Abstract
Background:
FAT4, a cadherin-related protein, was shown to function as a tumour suppressor; however, its role in human gastric cancer remains largely unknown. Here, we investigated the role of FAT4 in gastric cancer and examined the underlying molecular mechanisms.
Methods:
The expression of FAT4 was evaluated by immunohistochemistry, western blotting, and qRT–PCR in relation to the clinicopathological characteristics of gastric cancer patients. The effects of FAT4 silencing on cell proliferation, migration, and invasion were assessed by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium) assay, and migration and invasion assays in gastric cancer cell lines in vitro and in a mouse xenograft model in vivo.
Results:
Downregulation of FAT4 expression in gastric cancer tissues compared with adjacent normal tissues was correlated with lymph-node metastasis and poor survival. Knockdown of FAT4 promoted the growth and invasion of gastric cancer cells via the activation of Wnt/β-catenin signalling, and induced epithelial-to-mesenchymal transition (EMT) in gastric cancer cells, as demonstrated by the upregulation and downregulation of mesenchymal and epithelial markers. Silencing of FAT4 promoted tumour growth and metastasis in a gastric cancer xenograft model in vivo.
Conclusions:
FAT4 has a tumour suppressor role mediated by the modulation of Wnt/β-catenin signalling, providing potential novel targets for the treatment of gastric cancer.
Similar content being viewed by others
Main
Fat, a candidate tumour suppressor in Drosophila, is a member of the cadherin superfamily, which is a large group of Ca2+-dependent cell adhesion molecules that have critical roles in tissue differentiation and are characterised by an ectodomain with at least two consecutive calcium-binding cadherin repeats (Hulpiau and van Roy, 2009; Matis and Axelrod, 2013). More than 110 cadherins with different structural characteristics and functions form part of the superfamily (van Roy, 2014). Epithelial cadherin (E-cadherin or CDH1) suppresses tumorigenicity and invasion in part through the inhibition of proto-oncogenic molecules such as β-catenin and epidermal growth factor receptor. E-cadherin interacts with β-catenin and inhibits its translocation to the nucleus and thus the activation of Wnt signalling (Hulpiau and van Roy, 2009). However, whether other members of the cadherin superfamily have tumour-suppressing activity remains unclear (van Roy, 2014). Fat is a component of the Salvador–Warts–Hippo signalling pathway, which controls organ size by modulating cell growth, proliferation, and apoptosis (Harvey and Tapon, 2007; Berx and van Roy, 2009) and has a role in planar cell polarity (Qi et al, 2009). Four FAT proteins (FAT1–4) have been identified in mammals, of which FAT1 is the best studied (Sadeqzadeh et al, 2014). FAT1 regulates cell growth and migration through specific protein–protein interactions via its cytoplasmic tail, and deregulated FAT1 expression is associated with different human diseases including cancer. It suppresses cancer cell growth by binding β-catenin and antagonising its nuclear localisation, and silencing of FAT1 upregulates Wnt/β-catenin target genes such as c-myc and cyclin D1, promoting tumour growth. FAT4 was first proposed to have a tumour suppressor role by Qi et al (2009), who showed that biallelic FAT4 inactivation conferred tumorigenicity to a mouse mammary epithelial cell line and its ectopic expression suppressed the tumorigenicity of FAT4-deficient tumour cells. Mutations in the FAT4 gene have been identified in 5% of gastric cancers in association with well-differentiated and moderately differentiated tumours and occur more frequently in intestinal-type gastric cancers (Zang et al, 2012). Silencing of FAT4 promoted cell proliferation, invasion, and migration in gastric cancer cells expressing wild-type FAT4, providing further evidence of a tumour suppressor role for FAT4.
The Wnt/β-catenin signalling pathway has critical roles in the regulation of cell growth, development, and stem-cell differentiation, and constitutive activation of Wnt/β-catenin signalling is a hallmark of many cancers (Yao et al, 2011). In the canonical Wnt pathway, the inactivation of a destruction complex that targets β-catenin for proteasomal degradation results in the translocation of β-catenin to the nucleus, where it displaces Groucho from T-cell factor/lymphoid enhancer factor (TCF/LEF) to promote the transcription of Wnt target genes (Clevers, 2006). Decreased expression of E-cadherin and γ-catenin, which promote cell–cell contact, and upregulation of mesenchymal markers such as vimentin and N-cadherin and the activation of certain matrix metalloproteases (MMPs) are the characteristics of epithelial-to-mesenchymal transition (EMT), a process by which molecular reprogramming causes cells to switch from a polarised immobile epithelial phenotype to a motile mesenchymal phenotype leading to increased migration and invasion (Kalluri and Weinberg, 2009).
In the present study, we examined the role of FAT4 in gastric cancer and explored the mechanisms underlying its tumour suppressor role. We showed that FAT4 is downregulated in gastric cancer tissues and correlated with lymph-node metastasis, and we used silencing experiments in vitro and in vivo to show that FAT4 acts as a tumour suppressor via a mechanism involving the activation of Wnt/β-catenin signalling.
Materials and methods
Human gastric cancer samples and cell lines
Formalin-fixed, paraffin-embedded tissue samples were collected from 160 gastric cancer patients (120 men and 40 women with a mean age of 60 years) who underwent surgery at the 150 Central Hospital of the Chinese People's Liberation Army (PLA) (Luoyang, China) between 2007 and 2012. Primary gastric cancer tissues and paired adjacent non-cancerous tissues (>5–10 cm away from the primary site) were obtained from 30 patients who were being treated at the 150 Central Hospital of PLA and stored in liquid nitrogen until use. Informed consent was provided by all patients examined. The study protocol was approved by the ethics committee of the 150 Central Hospital of PLA.
Human gastric cancer cell lines (BGC-823 and HGC-27) were obtained from Shanghai Cell Biology, Chinese Academy of Sciences (Shanghai, China). BGC-823 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NY, USA) and HGC-27 cells were cultured in RPMI-1640 (Gibco) supplemented with 10% fetal bovine serum (FBS).
RNA isolation and quantitative RT–PCR
Total RNA from tissue samples and cultured cells was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Quantitative RT–PCR was performed using the PrimeScript RT reagent Kit (TaKaRa, Dalian, China) and SYBR Premix Ex Taq (TaKaRa). PCR primers were as follows: FAT4 forward 5′-TTGAAGGAAGGAGAACCCAT-3′ and reverse 5′-TCGTCCAATAGTAAAGAGGC-3′; E-cadherin forward 5′-TCCAGTGAACAACGATGGCA-3′ and reverse 5′-CCTGGGCAGTGTAGGATGTG-3′; γ-Catenin forward 5′-TCTTCAAGTCGGGTGGCATC-3′ and reverse 5′-TCAGCTTGCTCTCCTGGTTG-3′; N-cadherin forward 5′-AGTCAACTGCAACCGTGTCT-3′ and reverse 5′-AGCGTTCCTGTTCCACTCAT-3′; and vimentin forward 5′-GGACCAGCTAACCAACGACA-3′ and reverse 5′-AAGGTCAAGACGTGCCAGAG-3′.
Immunohistochemistry analysis
Formalin-fixed, paraffin-embedded gastric tissue samples were cut into 4-μm thick sections for immunohistochemical staining. Sections were incubated with rabbit polyclonal anti-FAT4 antibody (1 : 150, Novus Biologicals, Littleton, CO, USA) at 4 °C overnight, washed with PBS, and incubated with a biotinylated secondary antibody (Abcam, Cambridge, UK) for 30 min. Peroxidase reactivity was visualised with 3,3′-diaminobenzidine (DAB) (Boster, Wuhan, China) and counterstaining with haematoxylin. Evaluation of immunohistochemistry (IHC) was performed using a previously described method (Kusinska et al, 2009). The IHC intensity was scored as 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining). The percentage of positive cells was scored as 0 (negative), 1 (<10%), 2 (10–50%), or 3 (>50%). The final score was obtained by multiplying the scores for IHC intensity by the percentage of positive cells. A final score of ⩽1 was considered as negative, and a final score of 2–9 was considered as positive.
Western blot analysis
Lysates were prepared from cultured cells or tumour specimens using RIPA lysis buffer (Beyotime, Shanghai, China), clarified by centrifugation and the supernatant was stored until analysis. Nuclear extracts were prepared with the NE-PER nuclear extraction kit from Pierce Biotechnology (Rockford, IL, USA). Protein concentration was determined using the BCA Protein Assay kit (Bio-Rad, Hercules, CA, USA), and 30 μg of protein was separated by SDS–PAGE on 5% or 10% polyacrylamide gels and transferred electrophoretically onto polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). After blocking in 5% non-fat milk in Tris-buffered saline/Tween-20 buffer (TBST) for 2 h at room temperature or overnight at 4 °C, membranes were incubated with one of the following primary antibodies for 2 h at room temperature or overnight at 4 °C: FAT4 (1 : 500), c-Myc (1 : 200), cyclin D1 (1 : 1000) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), E-cadherin (1 : 1000), γ-catenin (1 : 1000) (all from Cell Signaling Technology, Danvers, MA, USA), N-cadherin (1 : 500), vimentin (1 : 500), β-catenin (1 : 4000), TCF4 (1 : 1000), MMP-14 (1 : 2000), and MMP-16 (1 : 1000) (all from Abcam). Antibodies against β-actin and GAPDH were used as internal controls. After washing three times in TBST, membranes were incubated with the corresponding horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature and visualised using the ECL Western Blotting Detection Reagent (Pierce Biotechnology).
Lentivirus production and oligonucleotide transfection
Lentivirus containing FAT4 short-hairpin RNA (shRNA) or scrambled oligonucleotides were obtained from GeneChem Inc. (Shanghai, China). The target sequences for the FAT4 shRNA were as follows: FAT4 shRNA1, 5′-GAGCGCTTTGTCTTAATGATT-3′ and FAT4 shRNA2, 5′-GCTTTGTATAAAGTGGAGATT-3′. BGC-823 and HGC-27 cells were infected with lentivirus to generate stable FAT4 silencing cell lines.
Small interfering RNA (siRNA) against β-catenin was purchased from GeneChem. Target cells were plated in six-well plates at a density of 1 × 105 cells per well and transfected with β-catenin siRNA using Lipofectamine 2000 (Life Technologies, Inc., Grand Island, NY, USA) according to the manufacturer’s instructions. TCF wild-type (Topflash) or mutated control (Fopflash) luciferase reporter plasmids were purchased from Upstate Biotechnology (Lake Placid, NY, USA), and transfected into FAT4-knockdown cells using Lipofectamine 2000. Luciferase activity was measured with the dual-luciferase reporter assay system 48 h after transfection and normalised to Renilla luciferase activity.
MTT assay
Cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay according to the manufacturer’s instructions. Briefly, cells were seeded in 96-well plates at a density of 1 × 104 cells per well and after 36 h of growth in a humidified atmosphere at 37 °C and 5% CO2, 20 μl of MTT dye solution (5 mg ml−1, Sigma, Grand Island, NY, USA) was added to each well and incubated for 4 h at 37 °C. After the addition of dimethyl sulphoxide (150 μl), plates were incubated for an additional 10 min and absorbance was measured using a microplate spectrophotometer (ELx800, Bio-TEK, Winooski, VT, USA) at a wavelength of 570 nm.
Colony-formation assays
Cells were seeded at a density of 500 cells per 35-mm culture dish and incubated at 37 °C in a 5% CO2 incubator for 14 days. The medium was replaced every 3 days. At the end of the incubation period, cells were fixed with 4% paraformaldehyde, stained with Giemsa solution (Sigma-Aldrich, St Louis, MO, USA), and colonies were counted. For soft agar colony-formation assay, BGC-823 and HGC-27 cells treated as indicated were resuspended in 0.3% soft agar in culture medium containing 10% FBS and layered onto 0.6% solidified agar in six-well plates at a density of 1 × 104 cells per well. After incubation for 2 weeks, colonies were stained with 0.005% crystal violet and photographed. Colony numbers were counted in five random fields at × 40 magnification.
In vitro invasion and migration assays
The invasive and migratory potential of cells was evaluated using Transwell chambers (8 μm pore; BD Biosciences, San Jose, CA USA). For invasion assays, 1 × 105 cells in 100 μl of serum-free medium were added to the upper chamber coated with Matrigel. Medium supplemented with 10% FBS was added to the lower chamber. After incubation for 24 h, non-invading cells on the upper surface were removed with a cotton swab and cells invading to the lower chamber were fixed with methanol, stained with 0.1% crystal violet, dried, and photographed. For migration assays, 5 × 104 cells in 100 μl serum-free medium were placed in the top chamber without Matrigel, and 500 μl of 10% FBS containing medium was added to the lower chamber. After 16 h, cells on the upper surface were removed and cells migrated to the lower chamber were fixed and stained as described above. The number of invading or migrating cells was counted under the microscope in five representative fields and expressed as the average per field.
In vivo tumorigenesis and metastasis
All animal experiments were performed according to protocols approved by the Experimental Animal Center of the 150 Central Hospital of PLA. BGC-823 cells infected with scramble shRNA or FAT4 shRNA (1 × 106 cells per mouse) were injected subcutaneously into the flanks of female athymic BALB/c nude mice (Shanghai Slac Laboratory Animal Co., Shanghai, China) aged 4–5 weeks. After 32 days, the animals were killed and tumours were dissected. Tumour volumes were calculated using the following formula: volume=(width)2 × length/2. Tumour tissues were fixed with 10% formalin and embedded in paraffin. Tumour sections were obtained and stained with specific antibodies against FAT4 (1 : 150) or Ki-67 (1 : 500, Abcam). To investigate the effect of FAT4 on tumour metastasis, BGC-823 cells (6 × 106 per mouse) treated as indicated were injected intravenously into the mouse tail vein. Four weeks post injection, the mice were killed and the lungs and livers were removed and paraffin embedded. Sections (4 μm) were prepared and stained with H&E. Metastatic lesions were counted under a dissecting microscope. All experiments involving animals were performed in accordance with the National Institutes of Health guidelines for the use of experimental animals.
Statistical analysis
Data represent the mean±s.d. from at least three independent experiments performed in triplicate. Statistical analysis of the differences in the expression levels of FAT4 mRNA in paired tumour and normal tissues was analysed by the Mann–Whitney U-test using Prism 5 (GraphPad Software Inc., La Jolla, CA, USA). A Chi-square test was used to determine the association between FAT4 expression and clinicopathological parameters. Kaplan–Meier analysis and log-rank tests were used to assess the survival rate, and to compare differences in survival curves. Data were analysed using SPSS17.0 software (SPSS Inc., Cary, NC, USA). Differences were considered as significant at P<0.05.
Results
FAT4 is downregulated in gastric cancer tissues
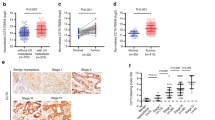
Expression of FAT4 was detected in 30 gastric cancer and paired adjacent normal tissues by qRT–PCR. FAT4 was downregulated in 43.3% (13 out of 30) of gastric cancer tissues compared with paired adjacent normal tissues (Figure 1A). Similar results were obtained by western blot analysis of FAT4 protein expression in gastric cancer and adjacent non-cancerous tissues. Representative blots and graph of the average of four samples are shown in Figure 1B (P<0.05). Full-size gels of FAT4 protein expression showed a cleaved product (Supplementary Figure S1). Immunohistochemistry showed strong FAT4 staining in normal gastric tissues and low or undetectable staining in tumour tissues in both the intestinal and diffuse pathological variants (Figure 1C). The Kaplan–Meier method and the log-rank test were used to examine the correlation of FAT4 expression with survival, which showed that negative FAT4 expression was significantly correlated with poor overall survival (P<0.05) (Figure 1D). To determine the relation between FAT4 expression and the clinicopathological characteristics of the 160 patients who provided formalin-fixed paraffin-embedded samples, patients were divided into two groups according to negative or positive FAT4 expression. The results showed that negative FAT4 expression was significantly associated with lymph-node invasion (P=0.0) and survival (P=0.017) (Table 1).
FAT4 expression in gastric cancer tissues. (A) FAT4 mRNA expression levels in 30 paired gastric cancer tissues and adjacent non-cancerous tissues expressed as relative expression normalised to the expression of β-actin and calculated by the 2−ΔΔct method. *P<0.05 by Mann–Whitney U-test. (B) FAT4 protein expression in representative samples of gastric cancer tissues (T) and paired adjacent non-cancerous tissues (N). *P<0.05 by Student’s t-test. (C) Immunohistochemical staining of FAT4 in gastric cancer tissues. Original magnification, × 200, × 400; bar=100 μm. (D) Kaplan–Meier survival analysis according to FAT4 expression in gastric cancer patients (N=160). The P-values from log-rank tests comparing the two KM curves are shown.
FAT4 knockdown promotes the growth and invasion of gastric cancer cells in vitro
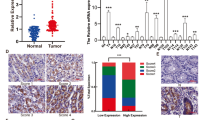
Two shRNA constructs were used to knockdown FAT4 expression in the gastric cancer cell lines BGC-823 and HGC-27, and the efficiency of shRNA-mediated knockdown of FAT4 expression was confirmed by western blotting and quantified by densitometric scanning of the bands (Figure 2A). The effect of FAT4 silencing on cell viability was assessed by the MTT assay, which showed that both FAT4 shRNA1 and shRNA2 significantly promoted cell growth compared with scramble shRNA-infected controls in both cell lines (P<0.05) (Figure 2B). Colony-formation and soft agar assays showed that silencing of FAT4 significantly increased the colony-forming ability of BGC-823 and HGC-27 cells (Figure 2C and D). FAT4 shRNA1 and shRNA2 significantly promoted the migration and invasion of BGC-823 and HGC-27 cells compared with scramble-infected cells (Figure 2E and F).
FAT4 knockdown promotes the proliferation and invasion of BGC-823 and HGC-27 cells. BGC-823 and HGC-27 cells were infected with FAT4 shRNA or scramble control shRNA. FAT4 protein expression was analysed by western blotting and quantified by densitometric scanning (A). Cell proliferation was measured by MTT assay (B). Colony formation was analysed by colony-formation (C) and soft agar (D) assays. Migration (E) and invasion (F) were analysed in BGC-823 and HGC-27 cells infected as indicated. Representative images of colony formation, migrating, and invading cells in scramble and FAT4-silenced gastric cancer cells are shown. *P<0.05, **P<0.01 by Student’s t-test.
FAT4 knockdown activates Wnt/β-catenin signalling
Based on previous studies showing an association of FAT1 with aberrant Wnt activation in cancer (Hou et al, 2006; Morris et al, 2013), we examined whether the effects of FAT4 on the growth and migratory abilities of gastric cancer cells were mediated by the Wnt/β-catenin pathway. Analysis of nuclear and cytosolic extracts of BGC-823 and HGC-27 cells by western blotting showed that silencing of FAT4 increased β-catenin levels in the nuclei of gastric cancer cells concomitant with a decrease in β-catenin levels in the cytosolic fraction (Figure 3A). To further examine the effect of FAT4 silencing on Wnt/β-catenin signalling, BGC-823 and HGC-27 cells were transfected with TOPflash and FOPflash constructs, which are commonly used to evaluate β-catenin-dependent activity driving the expression of TCF, and the Renilla pRL-TK plasmid as an internal control. TOPflash is a TCF reporter plasmid containing TCF binding sites upstream of a luciferase reporter gene, and FOPflash, which contains mutated TCF binding sites, is used as a negative control for TOPflash activity. The results of luciferase activity showed that silencing of FAT4 resulted in a 2.5- to 3-fold increase in the ratio of TOPflash to FOPflash activity in both cell lines, indicating a significant upregulation of β-catenin signalling in response to FAT4 knockdown in these cells (Figure 3B). To confirm the involvement of the Wnt/β-catenin pathway in the tumorigenic effects of FAT4, the expression of several Wnt target genes was examined by western blotting. The results showed that silencing of FAT4 upregulated the expression of TCF4, c-Myc, cyclin D1, MMP-14, and MMP-16 in BGC-823 and HGC-27 cells (Figure 3C).
Effect of FAT4 knockdown on Wnt/ β -catenin signalling. (A) Altered nuclear translocation of β-catenin in response to FAT4 knockdown. Nuclear and cytosolic extracts of BGC-823 and HGC-27 cells were analysed by western blotting with Lamin A and β-actin as a loading control. (B) Cells were transfected with TOPflash or FOPflash and Renilla pRL-TK plasmids and subjected to dual-luciferase assays 48 h after transfection. Reporter activity was normalised to Renilla luciferase activity. (C) Western blot analysis of Wnt target gene expression in response to FAT4 knockdown in BGC-823 and HGC-27 cells. β-Actin was used as the loading control. **P<0.01 by Student’s t-test.
FAT4 promotes tumour cell growth and invasion via β -catenin signalling
The involvement of the Wnt/β-catenin signalling pathway in the FAT4-mediated modulation of cell growth, migration, and invasion was further examined by assessing the effect of FAT4 knockdown and β-catenin silencing in the BGC-823 and HGC-27 cell lines. Transfection of FAT4 shRNA1-treated cells with siRNA against β-catenin did not affect β-catenin expression in whole-cell extracts (Figure 4A). MTT assays showed that FAT4 knockdown significantly increased cell growth over a period of 6 days, whereas silencing of β-catenin decreased the rate of cell growth compared with scramble-transfected cells (P<0.05, both) (Figure 4B). The siRNA-mediated silencing of β-catenin in FAT4-knockdown cells abrogated the effect of FAT4 on cell proliferation, colony formation, invasion, and migration (Figure 4C–E), confirming our previous results and indicating that the tumour suppressor role of FAT4 may be mediated by the inhibition of β-catenin signalling.
FAT4 knockdown-induced gastric cancer cell growth, invasion, and migration are mediated by the Wnt/ β -catenin signalling pathway. BGC-823 and HGC-27 cells were transfected with or without shRNA (shRNA1) against FAT4 and siRNA targeting β-catenin, and analysed by western blotting (A), MTT assay (B), colony-formation assay (C), and migration and invasion assays (D and E). *P<0.05, **P<0.01 by Student’s t-test.
FAT4 knockdown promotes EMT in gastric cancer cells
Because of the important role of EMT in tumorigenesis and metastasis, the effect of FAT4 knockdown on EMT was evaluated by measuring the changes in the mRNA and protein expression of the EMT markers E-cadherin, γ-catenin, N-cadherin, and vimentin. qRT–PCR analysis showed that FAT4 knockdown significantly downregulated the epithelial markers E-cadherin and γ-catenin and significantly upregulated the mesenchymal markers N-cadherin and vimentin (Figure 5A). Similar results were obtained at the protein level by western blotting (Figure 5B), indicating that FAT4 downregulation promotes EMT in gastric cancer cells in vitro.
FAT4 knockdown promotes gastric cancer tumorigenesis and metastasis in vivo
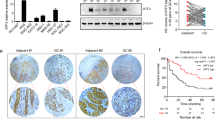
The effect of FAT4 on gastric cancer was examined in vivo in a gastric cancer xenograft mouse model established by injection of BGC-823 cells infected with shRNA against FAT4 or scramble-infected controls. Representative images of tumours excised from xenograft tumour-bearing mice after 32 days, and tumour volumes and weights are shown in Figure 6A and B. Tumours grew faster, and tumour volume and tumour weight were significantly higher in FAT4 shRNA tumours than in scramble control tumours. Figure 6C shows representative images of sections from xenograft tumours after IHC staining for FAT4 and Ki-67. The results showed markedly lower FAT4 staining and increased Ki-67 staining in FAT4 shRNA tumours than in scramble control tumours, indicating that FAT4 knockdown promoted cell proliferation in vivo. Assessment of the effect of FAT4 on the metastatic potential of gastric cancer cells showed the presence of metastatic lesions in the lungs and livers of mice injected with FAT4 shRNA bearing BGC-823 cells compared with scramble control-injected mice (Figure 6D). The numbers of visible surface metastatic lesions on the lungs and livers of individual mice (N=5 per group) were counted, which showed a significantly higher number of metastatic lesions in mice injected with FAT4-knockdown cells than in those receiving scramble control cells (Figure 6E). The presence of metastases was confirmed in H&E-stained sections of the lung and liver (Figure 6F).
Effect of FAT4 knockdown on tumorigenesis and metastasis in vivo. (A) Photographs of excised tumours and tumour growth curves in mice transplanted with BGC-823/Scramble or BGC-823/FAT4 shRNA cells (FAT4 shRNA1) (N=5 per group). (B) After 32 days, the mice were killed and tumours were weighed. (C) IHC staining for FAT4 and Ki-67 in sections of transplanted tumours. (D) BGC-823/Scramble and BGC-823/FAT4 shRNA cells (6 × 106 cells per mouse) were injected into the tail vein of two groups of nude mice. Four weeks post injection, the mice were killed and the lungs and livers were removed and paraffin embedded. Arrows indicate surface metastatic nodules. (E) Number of visible surface metastatic lesions on the lungs and livers of individual mice (N=5 per group) receiving an orthotopic injection of the indicated cells. (F) Representative images of haematoxylin and eosin-stained lung and liver sections of mice injected with the indicated cells. *P<0.05, **P<0.01 by Student’s t-test.
Discussion
The FAT cadherin family, which is structurally conserved from flies to vertebrates, consists of single pass transmembrane receptors with 32–34 cadherin repeats. Although the function of Fat in Drosophila has been studied extensively, the roles and interacting proteins of the four FAT proteins in vertebrates are only recently beginning to be understood. The members of the FAT cadherin family show similar extracellular domain structures; however, the intracellular cytoplasmic domains differ, which may reflect the specific functions of each FAT cadherin member (Saburi et al, 2012). FAT1, which is the most studied, has a role in the regulation of cell migration, polarity, cell–cell adhesion, neuronal function, and Wnt and Hippo signalling through interactions with different proteins including β-catenin, Atrophin, enable (ENA)/Vasodilator-stimulated phosphoprotein (VASP) and Homer, and scribble (Moeller et al, 2004; Hou et al, 2006; Schreiner et al, 2006; Hou and Sibinga, 2009; Skouloudaki et al, 2009). Deregulated expression of FAT1 has been associated with several diseases including cancer (Sadeqzadeh et al, 2014). FAT4, which is the most similar to Drosophila Fat in sequence of all the vertebrate FAT proteins, has been shown to interact with Dachsous1 and have a role in Hippo signalling and planar cell polarity (Ishiuchi et al, 2009; Mao et al, 2011); however, whether FAT4 regulates cell proliferation upstream of the Hippo pathway in vertebrates is controversial (Bossuyt et al, 2014). Although few studies have examined the association of FAT4 with cancer, a correlation between FAT4 expression and tumorigenesis was suggested in breast and gastric cancers (Qi et al, 2009; Zang et al, 2012). In the present study, we investigated the expression of FAT4 in gastric cancer and examined its potential tumour suppressor role in vitro and in vivo.
Analysis of FAT4 expression in gastric cancer and adjacent normal tissues showed that FAT4 was downregulated in gastric tumours, and negative FAT4 expression was significantly correlated with lymph-node metastasis and poor survival. Our present IHC results showing that >75% of tumours had negative FAT4 expression differs from the findings of Zang et al (2012), who detected FAT4 mutations in ∼5% of gastric cancer patients and no association between FAT4 expression and TNM status. FAT4 mRNA expression is downregulated by promoter hypermethylation in breast cancer and lung cancer (Qi et al, 2009; Rauch et al, 2012). Unlike FAT1, which has been shown to function as an oncogene or a tumour suppressor, FAT4 has only been shown to have a tumour suppressive role to date. FAT1 is downregulated in oral cancer and invasive breast cancer and loss of heterozygosity has been reported in a number of cases, whereas FAT1 upregulation has been detected in colon, breast, small intestine, and lung cancer, melanoma, and leukaemia (Nakaya et al, 2007; de Bock et al, 2012; Lee et al, 2012; Sadeqzadeh et al, 2014). FAT4 mutations have been detected in gastric cancer (Zang et al, 2012); however, the expression and function of FAT4 in gastric cancer and the underlying mechanisms remain unclear.
In the present study, gastric cancer cell lines were subjected to shRNA-mediated silencing of FAT4 to explore the role of FAT4 in gastric cancer in vitro. We showed that FAT4 knockdown promoted cell viability, increased colony formation, and induced the migration and invasion of gastric cancer cells. Further experiments showed that these effects were mediated by the Wnt/β-catenin signalling pathway, as evidenced by the nuclear translocation of β-catenin and the upregulation of Wnt target genes. Furthermore, FAT4 upregulated the expression of TCF4, which is the most prominently expressed TCF family member in the intestinal epithelium and has been implicated in the development of several cancers including colorectal and gastric cancers (Korinek et al, 1997; Ju et al, 2014). A luciferase-based assay in which a plasmid with TCF/LEF binding sites in the promoter region drives the expression of luciferase showed that shRNA FAT4 increased luciferase activity, further confirming that FAT4 inhibits β-catenin-mediated transcription and thus functions as a tumour suppressor. Similar results were reported for FAT1, which was shown to directly bind β-catenin and inhibit its nuclear translocation (Morris et al, 2013). A recent study suggested that the effect of FAT4 on the regulation of β-catenin signalling is mediated by its modulation of the activity of two members of the Hippo signalling pathway, YAP and TAZ (Das et al, 2013). In colorectal cancer cells, YAP was shown to negatively regulate Wnt target genes by directly binding to β-catenin and preventing its nuclear translocation (Imajo et al, 2012). In hepatoblastoma cells, knockdown of YAP inhibited its interaction with β-catenin and cell proliferation, whereas overexpression of YAP and β-catenin promoted tumorigenesis in the mouse liver, indicating that the interaction between YAP and β-catenin has a role in the pathogenesis of hepatoblastoma (Tao et al, 2014). These studies support our present findings and confirm the involvement of Hippo pathway components in carcinogenesis via their negative regulation of Wnt/β-catenin signalling. However, the mechanism by which FAT4 modulates Wnt signalling remains unclear. Future experiments should be aimed at testing whether FAT4 interacts directly with β-catenin and further examine the involvement of other Hippo signalling components such as YAP and TAZ. In addition, because the cytoplasmic domain of FAT1 has been shown to bind β-catenin (Hou et al, 2006), and the results of cytoplasmic and partly nuclear FAT4 staining in Figure 6C suggest the possibility that FAT4 is cleaved and translocated to the nucleus similar to FAT1 (Magg et al, 2005), the introduction of mutations in the cytoplasmic domain of FAT4 and their effect on Wnt/β-catenin signalling would be of interest.
The role of Wnt pathway activation in gastric carcinogenesis has been demonstrated extensively. Mutations in β-catenin and Wnt pathway activation have been associated with gastric adenocarcinomas (Clements et al, 2002; Ebert et al, 2002; Tsukamoto et al, 2003). The expression of β-catenin is downregulated in 47% of human gastric cancers, and activation of Wnt/β-catenin signalling has been reported in ∼30% of gastric cancer patients (Takayama et al, 1996; Mishra et al, 2005). Gastric cancer is characterised by early invasiveness into surrounding tissues and the peritoneal cavity and invasion and metastasis are the underlying causes of poor survival from this disease (Catalano et al, 2009). Several genes involved in tumour invasion and metastasis are Wnt pathway targets, including several MMPs such as MMP-7, MMP-26, MMP-16 (also known as MT3-MMP), and MMP-14 (also known as MT1-MMP) (Lowy et al, 2006). Wnt signalling-mediated upregulation of MMP-16 regulates the invasiveness of gastric cancer cells, and MMP-16 is overexpressed in human gastric cancers with activating β-catenin mutations (Lowy et al, 2006). MMP-14 is required for the regulation of cell polarity during the directed migration of cells and is essential for the degradation of extracellular matrix components (Coyle et al, 2008). The ability of tumour cells to modify the extracellular matrix and become motile is the basis of invasiveness, and upregulation of MMPs is a hallmark of EMT (Fanelli et al, 2012). In the present study, we showed that silencing of FAT4 upregulated the expression of MMP-14 and MMP-16 and promoted EMT, as shown by the downregulation of the epithelial markers E-cadherin and γ-catenin concomitant with the upregulation of the mesenchymal markers N-cadherin and vimentin. In vivo experiments confirmed the tumour suppressor role of FAT4: FAT4 silencing promoted tumour growth and metastasis in a gastric cancer xenograft mouse model. On the other hand, alternative splicing of FAT1 was shown to be associated with cell migration, with the +12 isoform of FAT1 detected in cancer cells undergoing EMT (Braun et al, 2007; Shapiro et al, 2011). Taken together, our in vitro and in vivo results provide evidence of an association between FAT4 expression and tumour growth and metastasis via the Wnt/β-catenin signalling pathway, which warrants further investigation into the role of FAT4 in gastric cancer.
Change history
22 December 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Berx G, van Roy F (2009) Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol 1: a003129.
Bossuyt W, Chen CL, Chen Q, Sudol M, McNeill H, Pan D, Kopp A, Halder G (2014) An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene 33: 1218–1228.
Braun GS, Kretzler M, Heider T, Floege J, Holzman LB, Kriz W, Moeller MJ (2007) Differentially spliced isoforms of FAT1 are asymmetrically distributed within migrating cells. J Biol Chem 282: 22823–22833.
Catalano V, Labianca R, Beretta GD, Gatta G, de Braud F, Van Cutsem E (2009) Gastric cancer. Crit Rev Oncol Hematol 71: 127–164.
Clements WM, Wang J, Sarniak A, Kim OJ, MacDonald J, Fenoglio-Preiser C, Groden J, Lowy AM (2002) β-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res 62: 3503–3506.
Clevers H (2006) Wnt/β-catenin signaling in development and disease. Cell 127: 469–480.
Coyle RC, Latimer A, Jessen JR (2008) Membrane-type 1 matrix metalloproteinase regulates cell migration during zebrafish gastrulation: evidence for an interaction with non-canonical Wnt signaling. Exp Cell Res 314: 2150–2162.
de Bock CE, Ardjmand A, Molloy TJ, Bone SM, Johnstone D, Campbell DM, Shipman KL, Yeadon TM, Holst J, Spanevello MD, Nelmes G, Catchpoole DR, Lincz LF, Boyd AW, Burns GF, Thorne RF (2012) The Fat1 cadherin is overexpressed and an independent prognostic factor for survival in paired diagnosis-relapse samples of precursor B-cell acute lymphoblastic leukemia. Leukemia 26: 918–926.
Das A, Tanigawa S, Karner CM, Xin M, Lum L, Chen C, Olson EN, Perantoni AO, Carroll TJ (2013) Stromal-epithelial crosstalk regulates kidney progenitor cell differentiation. Nat Cell Biol 15: 1035–1044.
Ebert MF, Fei G, Kahmann S, Müller O, Yu J, Sung JJ, Malfertheiner P (2002) Increased β-catenin mRNA levels and mutational alterations of the APC and β-catenin genes are present in intestinal-type gastric cancer. Carcinogenesis 23: 87–91.
Fanelli MF, Chinen LT, Begnami MD, Costa WL Jr, Fregnami JH, Soares FA, Montagnini AL (2012) The influence of transforming growth factor-α, cyclooxygenase-2, matrix metalloproteinase (MMP)-7, MMP-9 and CXCR4 proteins involved in epithelial-mesenchymal transition on overall survival of patients with gastric cancer. Histopathology 61: 153–161.
Harvey K, Tapon N (2007) The Salvador-Warts-Hippo pathway—an emerging tumour suppressor network. Nat Rev Cancer 7: 182–191.
Hou R, Liu L, Anees S, Hiroyasu S, Sibinga NE (2006) The Fat1 cadherin integrates vascular smooth muscle cell growth and migration signals. J Cell Biol 173: 417–429.
Hou R, Sibinga NE (2009) Atrophin proteins interact with the Fat1 cadherin and regulate migration and orientation in vascular smooth muscle cells. J Biol Chem 284: 6955–6965.
Hulpiau P, van Roy F (2009) Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol 41 (2): 649–669.
Imajo M, Miyatake K, Iimura A, Miyamoto A, Nishida E (2012) A molecular mechanism that links Hippo signalling to the inhibition of Wnt/β-catenin signalling. EMBO J 31: 1109–1122.
Ishiuchi T, Misaki K, Yonemura S, Takeichi M, Tanoue T (2009) Mammalian Fat and Dachsous cadherins regulate apical membrane organization in the embryonic cerebral cortex. J Cell Biol 185: 959–967.
Ju X, Ishikawa TO, Naka K, Ito K, Ito Y, Oshima M (2014) Context-dependent activation of Wnt signaling by tumor suppressor RUNX3 in gastric cancer cells. Cancer Sci 105: 418–424.
Kalluri R, Weinberg RA (2009) The basics of epithelial-mesenchymal transition. J Clin Invest 119: 1420–1428.
Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H (1997) Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275: 1784–1787.
Kusinska RU, Kordek R, Pluciennik E, Bednarek AK, Piekarski JH, Potemski P (2009) Does vimentin help to delineate the so-called 'basal type breast cancer'? J Exp Clin Cancer Res 28: 118.
Lee S, Stewart S, Nagtegaal I, Luo J, Wu Y, Colditz G, Medina D, Allred DC (2012) Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res 72: 4574–4586.
Lowy AM, Clements WM, Bishop J, Kong L, Bonney T, Sisco K, Aronow B, Fenoglio-Preiser C, Groden J (2006) β-catenin/Wnt signaling regulates expression of the membrane type 3 matrix metalloproteinase in gastric cancer. Cancer Res 66: 4734–4741.
Magg T, Schreiner D, Solis GP, Bade EG, Hofer HW (2005) Processing of the human protocadherin Fat1 and translocation of its cytoplasmic domain to the nucleus. Exp Cell Res 307: 100–108.
Mao Y, Mulvaney J, Zakaria S, Yu T, Morgan KM, Allen S, Basson MA, Francis-West P, Irvine KD (2011) Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development. Development 138: 947–957.
Matis M, Axelrod JD (2013) Regulation of PCP by the Fat signaling pathway. Genes Dev 27: 2207–2220.
Mishra L, Shetty K, Tang Y, Stuart A, Byers SW (2005) The role of TGF-beta and Wnt signaling in gastrointestinal stem cells and cancer. Oncogene 24: 5775–5789.
Moeller MJ, Soofi A, Braun GS, Li X, Watzl C, Kriz W, Holzman LB (2004) Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J 23: 3769–3779.
Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan Ş, Eng S, Kannan K, Zou Y, Peng L, Banuchi VE, Paty P, Zeng Z, Vakiani E, Solit D, Singh B, Ganly I, Liau L, Cloughesy TC, Mischel PS, Mellinghoff IK, Chan TA (2013) Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet 45: 253–261.
Nakaya K, Yamagata HD, Arita N, Nakashiro KI, Nose M, Miki T, Hamakawa H (2007) Identification of homozygous deletions of tumor suppressor gene FAT in oral cancer using CGH-array. Oncogene 26: 5300–5308.
Qi C, Zhu YT, Hu L, Zhu YJ (2009) Identification of Fat4 as a candidate tumor suppressor gene in breast cancers. Int J Cancer 124: 793–798.
Rauch TA, Wang Z, Wu X, Kernstine KH, Riggs AD, Pfeifer GP (2012) DNA methylation biomarkers for lung cancer. Tumour Biol 33: 287–296.
Saburi S, Hester I, Goodrich L, McNeill H (2012) Functional interactions between Fat family cadherins in tissue morphogenesis and planar polarity. Development 139: 1806–1820.
Sadeqzadeh E, de Bock CE, Thorne RF (2014) Sleeping giants: emerging roles for the fat cadherins in health and disease. Med Res Rev 34: 190–221.
Schreiner D, Muller K, Hofer HW (2006) The intracellular domain of the human protocadherin hFat1 interacts with Homer signalling scaffolding proteins. FEBS Lett 580: 5295–5300.
Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB (2011) An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 7: e1002218.
Skouloudaki K, Puetz M, Simons M, Courbard JR, Boehlke C, Hartleben B, Engel C, Moeller MJ, Englert C, Bollig F, Schafer T, Ramachandran H, Mlodzik M, Huber TB, Kuehn EW, Kim E, Kramer-Zucker A, Walz G (2009) Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc Natl Acad Sci USA 106: 8579–8584.
Takayama T, Shiozaki H, Shibamoto S, Oka H, Kimura Y, Tamura S, Inoue M, Monden T, Ito F, Monden M (1996) Beta-catenin expression in human cancers. Am J Pathol 148: 39–46.
Tao J, Calvisi DF, Ranganathan S, Cigliano A, Zhou L, Singh S, Jiang L, Fan B, Terracciano L, Armeanu-Ebinger S, Ribback S, Dombrowski F, Evert M, Chen X, Monga SP (2014) Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology 147: 690–701.
Tsukamoto T, Yamamoto M, Ogasawara N, Ushijima T, Nomoto T, Fujita H, Matsushima T, Nozaki K, Cao X, Tatematsu M (2003) β-Catenin mutations and nuclear accumulation during progression of rat stomach adenocarcinomas. Cancer Sci 94: 1046–1051.
van Roy F (2014) Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer 14: 121–134.
Yao H, Ashihara E, Maekawa T (2011) Targeting the Wnt/β-catenin signaling pathway in human cancers. Expert Opin Ther Targets 15: 873–887.
Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P (2012) Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet 44: 570–574.
Acknowledgements
This study was supported by the grant from the Natural Scientific Foundation of China (No. 81372670) and the grant from the Natural Scientific Foundation of Shanghai (No. 13ZR1413500).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Cai, J., Feng, D., Hu, L. et al. FAT4 functions as a tumour suppressor in gastric cancer by modulating Wnt/β-catenin signalling. Br J Cancer 113, 1720–1729 (2015). https://doi.org/10.1038/bjc.2015.367
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2015.367
Keywords
This article is cited by
-
FAT4 overexpression promotes antitumor immunity by regulating the β-catenin/STT3/PD-L1 axis in cervical cancer
Journal of Experimental & Clinical Cancer Research (2023)
-
Identification of FAT4 as a positive prognostic biomarker in DLBCL by comprehensive genomic analysis
Clinical and Experimental Medicine (2023)
-
FAT4 activation inhibits epithelial-mesenchymal transition (EMT) by promoting autophagy in H2228/Cer cells
Medical Oncology (2022)
-
FAT4 silencing promotes epithelial-to-mesenchymal transition and invasion via regulation of YAP and β-catenin activity in ovarian cancer
BMC Cancer (2020)
-
Down-regulated long non-coding RNA RNAZFHX4-AS1 suppresses invasion and migration of breast cancer cells via FAT4-dependent Hippo signaling pathway
Cancer Gene Therapy (2019)