in mammalian MARKs and how this inhibition is relieved.
In this study, we report that DAPK DD binds to the spacer regions of MARK1 and MARK2, thereby relieving an autoinhibitory interaction between their N- and C-terminal segments. This activation of MARK1/2 mediates the effect of DAPK on regulating MT dynamics and DAPK promotes the effect of MARK on modulating polarized neurite outgrowth. Furthermore, DAPK promotes PAR-1-induced tau toxicity in a Drosophila tauopathy model. This study identifies a novel regulatory mode for MARK1/2, and suggests a potential role of DAPK in neurodegenerative diseases.
Results
DAPK inhibits MT assembly
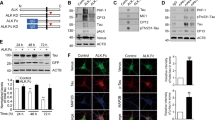
To determine the effect of DAPK on MT, we examined the steady-state MT networks in DAPK-transfected MCF7 cells. Immunostaining with anti-tubulin antibody did not reveal a pronounced effect of DAPK on MT organization, although DAPK-expressing cells displayed a modest reduction in anti-tubulin staining (Supplementary Figure S1). We then undertook a more sensitive approach by assaying MT regrowth after recovery from nocodazole treatment, and revealed a substantial delay of MT regrowth induced by DAPK (Figures 1a and b). After 40 min of recovery, only 59% of the DAPK-expressing cells displayed MT reappearance, whereas 90% of the control cells did so. Similar results were obtained in HepG2 and HCC36 cells (Figure 1b). To determine whether this delay of MT regrowth was due to a reduction in MT growth rate or a defect in the MT nucleation function of centrosome, HCC36 cells transfected with EB1-GFP, an MT plus-end binding protein, were examined by time-lapse microscopy. Nucleation rate was determined by the number of EB1-GFP comets emerged from the centrosome over time, whereas MT growth velocity was calculated by superimposing successive images and measuring the displacement of MT tip. Importantly, DAPK induced a significant decrease of MT growth velocity (Supplementary Movie 1 and Figures 1c and d) without affecting MT nucleation rate (Supplementary Movies 2 and 3 and Supplementary Figures S2a and b). Expression of DAPK in differentiated neuronal cell line N2a similarly reduced MT growth velocity (Supplementary Figure S3). These data thus indicate an inhibitory role of DAPK in MT assembly.
DAPK inhibits MT assembly. (a) MCF7 cells transfected with DAPK or control vector together with GFP were assayed for MT regrowth at indicated time points after washing out nocodazole. MT morphology was monitored by immunofluorescent staining with anti-tubulin antibody and examined by confocal microscopy. GFP-positive cells in the DAPK panel are marked by white contours. Bar, 10 μm. (b) Cells as indicated were transfected and assayed for MT regrowth as in a. GFP-positive cells displaying MT regrowth at 40 min (for MCF7), 30 min (for HepG2) or 10 min (for HCC36) after recovery from nocodazole treatment were quantified and plotted. Data represent mean±S.E.M. from three independent experiments, and at least 60 cells were analyzed for each transfection. *P<0.05. (c) Measurement of MT growth velocities in HCC36 cells expressing DAPK or control vector. Shown are overlays of three successive time-lapse images taken at 3.6-s intervals. The first, second, and third images are pseudocolored in red, green, and blue, respectively. The boxed regions are shown at higher magnification on the right and two representative tracks of EB1-GFP comets in each image are marked. Bars, 10 μm (left panel) and 3 μm (right panel). (d) Average MT growth velocities in indicated cells (see Materials and Methods section for calculation). Data represent mean±S.E.M. from measurements of six cells, five MTs per cell. *P<0.05
DAPK activates MARK1 and MARK2
We next investigated the mechanism through which DAPK regulates MT dynamics. As DAPK is mainly localized on actin rather than MT cytoskeletons,5, 24 we postulated that an MT-regulating protein mediates this effect of DAPK. We therefore tested whether DAPK could interact with an MT-regulating protein. Immunoprecipitation analysis on co-transfected cells demonstrated that Flag-DAPK interacted with MARK1 and MARK2, but not with closely related MARK3 and MARK4 or with several other MT-regulating proteins (Figure 2a). Reciprocally, Flag-MARK1 and Flag-MARK2 associated with endogenous DAPK (Figure 2b). These data revealed MARK1/2 as DAPK-binding proteins.
DAPK binds and activates MARK1 and MARK2. (a) Flag-DAPK interacts with endogenous MARK1 and MARK2. MCF7 cells transfected with Flag-DAPK or control vector were lysed for immunoprecipitation with anti-Flag antibody. The cell lysates and immunoprecipitates were analyzed by immunoblot with various antibodies as indicated. (b) Flag-MARK1 and Flag-MARK2 interact with endogenous DAPK. 293T cells transfected with Flag-MARK1 or Flag-MARK2 were subjected to immunoprecipitation with anti-Flag antibody, followed by immunoblot with anti-DAPK antibody. (c) DAPK activates MARK1 and MARK2. 293T cells were transfected with myc-DAPK and/or various Flag-MARK isoforms. Flag-MARK isoforms were isolated by immunoprecipitation and then assayed for their kinase activities with tau-C as the substrate. MARK4L and MARK4S indicate the long and short forms of MARK4, respectively. (d) DAPK promotes MARK2-induced tau phosphorylation at S262 in vivo. 293T cells transfected with various constructs were analyzed by immunoblot with antibodies as indicated. (e) DAPK siRNA downregulates MARK2-induced tau S262 phosphorylation. 293T cells stably expressing DAPK siRNA or control siRNA were transfected with MARK2 and tau and then analyzed by immunoblot with indicated antibodies
MARK family kinases are known to induce MT destabilization by phosphorylating tau.22 To evaluate whether DAPK influenced the catalytic activities of MARK1/2, Flag-MARK isolated from cells co-transfected with DAPK was tested in vitro for its ability to phosphorylate tau C-terminal fragment (tau-C). This analysis demonstrated that DAPK stimulated the activities of MARK1/2 but not MARK3/4 (Figure 2c), consistent with its inability to bind MARK3/4. Surprisingly, DAPK kinase-defective mutant (K42A) activated MARK1/2 as efficiently as wild-type (WT) DAPK, indicating that DAPK kinase activity is dispensable for MARK activation. Notably, MARK2 kinase-defective mutant (K82R) isolated from cells co-transfected with DAPK could not induce tau phosphorylation (Figure 2c, upper panel), indicating that the phosphorylation signal detected in our in vitro kinase assay was not due to the activity of co-precipitated DAPK. Accordingly, DAPK could not phosphorylate tau in vitro (Supplementary Figure S4). In the reciprocal experiment, we showed that DAPK catalytic activity was not affected by MARK1/2 overexpression (Supplementary Figure S5). As MARK1 and MARK2 are highly related, we focused mainly on the better-characterized MARK2 in the following studies. To validate the ability of DAPK to activate MARK2 in intact cells, we introduced full-length tau into 293T cells and its phosphorylation at S262, the primary residue targeted by MARK, was detected by a specific antibody. Although MARK2 overexpression increased tau S262 phosphorylation, co-expression of DAPK or DAPK K42A further augmented this phosphorylation (Figure 2d). Conversely, DAPK silencing by two independent siRNAs impaired MARK2-induced tau phosphorylation (Figure 2e). Together, our data indicate that DAPK activates MARK1/2 through a phosphorylation-independent mechanism.
DAPK activates MARK2 to promote tau phosphorylation in neurons
Next, we assessed the interplay of DAPK with MARK2 in a physiological setting. Using mice-brain extracts, we detected an interaction between endogenous DAPK and endogenous MARK2 by immunoprecipitation (Figure 3a). Although DAPK is mainly localized on actin cytoskeletons,5, 24 a small portion is associated with MT6 and was also found in soluble cytosol, where a great portion of MARK2 was distributed (Figure 3b). Consistent with these subcellular localizations, DAPK–MARK2 association was detected in both soluble and insoluble fractions of cell extracts (Figure 3b). In differentiated rat hippocampal neurons, DAPK and MARK2 partially co-localized in cell body, axon, and dendrites and both were slightly enriched in the axon tip (Figure 3c). When these neurons were transfected with MARK2 together with DAPK siRNA or control siRNA, we found that DAPK siRNA caused a significant reduction of MARK2-induced S262 phosphorylation on endogenous tau, as revealed by quantifying the pS262 tau immunofluorescent signals (Supplementary Figure S6a) and by immunoblot analysis (Figure 3d). Furthermore, silencing of both MARK1 and MARK2 blocked DAPK-induced tau S262 phosphorylation (Figure 3e). More importantly, a decrease of pS262 tau, but not total tau, was observed in brain extracts derived from DAPK−/− mice, compared with that from DAPK+/+ mice (Figure 3f). These results strongly suggest a role of endogenous DAPK in stimulating the activity of endogenous MARK, which in turn phosphorylates tau in neurons.
DAPK is a physiological activator of MARK2. (a) DAPK interacts with MARK2 endogenously. Mouse-brain homogenates were lysed for immunoprecipitation with anti-MARK2 antibody or a control antibody (IgG). The immunoprecipitates and lysates were analyzed by immunoblot with antibodies as indicated. (b) Detection of DAPK and MARK2 interaction in both soluble and insoluble compartments of cells. HCC36 cells were lysed for separating soluble and insoluble fractions as described in the Materials and Methods section. Equal amount of fractions was used for immunoprecipitation (IP) and immunoblot (lysate) analyses with antibodies as indicated. (c) Localization of DAPK and MARK2 in hippocampal neuron. Primary rat hippocampal neurons at DIV3 were immunostained with antibodies to DAPK, MARK2, and tau and examined by confocal microscopy. Bar, 20 μm. The specificities of MARK2 and DAPK antibodies are shown in Supplementary Figure S6b. (d) DAPK knockdown impairs MARK2-induced tau S262 phosphorylation (d) and MARK1/2 knockdown blocks DAPK-induced tau S262 phosphorylation (e) in primary neurons. Primary rat cortical neurons were transfected with indicated siRNA and/or cDNA at DIV0. The neurons were lysed at DIV3 for immunoblot analysis with indicated antibodies. (f) Reduced tau S262 phosphorylation in DAPK−/− mice. Brain extracts from DAPK−/− and DAPK+/+ mice were analyzed by immunoblot with indicated antibodies
DAPK relieves MARK1/2 autoinhibition by interacting with their spacer regions
To investigate the mechanism through which DAPK activates MARK2, we mapped DAPK region that is responsible for this activation. By incubating recombinant WT MARK2 with recombinant DAPK fragments in a kinase reaction with tau-C as the substrate, we found that all DAPK fragments lacking DD were incapable of activating MARK2. Conversely, DD alone promoted MARK2 catalytic activity efficiently (Figure 4a). Thus, DAPK DD is both necessary and sufficient for MARK2 activation. Next, we mapped MARK2 region that is required for its activation by DAPK. Although WT MARK2 and ΔKA mutant could be stimulated by DAPK DD, further deletion to remove the spacer region abolished this activation (Figure 4b). Notably, the spacer deletion mutant exhibited a significantly higher activity than WT MARK2 and ΔKA mutant, suggesting the existence of an autoinhibitory mechanism involving this region. In the following experiments, we determined whether DAPK exerts an effect through MARK2 spacer region to relieve this autoinhibition. Using MARK2 fragments, we showed that DAPK DD was able to interact with all MARK2 fragments containing the spacer region, with the highest affinity to the spacer region alone (Figure 4c). Furthermore, an intramolecular interaction between MARK2 N-terminal segment and its spacer region was detected. This interaction, however, was disrupted by introducing DAPK, but not its DD deletion mutant (ΔDD) (Figure 4d). Finally, we showed that pre-incubation of MARK2 N-terminal fragment with its C-terminal fragment diminished the catalytic activity of N-terminal fragment, and this effect was rescued by introducing DAPK but not DAPKΔDD (Figure 4e). Together, our data indicate that DAPK DD binds to the spacer region of MARK2, thereby disrupting an autoinhibitory interaction between the spacer and catalytic regions of MARK2. The same mechanism likely accounts for MARK1 activation by DAPK, as DAPK DD similarly interacted with the spacer region of MARK1 (Supplementary Figure S7). Mutation of the MARK2 T595 residue to A or E did not affect its interaction of DAPK, indicating that aPKC phosphorylation has no effect on the DAPK–MARK2 complex (Supplementary Figure S8). Notably, DAPK did not affect MARK1/2 phosphorylation at the conserved Thr residue in their activation loops (Supplementary Figure S9). Furthermore, T208 mutation that blocks or mimics the phosphorylation event did not affect MARK2 interaction with DAPK (Supplementary Figure S10). Thus, disruption of the intramolecular interaction and phosphorylation of the activation loop exert an effect independently to stimulate MARK1/2 activity.
DAPK relieves MARK2 autoinhibition by binding to MARK2 spacer region. (a) DAPK DD is responsible for MARK2 activation. Various DAPK fragments and full-length MARK2 purified from baculovirus were incubated in the kinase reaction with tau-C as the substrate. Substrate phosphorylation was detected by autoradiography (upper panel) and the input amounts of MARK2 and DAPK fragments were detected by immunoblot (lower panels). Schematics of DAPK mutants and their ability to activate MARK2 are shown on the right. (b) The spacer region is required for MARK2 activation by DAPK. Baculovirally purified DAPK DD (DD) was incubated with baculovirally purified MARK2 fragments and assayed for MARK2-mediated tau-C phosphorylation as in a. Schematics of MARK2 mutants and their responsiveness to DAPK DD are shown on the right. (c) DAPK DD interacts with MARK2 spacer region. 293T cells were co-transfected with GFP-DD (DAPK DD fused with GFP) and various Flag-MARK2 fragments as indicated. Cell lysates were used for immunoprecipitation with anti-Flag antibody. The immunoprecipitates and cell lysates were analyzed by immunoblot with antibodies as indicated. Schematics of MARK2 mutants and their ability to bind DAPK DD are shown on the right. (d) DAPK DD disrupts MARK2 intramolecular interaction. 293T cells transfected with Flag-tagged DAPK, DAPKΔDD (ΔDD), and/or MARK2 spacer region (spacer) were lysed and cell lysates were incubated with bacterially purified GST or GST-MARK2-N. Bound proteins were analyzed by immunoblot with anti-Flag antibody. The input amounts of GST fusion proteins and the expression of Flag-tagged proteins are shown on the lower panels. (e) DAPK relieves MARK2 autoinhibition. GST-MARK2-N purified from bacteria and various Flag-tagged proteins purified from baculovirus were incubated in the kinase reaction as in a. Tau phosphorylation was detected by immunoblot with pS262 tau antibody and the equal inputs of various proteins are shown on the lower panels
DAPK–MARK signaling axis inhibits MT assembly and stability
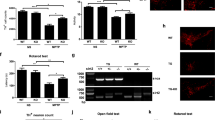
Having elucidated a MARK1/2 activation mode by DAPK, we next investigated the biological functions of DAPK–MARK pathway. First, we determined whether DAPK could enhance the MT destabilizing effect of MARK2. Indeed, although MARK2 overexpression caused MT disruption in 32% of the cells, this effect was further promoted by co-expression of DAPK, but not DAPKΔDD (Figure 5a). DAPK, but not DAPKΔDD, also synergized with MARK2 to suppress MT regrowth in cells recovered from nocodazole (Figure 5b). To determine whether the inhibitory role of DAPK in MT assembly was mediated by MARK1/2, we simultaneously silenced MARK1 and MARK2 using two different sets of siRNAs. These siRNAs were all capable of downregulating the corresponding MARK isoforms (Figure 5c, left panel). Importantly, depletion of MARK1/2 reversed the inhibitory effect of DAPK on MT regrowth (Figure 5c, right panel). These results indicate that the DAPK–MARK signaling axis inhibits MT assembly and stability.
DAPK coordinates with MARK1/2 to modulate MT assembly and stability. (a) MCF7 cells transfected with indicated constructs together with GFP were fixed and immunostained by anti-tubulin antibody. MT networks were visualized by confocal microscopy (left panel). Bar, 10 μm. GFP-positive cells with disrupted MT networks were quantified and plotted (right panel). (b) MCF7 cells transfected with indicated constructs together with GFP were treated with nocodazole and then assayed for MT regrowth as in Figure 1a. GFP-positive cells displaying MT regrowth at 40 min after washing out nocodazole were quantified and plotted. (c) MCF7 cells transfected with indicated siRNA and/or DAPK constructs, together with GFP construct were assayed for MT regrowth as in b. The abilities of various MARK siRNAs to downregulate their corresponding MARK proteins were determined by immunoblot (left panel). Data shown in panels a, b, and c represent mean±S.E.M. from three independent experiments and 100 GFP-positive cells were analyzed for each transfection. *P<0.05; **P<0.005
DAPK enhances the effect of MARK2 on axon formation
MARK2 elicits profound effects on neuronal differentiation. Overexpression of MARK2, but not its kinase-defective mutant, in undifferentiated primary hippocampal culture impairs the establishment of neuronal polarity, thereby blocking axon formation.25 To investigate the influence of DAPK on this function of MARK2, hippocampal neurons at DIV0 were co-transfected with MARK2 and DAPK, together with a GFP plasmid. These neurons were examined at DIV3 for axon formation by assaying the expression of axon marker Tau1. Remarkably, the inhibitory effect of MARK2 on axon formation was enhanced by co-expression of DAPK (Figure 6a), as evident by an increased percentage of neurons that contained no axon. DAPKΔDD, however, failed to promote this effect. Furthermore, siRNA-mediated silencing of endogenous DAPK rescued the axon-formation defect induced by MARK2 (Figure 6b). Together, our data support a physiological role of DAPK in promoting the effect of MARK2 on neuronal differentiation.
DAPK enhances the inhibitory effect of MARK2 on axon formation. (a and b) Primary hippocampal neurons transfected with indicated constructs or siRNAs together with GFP were stained at DIV3 with Tau1 (an axon marker) and Tuj1 (a neuron marker) antibodies and examined by confocal microcopy (left panels). Bar, 20 μm. The axon number in each GFP/Tuj1-double positive neuron was quantified and plotted (right panels). Axon is defined as Tau1-positive processes >100 μm. Data represent mean±S.E.M. from three independent experiments, and at least 30 GFP-positive neurons were analyzed for each transfection
DAPK induces rough eye and photoreceptor loss through PAR-1
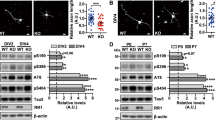
The involvement of DAPK in neuronal damage and its implication in AD prompted us to test a possible genetic interaction between human DAPK and Drosophila MARK2 ortholog PAR-1 using a Drosophila tauopathy model.26 In this model, overexpression of PAR-1 in retina induces tau toxicity, characterized by loss of photoreceptor neurons, leading to a rough-eye phenotype.15 Transgenic expression of DAPK or DAPK K42A by the pan-retinal GMR-GAL4 driver using the binary GAL4/UAS system27 resulted in a severe rough-eye phenotype with loss of ommatidia and disorganization of ommatidial array (Figure 7a, upper panel). No obvious difference could be observed between DAPK and DAPK K42A overexpression. Deletion of DD in the DAPK K42A-ΔDD mutant, however, failed to induce rough-eye phenotype when expressed by the same GMR-GAL4 driver (Figure 7a, upper panel), even though its expression level was comparable to that of DAPK and DAPK K42A (Figure 7b). These results suggest a critical role of DAPK DD in eye degeneration. The eye-ablation phenotype induced by DAPK and DAPK K42A was likely mediated by PAR-1 activation, as first indicated by the physical interaction between DAPK DD and the spacer region of Drosophila PAR-1 (Figure 7c). More importantly, introducing one allele of par-1 mutant (par-1Δ16)28 alleviated the rough-eye phenotype induced by DAPK or DAPK K42A, as evident by the reappearance of ommatidia and better-organized ommatidial arrays (Figure 7d, upper panel). DAPK or DAPK K42A expression level, however, was not affected by the par-1 mutant (Supplementary Figure S11). Staining of eye sections revealed normal numbers and trapezoidal arrangement of photoreceptors in the LacZ transgenic fly. However, photoreceptor neurons were completely absent in sections derived from DAPK and DAPK K42A transgenic flies, and this neuron-loss effect was reversed in the DAPK K42A-ΔDD transgenic fly (Figure 7a, lower panel and Figure 7e). The severe phenotype induced by DAPK or DAPK K42A was partially rescued in the par-1 heterozygous background, in which photoreceptors were occasionally observed, albeit with abnormal numbers and arrangements (Figure 7d, lower panel and Figure 7e). To further study the genetic interaction between h-DAPK and PAR-1, we evaluated the effect of DAPK and PAR-1 co-expression under the GMR-GAL4 driver. To observe a synergism between DAPK and PAR-1 functions, low-expression DAPK and DAPK K42A transgenes were selected (Figure 8b). Expressions of these transgenes induced milder rough eyes than the high-expression transgenes (comparing Figure 8a with Figure 7a). Introduction of the low-expression DAPK or DAPK K42A transgene together with PAR-1 transgene, which by itself had a mild rough-eye phenotype, produced a synergistic interaction between DAPK and PAR-1 in causing eye ablation, as shown by markedly reduced eye size with fewer ommatidia present in these smaller eyes (Figure 8a). These results support the role of DAPK in promoting PAR-1 activity, thereby enhancing PAR-1-induced rough-eye phenotype.
DAPK and DAPK K42A promote eye degeneration partly through Drosophila PAR-1. (a, d, and f) SEM images of indicated transgenic fly eyes (upper panel) and toluidine blue staining of photoreceptor neurons (lower panel). Arrowheads indicate photoreceptors. DAPKwtH and DAPKK42AH indicate the high-expression transgenes of DAPK and DAPK K42A, respectively. Bars, 100 μm (upper panel) and 10 μm (lower panel). (b) The expression levels of various DAPK derivatives in adult head were analyzed by immunoblot. (c) DAPK DD binds to the spacer region of PAR-1. 293T cells transfected with GFP-DD and Flag-PAR-1 spacer region (Flag-SR) were analyzed by immunoprecipitation followed by immunoblot with antibodies as indicated. (e) Quantitative analysis of photoreceptor neurons in indicated transgenic flies. The number observed in the LacZ transgenic fly was defined as 100%. Noted that the DAPK K42A-ΔDD transgenic fly exhibited a modest increase in photoreceptor neurons within each ommatidia but a slight reduction of photoreceptor clusters in a field. As a result, the total number of photoreceptor neurons in a field does not differ significantly from that of the LacZ transgenic fly. (g) Immunoblot analysis demonstrates RNAi knockdown of tau expression
DAPK enhances the eye degeneration phenotypes of PAR-1 and tau. (a and c) SEM images of indicated transgenic fly eyes. DAPKwtL and DAPKK42AL indicate the low-expression transgenes of DAPK and DAPK K42A, respectively. Bar, 100 μm. (b) Immunoblot analysis of the expression levels of DAPK and DAPK K42A in indicated transgenic flies. (d) Immunoblot analysis demonstrates that DAPK and DAPK K42A overexpression increase tau S262 phosphorylation in flies. (e) Immunoblot analysis of the expression levels of WT h-tau and its S2A mutant in transgenic flies
DAPK enhances tau toxicity through a tau S262-dependent mechanism
To further elucidate the mechanism underlying DAPK-induced eye ablation, we tested the involvement of tau in this process. Expression of tau RNAi under the GMR-GAL4 driver caused a substantial reduction in tau expression (Figure 7g). Importantly, tau knockdown did not affect the expression of DAPK or DAPK K42A (Supplementary Figure S11), but alleviated the rough-eye phenotype induced by these transgenes (comparing Figure 7f with Figure 7a), indicating that this function of DAPK is mediated at least in part through tau. Next, we determined the effect of PAR-1-dependent tau phosphorylation on DAPK-induced eye phenotype. Importantly, when low-expression DAPK or DAPK K42A transgene was co-expressed with the (4R, 0) isoform of human tau (h-tau) by the GMR-GAL4 driver, an elevation of h-tau phosphorylation at S262 was observed (Figure 8d). Furthermore, although expression of h-tau alone caused a moderate rough-eye phenotype, co-expression of the low-expression DAPK or DAPK K42A transgene dramatically enhanced h-tau toxicity (Figure 8c). The PAR-1 phosphorylation-defective mutant h-tauS2A,15 however, failed to promote the mild rough-eye phenotype induced by low-expression DAPK or DAPK K42A transgene (comparing Figure 8c with Figure 8a), even though h-tauS2A and h-tau were expressed at similar levels (Figure 8e). These results indicate that DAPK promotes tau toxicity and this effect is dependent on PAR-1-mediated tau phosphorylation.
Discussion
In this study, we report a novel function of DAPK in modulating MT assembly by activating MARK1/2. We show that MARK is autoinhibited by an intramolecular interaction between its catalytic and spacer regions. DAPK, functioning through its DD, is capable of relieving this autoinhibition via an interaction with MARK spacer region. Through this effect, DAPK promotes the functions of MARK2 on MT destabilization and neuronal differentiation. Furthermore, DAPK potentiates PAR-1-induced tau toxicity in a Drosophila tauopathy model. Although we cannot exclude the possibility that the eye-ablation phenotype induced by DAPK was mediated in part through developmental defects, the genetic interactions of DAPK with PAR-1 and tau imply that DAPK functions as an upstream regulator in the pathophysiological cascades leading to tauopathy in neurodegenerative diseases, including AD.
The uncovering of DAPK function in promoting tau phosphorylation and tauopathy is intriguing. DAPK is abundantly expressed in brain regions severely affected in AD, such as cortex and hippocampus.29 A genome-wide SNP study reveals the association of DAPK with LOAD.9 Furthermore, mice with a deletion in DAPK kinase domain exhibit an enhanced spatial memory.30 Although these studies implicate a potential impact of DAPK on AD, our finding that DAPK enhances MARK-induced tau phosphorylation and tau toxicity provides a molecular linkage of DAPK to tauopathy-related neurodegenerative disorders, such as AD. Thus, besides promoting acute neuronal cell death, elevated DAPK expression caused by genetic variation or environmental stress1, 7, 8, 9 might increase the risk of AD through aberrant MARK activation. As DAPK null mice displays a reduced tau phosphorylation, it would be important to determine whether DAPK knockout could alleviate tau pathology and behavior abnormalities in tau transgenic mouse models.
The multi-domain feature of DAPK suggests that this protein exerts certain biological functions through a kinase activity-independent mechanism. However, the previously identified DAPK functions are all mediated by its kinase activity.1, 31 The activation of MARK1/2 and consequent regulation of tau phosphorylation, MT dynamics, and neuronal differentiation thus represent the first DAPK downstream pathway that does not require its catalytic activity. In line with the MARK activation effect of DAPK, it is intriguing to note a correlation between DAPK expression29 and tau S262 phosphorylation32 during mouse-brain development; both levels are highest in fetal brain and progressively decline as the brain mature.
The developmentally regulated DAPK expression in brain implies an effect of DAPK on neuronal development. Accordingly, we show that DAPK modulates polarized neurite outgrowth. The inhibitory effect of MARK2 on axon formation is enhanced by DAPK overexpression and attenuated by DAPK knockdown. However, DAPK knockdown is insufficient to recapitulate the effect of MARK2 silencing, that is, induction of multiple axons. We postulate that this difference may be attributed to the inability of DAPK siRNA to completely abrogate MARK2 catalytic activity. In addition to modulating axon formation, MARK2 inhibits dendrite development in polarized neurons33 and promotes neurite outgrowth in neuronal cell lines that do not display axon-dendrite specification,19 thus highlighting its pleiotropic effects on neuronal differentiation.
Our study indicates that DAPK binding and T208 phosphorylation exert an effect independently to activate MARK2. Although the T208 kinase LKB1 broadly activates AMPK family kinases, DAPK selectively activates MARK1/2 but not other MARK isoforms. This selectivity is presumably due to the limited sequence conservation of spacer regions among MARK isoforms. Interestingly, DAPK DD displays a higher affinity to spacer region alone than a segment containing spacer and KA1 motif and MARKΔKA binds DAPK DD better than WT protein (Figure 4c), implying an inhibitory role of KA1 motif in DAPK binding. This inhibition might be mediated by a membrane targeting function of KA1 region,34 thereby separating MARK2 from DAPK. Notably, an inhibitory intramolecular interaction is observed in the MARK yeast homolog Kin2, in which the KA1 region is critical for the interaction of C-terminal regulatory region with the kinase domain.23 Furthermore, in the yeast AMPK homolog SNF135 and mammalian AMPK-related kinase MELK,36 a segment equivalent to the spacer region of MARK is characterized as the autoinhibitory domain by binding and inactivating the catalytic domain. Thus, the intramolecular interaction between C-terminal regulatory region and N-terminal catalytic region may be a common mechanism for suppressing the activities of MARK isoforms and related AMPK family kinases.
Materials and Methods
Cell culture and transfection
293T, HepG2, and N2a cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). MCF7 cells were cultured in DMEM/F12 medium supplemented with 10% FCS, non-essential amino acids and 2 mM L-glutamine, whereas HCC36 cells were maintained in DMEM supplemented with 10% FCS, non-essential amino acids and 2 mM L-glutamine. Primary hippocampal and cortical neurons were prepared from embryonic day 18 S.D. rat pups. The neurons were seeded on poly-D-lysine-coated glass coverslips and maintained in neurobasal medium supplemented with 2% B27 (Invitrogen, Carlsbad, CA, USA) and 0.5 mM L-glutamine. For induction of neuronal differentiation, N2a cells were cultured for 24 h in DMEM medium supplemented with 1% FCS and 10 μM retinoic acid. 293T cells were transfected using the calcium-phosphate method as described,37 whereas MCF7 cells and rat hippocampal neurons (at DIV3) were transfected using the Lipofectamine 2000 reagent (Invitrogen) according to manufacturer's protocol. Transfection of freshly isolated hippocampal and cortical neurons was performed by nucleofection. Briefly, neurons were suspended in 100 μl transfection solution (Amaxa Biosystems, Gaithersburg, MD, USA) containing 4 μg of cDNA and/or 150 pmol of siRNA oligonucleotides. Nucleofection was performed using the Amaxa electroporator (Amaxa Biosystems) setting in the program G-13. After the addition of neurobasal medium supplemented with 2% B27, 0.5 mM L-glutamine and 25 nM glutamate, the neurons were plated on poly-D-lysine-coated dishes. The medium was replaced with culture medium at 2 h after plating.
Plasmids
The mammalian expression constructs for WT and various DAPK mutants with a C-terminal myc or Flag tag were described previously.5, 37 The DAPK DD was cloned to pEGFP to generate GFP–DD construct. The full-length cDNAs for human MARK1, MARK2, MARK3, and MARK4 were purchased from Open Biosystems (Huntsville, AL, USA) and then subcloned to pRK5-Flag. For generating GST-MARK2-N, the MARK2 N-terminal segment (residues 1–362) was cloned to pGEX-4T. For generating baculovirus expression constructs, the full-length MARK2, full-length DAPK, and their various mutants were cloned to pVL1392, or pVL1393. cDNAs for both human full-length tau and tau C-terminal fragment (tau-C; residues 245–441) were obtained from Dr. Pei-Jung Lu (National Cheng Kung University, Taiwan). The full-length tau was subcloned to pRK5-Flag. For generating UAS-driven transgenes, the cDNAs for human DAPK, DAPK K42A, DAPK K42A-ΔDD, and h-tauS2A were cloned to pUAST. The Drosophila PAR-1 cDNA was obtained from Dr. Anne Ephrussi (EMBL, Germany).
RNA interference
Lentiviruses carrying DAPK siRNAs (siDAPK 1: 5′-CAAGAAACGTTAGCAAATG-3′; siDAPK 2: 5′-GGTCAAGGATCCAAAGAAG-3′) were described previously.38 These viruses were used to infect 293T cells and infected cells were selected by puromycin. Knockdown of DAPK in rat hippocampal neurons was performed by transfection of neurons with siRNA oligonucleotides (siDAPK 2: 5′-GGUCAAGGAUCCAAAGAAG-3′; matching the sequences of both human and rat DAPKs). Knockdown of human MARK1 and MARK2 was performed with siRNA oligonucleotides (MARK1 siRNA 1: 5′-GACCACAGAUCGAUACGUA-3′; MARK1 siRNA 2: 5′-UAACUGUGAUUAUGAGCAA-3′; MARK2 siRNA 1: 5′-GAAACUAUUCCGCGAAGUA-3′; MARK2 siRNA 2: 5′-GAAGUUUAUUGUCCAUAGA-3′). Knockdown of rat MARK 1 and MARK2 were performed with the following siRNAs (rat MARK1 siRNA: 5′-GGAUAUACUGAAACGCAUA-3′; rat MARK2 siRNA: 5′-GAAUGAACCUGAAAGCAAA-3′). All siRNA oligonucleotides were purchased from Dharmacon Inc. (Lafayette, CO, USA).
Antibodies
The rabbit anti-DAPK antiserums using GST-DAPK (275–636) fragment (for immunoblot) and His6-DAPK(843–1431) fragment (for immunostaining) as immunogens were described previously.5, 38 Antibodies to tubulin (DM1A), MARK3, MARK4, lamin A, and Tau-1 were purchased from Millipore (Bedford, MA, USA). Antibodies to Flag, CRMP1, DAPK (DAPK-55: for detecting mouse DAPK), Tuj1, and the anti-Flag M2 agarose were obtained from Sigma (St. Louis, MO, USA). Antibodies to MARK1, APC, GSK3β, EB1 (1A11/4), Stathmin, and phospho-MARK (activation loop) were from Cell Signaling (Danvers, MA, USA). Antibodies to total tau (TAU-5) and pS262 tau were from Invitrogen. Antibodies to GFP (B-2) and CrkII (C-18) were from Santa Cruz (Santa Cruz, CA, USA). The MARK2 antibody for immunoprecipitation was generated by Kelowna International Scientific Inc. (New Taipei, Taiwan), using the GST-hMARK2 (residues 531–670) as an immunogen. The goat anti-MARK2 antibody was from Novus (Littleton, CO, USA, for immunoblot and immunostaining).
Generation of baculovirus
Monolayers of Sf-21 cells cultured in TNM-FH insect medium (AppliChem, Darmstadt, Germany) supplemented with 10% FCS were co-transfected with linearized baculovirus DNA (BD Biosciences, San Diego, CA, USA) and pVL1392/1393-based vector. The recombinant virus was harvested, amplified twice, and then used to infect monolayers of Sf-21 cells in TNM-FH medium (AppliChem). After a 3-day incubation, cells were harvested for immunoblot to detect the expression of recombinant protein.
Detergent extraction for separating soluble and insoluble proteins
Cells were extracted with 500 μl of MES buffer containing 0.5% Triton X-100 as described.39 Cell extract was centrifuged at 16 000 × g for 10 min and then the supernatant (soluble fraction) was harvested. The pellet, together with the detergent-insoluble matrix remaining on the plate, was extracted with 500 μl RIPA lysis buffer (insoluble fraction). Equal volume of the soluble and insoluble fractions was used for immunoprecipitation.
Immunoprecipitation
Immunoprecipitation using cell lysates containing equal amount of proteins was performed as described.37 Briefly, cells were lysed in RIPA lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mM sodium vanadate, 4 mM sodium pyrophosphate, and 20 mM NaF. Total cell lysates were pre-cleared for 1 h with Protein A-sepharose and then incubated for 2 h with various antibodies at 4°C. The immunocomplex was captured by Protein A-sepharose. Alternatively, the cell lysates were pre-cleared and then incubated with anti-Flag M2 agarose.
GST pull down
Cell lysates were prepared by RIPA lysis buffer. Equal amounts of GST fusion proteins immobilized on glutathione-sepharose beads were incubated with cell lysates at 4°C for 2 h. The beads were washed and proteins bound on beads were analyzed by immunoblot.
Kinase assay
For assaying proteins phosphorylated by DAPK, 100 ng of Flag-DAPK purified from insect cells or mammalian cells and 5 μg of bacterially purified His-tau, GST-tau-C, or GST-MLC were incubated in 50 μl of kinase buffer containing 50 mM Hepes (pH 7.5), 8 mM MgCl2, 2 mM MnCl2, 0.1 mg/ml BSA, 1 μM bovine calmodulin (Sigma), 0.5 mM CaCl2, 50 μM ATP, and 10 μCi γ-[32P] ATP at 25°C for 15 min. For assaying the kinase activities of various MARK isoforms, Flag-MARKs were immunoprecipitated from 293T or Sf21 cell lysates and assayed in 20 μl of kinase buffer containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM EGTA, 0.5 mM PMSF, 0.5 mM DTT, 0.5 mM benzamidine, 10 μM ATP, 10 μCi γ-[32P] ATP, and 5 μg GST-tau-C at 30°C for 15 min. Following kinase reaction, the proteins were resolved by SDS-PAGE and substrate phosphorylation was detected by autoradiography or western blot with pS262 tau antibody.
Immunofluorescence analysis
For monitoring MT networks, cells were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 20 min, permeabilized with extraction buffer containing 50 mM NaCl, 300 mM sucrose, 10 mM PIPES (pH 6.8), 3 mM MgCl2, and 0.5% Triton X-100 for 5 min, and then blocked with PBS supplemented with 1% goat serum, 1% BSA, and 0.1% Triton X-100 for 1 h. For immunofluorescence staining using hippocampal neurons, cells were permeabilized with 0.5% Triton X-100 in PBS for 15 min and then blocked with 5% donkey serum and 0.1% Triton X-100 in PBS for 1 h. The cells were incubated with various primary antibodies diluted in blocking solution at 4°C for overnight, and then with FITC-, Cy5-, rhodamine-, or Alexa Fluo 546-conjugated secondary antibody for 1 h. The cells were washed, mounted, and examined with a Carl Zeiss LSM510 confocal laser-scanning microscope (Carl Zeiss, Jena, Germany) equipped with a 63 × objective lens. Fluorescent images were captured using the LSM510 software (Carl Zeiss).
For comparative pS262 tau, DAPK, and MARK2 immunostaining, neurons were transfected, cultured, and immunostained in parallel under identical conditions. Images were obtained at the same day using the same exposure settings and were processed with the same parameters. To quantify the pS262 tau, DAPK or MARK2 signal in each neuron, the region showing GFP signal at the GFP channel was selected by ImageJ software (NIH, Bethesda, MD, USA), and the average pixel intensity (API) in the corresponding region at pS262 tau, DAPK, or MARK2 channel was measured. A total of 30 neurons for each cell population were randomly selected for measurement and the mean API was calculated and presented as the value relative to that of cells carrying control siRNA.
Live-cell image and data analysis
Live-cell images were observed with an Olympus IX71 microscope equipped with a DeltaVision Image System (Applied Precision, Inc., Issaquah, WA, USA), consisting of a 100 × /1.40 objective lens, a Weather Station Environmental Chamber, and a Cascade2_512 EMCCD camera with a softWoRx-Acquire Version software (Applied Precision, Inc.). Optical Axis Integration command was applied for visualizing MT nucleation event. For EB1-GFP recording, cells at 1 day after transfection with EB1-GFP were placed under microscope. Only cells with EB1-GFP restricted to the plus-ends of MT were chosen for recordings. Time-lapse sequences of GFP images were collected with an exposure time of 25 to 150 msec (for HCC36 cells) or 200 msec (for N2a cells) at an interval of 0.3 s. For calculating MT nucleation events, the number of EB1-GFP comets that originated from the centrosome was quantified over time and expressed as nucleation events/min. For measuring MT growth rates, successive images from time-lapse recordings were merged by the ImageJ software and the relative displacement of MT tips during the specified time period was visualized and quantified.
Fly stocks and genetics
All general fly stocks and GAL4 lines were obtained from the Bloomington Drosophila Stock Center. The Drosophila par-1 heterozygous mutant28 was provided by Dr. Yuh Nung Jan (University of California San Francisco, San Francisco, CA, USA), whereas the UAS-dpar-1 transgenic fly15 was obtained from Dr. Bingwei Lu (Stanford University School of Medicine, Palo Alto, CA, USA). The UAS-h-tau transgenic line26 was a gift of Dr. Mel Feany (Harvard University, Cambridge, MA, USA). To generate transgenic flies, the pUAST-based constructs were injected into Drosophila embryos. Stocks and crosses were maintained on standard cornmeal-based Drosophila medium at 25°C.
Histology and immunohistochemistry
Scanning electron microscopy (SEM) was performed for visualizing the external morphology of fly compound eye. Briefly, adult flies were fixed in PBS containing 2.5% glutaraldehyde and 4% paraformaldehyde at 4°C for overnight, followed by PBS containing 1% OsO4 at room temperature for 4 h. The flies were then subjected to gradient dehydration and critically point drying using a Pelco CPD 2400 critical point dryer (Ted Pella, Inc., Redding, CA, USA). After coating (with a Cressington 108 sputter coater, Ted Pella, Inc.), samples were viewed and photographed with a FEI Quanta 200 scanning electron microscope (FEI, Eindhoven, The Netherlands) at 15 kV. For analyzing photoreceptor neurons, 1 μm section of adult retina was stained with toluidine blue, and examined with an Olympus BX51 microscope (Olympus, Tokyo, Japan) equipped with a 100 × objective lens.
Analysis of protein expression in mice and Drosophila
DAPK−/− and DAPK+/+ mice were described previously.40 Cerebral cortical tissues of 6-month mice were dissected and homogenized in RIPA lysis buffer and then centrifuged at 14 500 r.p.m. for 20 min at 4°C. For detecting tau expression, the supernatant was heated at 95°C for 5 min and then centrifuged again to remove debris. Mouse experiments were conducted with approval from the Experimental Animal Committee, Academia Sinica. To analyze protein expression in flies, dissected fly heads were sonicated twice in RIPA lysis buffer and then centrifuged. Supernatants containing equal amount of proteins were resolved by SDS-PAGE, transferred to PVDF membrane and probed with various antibodies.
Abbreviations
- DAPK:
-
Death-associated protein kinase
- AD:
-
Alzheimer's disease
- MT:
-
microtubule
- DD:
-
death domain
- SNPs:
-
single nucleotide polymorphisms
- LOAD:
-
late-onset Alzheimer's disease
- MARK:
-
microtubule-affinity regulating kinase
- WT:
-
wild type
- h-tau:
-
human tau
- DMEM:
-
Dulbecco's modified Eagle's medium
- FCS:
-
fetal calf serum
- SEM:
-
scanning electron microscopy
References
Bialik S, Kimchi A . The death-associated protein kinases: structure, function, and beyond. Annu Rev Biochem 2006; 75: 189–210.
Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A . DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol 2002; 157: 455–468.
Eisenberg-Lerner A, Kimchi A . DAP kinase regulates JNK signaling by binding and activating protein kinase D under oxidative stress. Cell Death Differ 2007; 14: 1908–1915.
Bialik S, Bresnick AR, Kimchi A . DAP-kinase-mediated morphological changes are localization dependent and involve myosin-II phosphorylation. Cell Death Differ 2004; 11: 631–644.
Kuo J-C, Lin J-R, Staddon JM, Hosoya H, Chen R-H . Uncoordinated regulation of stress fibers and focal adhesions by DAP-kinase. J Cell Sci 2003; 116: 4777–4790.
Harrison B, Kraus M, Burch L, Stevens C, Craig A, Gordon-Weeks P et al. DAPK-1 binding to a linear peptide motif in MAP1B stimulates autophagy and membrane blebbing. J Biol Chem 2008; 283: 9999–10014.
Fujita Y, Taniguchi J, Uchikawa M, Endo M, Hata K, Kubo T et al. Neogenin regulates neuronal survival through DAP kinase. Cell Death Differ 2008; 15: 1593–1608.
Tu W, Xu X, Peng L, Zhong X, Zhang W, Soundarapandian MM et al. DAPK1 interaction with NMDA receptor NR2B subunits mediates brain damage in stroke. Cell 2010; 140: 222–234.
Li Y, Grupe A, Rowland C, Nowotny P, Kauwe JS, Smemo S et al. DAPK1 variants are associated with Alzheimer′s disease and allele-specific expression. Hum Mol Genet 2006; 15: 2560–2568.
Mi K, Johnson GV . The role of tau phosphorylation in the pathogenesis of Alzheimer′s disease. Curr Alzheimer Res 2006; 3: 449–463.
Drewes G, Trinczek B, Illenberger S, Biernat J, Schmitt-Ulms G, Meyer HE et al. Microtubule-associated protein/microtubule affinity-regulating kinase (p110mark). A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J Biol Chem 1995; 270: 7679–7688.
Augustinack JC, Schneider A, Mandelkow EM, Hyman BT . Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer′s disease. Acta Neuropathol 2002; 103: 26–35.
Perez M, Ribe E, Rubio A, Lim F, Morán MA, Ramos PG et al. Characterization of a double (amyloid precursor protein-tau) transgenic: tau phosphorylation and aggregation. Neuroscience 2005; 130: 339–347.
Chin JY, Knowles RB, Schneider A, Drewes G, Mandelkow EM, Hyman BT . Microtubule-affinity regulating kinase (MARK) is tightly associated with neurofibrillary tangles in Alzheimer brain: a fluorescence resonance energy transfer study. J Neuropathol Exp Neurol 2000; 59: 966–971.
Nishimura I, Yang Y, Lu B . PAR-1 kinase plays an initiator role in a temporally ordered phosphorylation process that confers tau toxicity in Drosophila. Cell 2004; 116: 671–682.
Matenia D, Mandelkow EM . The tau of MARK: a polarized view of the cytoskeleton. Trends Biochem Sci 2009; 34: 332–342.
Lizcano JM, Göransson O, Toth R, Deak M, Morrice NA, Boudeau J et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J 2004; 23: 833–843.
Timm T, Li XY, Biernat J, Jiao J, Mandelkow E, Vandekerckhove J et al. MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J 2003; 22: 5090–5101.
Timm T, Balusamy K, Li X, Biernat J, Mandelkow E, Mandelkow EM . Glycogen synthase kinase (GSK) 3beta directly phosphorylates Serine 212 in the regulatory loop and inhibits microtubule affinity-regulating kinase (MARK) 2. J Biol Chem 2008; 283: 18873–18882.
Hurov JB, Watkins JL, Piwnica-Worms H . Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr Biol 2004; 14: 736–741.
Suzuki A, Hirata M, Kamimura K, Maniwa R, Yamanaka T, Mizuno K et al. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr Biol 2004; 14: 1425–1435.
Matenia D, Griesshaber B, Li XY, Thiessen A, Johne C, Jiao J et al. PAK5 kinase is an inhibitor of MARK/Par-1, which leads to stable microtubules and dynamic actin. Mol Biol Cell 2005; 16: 4410–4422.
Elbert M, Rossi G, Brennwald P . The yeast par-1 homologs kin1 and kin2 show genetic and physical interactions with components of the exocytic machinery. Mol Biol Cell 2005; 16: 532–549.
Cohen O, Feinstein E, Kimchi A . DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. EMBO J 1997; 16: 998–1008.
Chen YM, Wang QJ, Hu HS, Yu PC, Zhu J, Drewes G et al. Microtubule affinity-regulating kinase 2 functions downstream of the PAR-3/PAR-6/atypical PKC complex in regulating hippocampal neuronal polarity. Proc Natl Acad Sci USA 2006; 103: 8534–8539.
Wittmann CW, Wszolek MF, Shulman JM, Salvaterra PM, Lewis J, Hutton M et al. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science 2001; 293: 711–714.
Phelps CB, Brand AH . Ectopic gene expression in Drosophila using GAL4 system. Methods 1998; 14: 367–379.
Cox DN, Lu B, Sun TQ, Williams LT, Jan YN . Drosophila par-1 is required for oocyte differentiation and microtubule organization. Curr Biol 2001; 11: 75–87.
Yamamoto M, Takahashi H, Nakamura T, Hioki T, Nagayama S, Ooashi N et al. Developmental changes in distribution of death-associated protein kinase mRNAs. J Neurosci Res 1999; 58: 674–683.
Yukawa K, Tanaka T, Bai T, Li L, Tsubota Y, Owada-Makabe K et al. Deletion of the kinase domain from death-associated protein kinase enhances spatial memory in mice. Int J Mol Med 2006; 17: 869–873.
Chuang YT, Fang LW, Lin-Feng MH, Chen RH, Lai MZ . The tumor suppressor death-associated protein kinase targets to TCR-stimulated NF-kappa B activation. J Immunol 2008; 180: 3238–3249.
Seubert P, Mawal-Dewan M, Barbour R, Jakes R, Goedert M, Johnson GV et al. Detection of phosphorylated Ser262 in fetal tau, adult tau, and paired helical filament tau. J Biol Chem 1995; 270: 18917–18922.
Terabayashi T, Itoh TJ, Yamaguchi H, Yoshimura Y, Funato Y, Ohno S et al. Polarity-regulating kinase partitioning-defective 1/microtubule affinity-regulating kinase 2 negatively regulates development of dendrites on hippocampal neurons. J Neurosci 2007; 27: 13098–13107.
Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA et al. Kinase associated-1 domain drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell 2010; 143: 966–977.
Jiang R, Carlson M . Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev 1996; 10: 3105–3115.
Beullens M, Vancauwenbergh S, Morrice N, Derua R, Ceulemans H, Waelkens E et al. Substrate specificity and activity regulation of protein kinase MELK. J Biol Chem 2005; 280: 40003–40011.
Chen CH, Wang WJ, Kuo JC, Tsai HC, Lin JR, Chang ZF et al. Bidirectional signals transduced by DAPK-ERK interaction promote the apoptotic effect of DAPK. EMBO J 2005; 24: 294–304.
Wang WJ, Kuo JC, Ku W, Lee YR, Lin FC, Chang YL et al. The tumor suppressor DAPK is reciprocally regulated by tyrosine kinase Src and phosphatase LAR. Mol Cell 2007; 27: 701–716.
Lin YM, Chen YR, Lin JR, Wang WJ, Inoko A, Inagaki M et al. eIF3k regulates apoptosis in epithelial cells by releasing caspase 3 from keratin-containing inclusions. J Cell Sci 2008; 121: 2382–2393.
Gozuacik D, Bialik S, Raveh T, Mitou G, Shohat G, Sabanay H et al. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ 2008; 15: 1875–1886.
Acknowledgements
We thank Pei-Jung Lu, Anne Ephrussi, Yuh Nung Jan, Bingwei Lu, and Mel Feany for reagents, Chiou-Yang Tang for Drosophila embryo injection, Chin-Chun Hung for confocal analysis, and Sue-Ping Lee for video microscopy analysis. This work was supported by National Science Council Grant 98-2320-B-001-021. RH Chen is an Academia Sinica Investigator awardee.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by E Baehrecke
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Wu, PR., Tsai, PI., Chen, GC. et al. DAPK activates MARK1/2 to regulate microtubule assembly, neuronal differentiation, and tau toxicity. Cell Death Differ 18, 1507–1520 (2011). https://doi.org/10.1038/cdd.2011.2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2011.2
Keywords
This article is cited by
-
Death-associated protein kinase 1 as a therapeutic target for Alzheimer's disease
Translational Neurodegeneration (2024)
-
Role of MARK2 in the nervous system and cancer
Cancer Gene Therapy (2024)
-
Targeting a vulnerable septum-hippocampus cholinergic circuit in a critical time window ameliorates tau-impaired memory consolidation
Molecular Neurodegeneration (2023)
-
Tau Toxicity in Neurodegeneration
Molecular Neurobiology (2022)
-
Tumor suppressor death-associated protein kinase 1 inhibits necroptosis by p38 MAPK activation
Cell Death & Disease (2020)