Abstract
Background:
Breast-cancer metastasis suppressor 1 (BRMS1) gene encodes for a predominantly nuclear protein that differentially regulates the expression of multiple genes, leading to suppression of metastasis without blocking orthotropic tumour growth. The aim of the present study was to evaluate for the first time the prognostic significance of BRMS1 promoter methylation in cell-free DNA (cfDNA) circulating in plasma of non-small cell lung cancer (NSCLC) patients. Towards this goal, we examined the methylation status of BRMS1 promoter in NSCLC tissues, matched adjacent non-cancerous tissues and corresponding cfDNA as well as in an independent cohort of patients with advanced NSCLC and healthy individuals.
Methods:
Methylation of BRMS1 promoter was examined in 57 NSCLC tumours and adjacent non-cancerous tissues, in cfDNA isolated from 48 corresponding plasma samples, in cfDNA isolated from plasma of 74 patients with advanced NSCLC and 24 healthy individuals.
Results:
The BRMS1 promoter was highly methylated both in operable NSCLC primary tissues (59.6%) and in corresponding cfDNA (47.9%) but not in cfDNA from healthy individuals (0%), while it was also highly methylated in cfDNA from advanced NSCLC patients (63.5%). In operable NSCLC, Kaplan–Meier estimates were significantly different in favour of patients with non-methylated BRMS1 promoter in cfDNA, concerning both disease-free interval (DFI) (P=0.048) and overall survival (OS) (P=0.007). In advanced NSCLC, OS was significantly different in favour of patients with non-methylated BRMS1 promoter in their cfDNA (P=0.003). Multivariate analysis confirmed that BRMS1 promoter methylation has a statistical significant influence both on operable NSCLC patients’ DFI time and OS and on advanced NSCLC patients’ PFS and OS.
Conclusions:
Methylation of BRMS1 promoter in cfDNA isolated from plasma of NSCLC patients provides important prognostic information and merits to be further evaluated as a circulating tumour biomarker.
Similar content being viewed by others
Main
Lung cancer is the leading cause of cancer death worldwide, with over 1 million deaths each year (Parkin et al, 2005; Anglim et al, 2008). Patient’s survival depends significantly on early detection, and for patients with operable stage IA, the 5-year survival can be as high as 55–80% (Wang et al, 2010). Early detection and precise diagnosis are critical for patients to receive proper therapeutic treatment as early as possible and thus could improve survival rates in non-small cell lung cancer (NSCLC) patients. Despite recent advancements in lung cancer therapies, the prognosis for patients with advanced NSCLC remains poor, so innovative, non-invasive, sensitive and reliable biomarkers still need to be discovered and exploited. Tumour biomarkers can have an important role in cancer screening, diagnosis, prognosis and therapeutic monitoring. Discovery and validation of novel biomarkers for early characterisation of carcinomas is one of the main aims of contemporary cancer research (Diamandis et al, 2013; Pavlou et al, 2013).
Methylation of DNA is one of the most frequently occurring epigenetic events taking place in mammalian genome and alterations in DNA methylation are very common in cancer cells. In particular, hypermethylation has been reported as an early event in carcinogenesis and progression to malignancy, frequently leading to gene silencing through methylation of CpG-rich regions near the transcriptional start sites of genes that regulate important cell functions (Laird, 2003). Methylation of specific genes appears to be an early event that has a fundamental role in the development and progression of cancer (Heyn and Esteller, 2012). Epigenetic changes such as individual gene promoter methylation are now under intensive evaluation as lung cancer biomarkers and present a strong potential to advance our understanding of its aetiology as well as provide novel early detection biomarkers (Zöchbauer-Müller et al, 2001; Anglim et al, 2008; Brock et al, 2008; Heyn and Esteller, 2012).
The genetic profile of solid tumours is currently obtained in an invasive way from surgical or biopsy specimens; moreover, information acquired from a single biopsy might fail to reflect tumour heterogeneity and reflects a limited snap-shot of a tumour that is continuously evolving and can acquire resistance to systemic treatment as a result of clonal evolution and selection. A ‘liquid biopsy’, or blood sample, can provide the genetic landscape of all cancerous lesions (primary and metastases) as well as offering the opportunity to systematically track genomic evolution (Crowley et al, 2013). Additionally, blood-based diagnostics can classify tumours into distinct molecular subtypes and monitor disease relapse and response to treatment (Hanash et al, 2011).
Circulating cell-free DNA (cfDNA) is an emerging non-invasive blood-based biomarker utilised to assess tumour progression and to evaluate prognosis, diagnosis and response to treatment (Marzese et al, 2013) and monitoring of the efficacy of anticancer therapies (Schwarzenbach et al, 2011). It was very recently shown that sequencing of cancer exomes in serial plasma samples can track genomic evolution of metastatic cancers in response to therapy (Murtaza et al, 2013).
Cancer cell-specific methylated DNA has been found in the blood of cancer patients, indicating that cfDNA is a tumour-associated DNA marker that can be used as a minimally invasive diagnostic test. Esteller et al have shown already in 1999 that detection of aberrant promoter hypermethylation of tumour suppressor genes could be detected in serum DNA from NSCLC patients (Esteller et al, 1999). Since then, many studies have described methylation of tumour suppressor genes in serum or plasma samples and in the corresponding primary tumours (Usadel et al, 2002; Hoque et al, 2006; Hsu et al, 2007). In the majority of these studies, the frequencies of methylation in plasma were lower in respect to those of the primary tumours (Hoque et al, 2006), while notably, in the majority of cases, DNA methylation was not detected in plasma or serum of healthy donors (Usadel et al, 2002; Hoque et al, 2006). Especially in lung cancer, DNA methylation of various genes has been detected in cfDNA circulating in plasma or serum, in sputum and in bronchoalveolar lavage samples (Palmisano et al, 2000; Fujiwara et al, 2005; Hsu et al, 2007).
Breast cancer metastasis suppressor 1 (BRMS1) is a predominantly nuclear protein that differentially regulates expression of multiple genes leading to suppression of metastasis without blocking orthotropic growth (Vaidya and Welch, 2007).This gene is significantly downregulated in some breast tumours, especially in metastatic disease, by epigenetic silencing (Metge et al, 2008). We have recently shown that BRMS1 promoter was methylated in DNA extracted from circulating tumour cells (CTCs) isolated from peripheral blood of breast cancer patients (Chimonidou et al, 2011, 2013). We have also recently shown that BRMS1 promoter methylation was not detected in non-cancerous breast tissues or benign fibroadenomas, while in breast cancer primary tumours it was significantly associated with reduced disease-free survival (Chimonidou et al, 2013). Although the role of BRMS1 in NSCLC has being recently studied in primary tumour tissues (Smith et al, 2009; Nagji et al, 2010; Yang et al, 2011), there is no information concerning the prognostic significance of BRMS1 gene promoter methylation in cfDNA circulating in plasma.
The aim of the present study was to evaluate for the first time the prognostic significance of BRMS1 promoter methylation in cfDNA circulating in plasma of NSCLC patients. Towards this goal, we examined the methylation status of BRMS1 promoter-associated CpG island in NSCLC tissues, matched adjacent non-cancerous tissues and cfDNA as well as in healthy individuals.
Patients and methods
The outline of the workflow of our study is shown in Figure 1.
Clinical samples
The study material consisted of three different sets of clinical samples: (a) Training set: this set consisted of 57 NSCLC fresh-frozen tissues and corresponding adjacent non-neoplastic tissues and 48 corresponding plasma samples. There were 46 men and 11 women (median age: 61 years), all diagnosed with operable (stage I–III) NSCLC; 27 patients were diagnosed with adenocarcinoma (AD), 25 had squamous cell carcinoma (SQ) and 5 were diagnosed with undifferentiated NSCLC; in this group, the majority of patients (91.5%) were smokers and suffered from mild-to-moderate chronic obstructive pulmonary disease according to pulmonary function tests that were included as a part of the standardised preoperative evaluation of the patients. All patients were treatment naïve when the samples were collected, but after surgery all patients received standard chemotherapy protocols for adjuvant NSCLC, such as gemcitabine plus taxanes (90%) or platinum-based chemotherapy (10%). The majority of patients changed stage after the disease relapse to IIIB, (b) Independent validation cohort: this set consisted of 74 cfDNA samples isolated from plasma of advanced (stage IV) NSCLC patients. In this group, blood was obtained at diagnosis and before the initiation of any systemic treatment. Fifty patients had a non-squamous histology and 53 had distant metastases whereas 21 had inoperable stage IIIB disease. Twenty-three patients were treated with single agent chemotherapy in the context of geriatric chemotherapy protocols of the Hellenic Oncology Research Group (HORG), namely docetaxel or gemcitabine whereas the remaining 51 patients received chemotherapy combinations associating a taxane with a platinum compound. Among the evaluable for response patients, 18 achieved an objective response (CR: n=1; PR: n=17) and 11 stable disease. At the time of the present analysis, all but one patient were dead because of disease progression and (c) Control population: this set consisted of 24 cfDNA samples isolated from plasma of healthy donors. The tumour type and stages were analysed histologically and tissue sections containing >80% of tumour cells were used for DNA extraction and methylation-specific PCR (MSP) analysis. All patients gave their informed consent to participate in the study, which has been approved by the Ethical and Scientific Committees of our Institutions. At the time of surgery, all tissue samples were immediately flash frozen in liquid nitrogen and stored at −80 °C until use. Immediately after venipuncture, peripheral blood in EDTA was centrifuged at 2000 g for 10 min at room temperature and 1 ml aliquots of plasma samples were stored at −80 °C until use.
Isolation of genomic DNA from tumour tissues
Genomic DNA (gDNA) from NSCLC tissues and corresponding adjacent tissues was isolated using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Isolation of cfDNA circulating in plasma
Cell-free DNA was isolated from plasma samples using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions. In all, 200 μl of plasma was mixed with 200 μl of working solution and 50 μl proteinase K (18 mg ml−1) and incubated for 10 min at 72 °C. DNA isolation was then processed as described in the manufacturer’s protocol.
Sodium bisulfite conversion
The concentration of DNA was determined in the Nanodrop ND-100 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). In all, 1 μg of extracted DNA was modified with sodium bisulfite (SB), to convert all non-methylated cytosines to uracil, while methylated cytosines were not converted. Bisulfite conversion was carried out in 1 μg of denaturated DNA using the EZ DNA Methylation Gold Kit (ZYMO Research Co., Orange, CA, USA), according to the manufacturer’s instructions and as previously described (Parkin et al, 2005; Pavlou et al, 2013). The converted DNA was stored at –70 °C until use. In each SB conversion reaction, dH2O and DNA isolated from the MCF-7 cell line were included as a negative and a positive control, respectively.
Methylation-specific PCR
The methylation status of BRMS1 gene in tissue samples was detected by conventional MSP by using specific primer pairs for both the methylated and unmethylated promoter sequences as previously described (Chimonidou et al, 2013). Each MSP reaction was performed in a total volume of 25 μl. In all, 1 μl of SB-converted DNA was added into a 24 μl reaction mixture that contained 0.1 μl of Taq DNA polymerase (5 U μl−1, hot start GoTaq Polymerase; Promega, Madison, WI, USA), 5 μl of the supplied 10 × PCR buffer, 2.0 μl of MgCl2 (50 mmol l−1), 0.5 μl of dNTP’s (10 mmol l−1; Fermentas, Carlsbad, CA, USA) and 1 μl of the corresponding forward and reverse primers (10 μmol l−1); dH2O was added to a final volume of 25 μl. Sodium bisulfite-treated DNA was amplified in two separate MSP reactions, one with a set of primers specific for the methylated and one for the unmethylated BRMS1 promoter sequences. Methylation-specific PCR products for methylated and unmethylated BRMS1 promoter were fractionated on 2% agarose gels containing 40 mM Tris-acetate/1.0 mM EDTA (pH=8) and visualised by ethidium bromide staining.
Real-time MSP
The methylation status of BRMS1 gene in cfDNA samples was detected by a newly designed and more sensitive real-time MSP assay based on the same set of MSP-specific primer pairs as previously described (Chimonidou et al, 2013) and a newly designed hydrolysis (Taqman) LNA probe that is hybridising to a methylation-independent region (Table 1). Each reaction was performed in a total volume of 10 μl in the LightCycler 2.0 real time PCR instrument (Roche, Mannheim, Germany). One microlitre of SB-converted DNA was added into a 9-μl reaction mixture that contained 0.1 μl of Taq DNA polymerase (5 U μl−1, DNA polymerase; Promega), 2 μl of the supplied PCR buffer (5 × ), 1.0 μl of MgCl2 (25 mmol l−1), 0.2 μl of dNTPs (10 mmol l−1; Fermentas) and 0.2 μl of the forward and reverse primers (10 μmol l−1), 0.15 μl BSA (10 μg μl−1), 1 μl hydrolysis LNA probe (3 μmol l−1); finally, dH2O was added to a final volume of 10 μl. Similar thermocycling conditions were used: 1 cycle at 95 °C for 2 min, followed by 45 cycles at 95 °C for 10 s and 60 °C for 1 min. Sodium bisulfite-converted DNA from the DNA methylation standard (100%) was included in every run as a positive control.
In both cases (MSP and real-time MSP), human placental gDNA (Sigma-Aldrich, St Louis, MO, USA) methylated in vitro with SssI methylase (NEB, Ipswich, MA, USA) was used, after SB conversion, as fully methylated (100%) MSP-positive control; the same unmethylated placental gDNA, was used, after SB conversion, as a negative MSP control. The specificity and sensitivity of the MSP assay for BRMS1 promoter methylation has been previously verified (Chimonidou et al, 2011, 2013).
Statistical analysis
Correlations between methylation status and clinico-pathological features of the patients were assessed by using the Chi-square test. Disease-free interval (DFI), progression-free survival (PFS) and overall survival (OS) curves were calculated by using the Kaplan–Meier method and comparisons were performed using the log-rank test. P-values <0.05 were considered as statistically significant. Statistical analysis was performed by using the SPSS Windows version 17.0 (SPSS Inc., Chicago, IL, USA).
Results
Concordance between BRMS1 promoter methylation in operable NSCLC tissues and corresponding cfDNA
We first compared our results on BRMS1 promoter methylation in 48 operable NSCLC fresh tissues and corresponding cfDNA circulating in plasma of these patients. According to our findings, in 14 out of 48 (29.2%) NSCLC patients BRMS1 promoter was found methylated both in tumour tissues and in cfDNA samples. In 16 out of 48 (33.3%) NSCLC patients, BRMS1 promoter was not methylated both in tumour tissues and in cfDNA samples. The concordance between BRMS1 promoter methylation in NSCLC fresh-frozen tissues and cfDNA was 30 out of 48 (62.5%). In 9 out of 48 (18.8%) cases, BRMS1 promoter was not methylated in cfDNA, while it was methylated in corresponding primary tumours. In another 9 out of 48 (18.8%) cases, BRMS1 promoter was methylated in cfDNA, while the corresponding tumours were found to be negative for methylation.
Evaluation of the prognostic significance of BRMS1 promoter methylation in operable NSCLC fresh-frozen tissues
The methylation status of BRMS1 promoter was first assessed in 57 pairs of fresh-frozen NSCLC tissues and their adjacent non-cancerous tissues, using the BRMS1 conventional MSP assay. The BRMS1 promoter was found to be methylated in 34 out of 57 (59.6%) tumour tissues and in 31 out of 57 (54.3%) of the corresponding adjacent non-cancerous tissues. The patient’s characteristics according to the methylation status of BRMS1 in NSCLC are presented in Table 2. Chi-square analysis did not reveal any statistically significant correlation between BRMS1 promoter methylation and the clinico-pathological features of these patients.
During the follow-up period (73 months), 5 out of 57 patients without disease relapse died from other reasons and were thus not included in the survival analysis. In the remaining 52 patients, 36 out of 52 (69.2%) relapsed and 31 out of 52 (59.6%) died from the disease, during a median follow-up period of 45 months (range 1–73 months). Methylation of BRMS1 was detected in 24 out of 36 (66.7%) of patients who relapsed, and in 20 out of 31 (64.5%) of patients who died. The incidence of relapses was similar between patients with methylated (24 out of 32, 75.0%) and non-methylated BRMS1 promoter (12 out of 20, 60.0%) (P=0.202) while the incidence of deaths was also similar between patients with methylated (20 out of 32, 62.5%) and non-methylated BRMS1 promoter (11 out of 20, 55.0%) (P=0.402) (Table 3).
Kaplan–Meier estimates of the cumulative DFI and OS for NSCLC patients with methylated and non-methylated BRMS1 promoter in tumour tissues were not significantly different (P=0.106 and P=0.376, respectively; log-rank test, data not shown).
Evaluation of the prognostic significance of BRMS1 promoter methylation in corresponding cfDNA
We further evaluated the prognostic significance of BRMS1 methylation in corresponding cfDNA circulating in plasma of 48 out of the initial 57 enrolled patients. In addition, plasma samples from 24 control healthy individuals were also analysed.
The BRMS1 promoter was methylated in 23 out of 48 (47.9%) plasma samples of operable NSCLC patients but not in any of the control plasma samples (0%). During follow-up, 4 out of these 48 patients died from reasons other than cancer and were thus not included in the survival analysis. In the remaining group of 44 patients, after a median follow-up period of 45 months (range 1–73 months), 32 out of 44 (72.7%) patients relapsed and 27 out of 44 (61.4%) died from the disease. Methylation of BRMS1 promoter was detected in 19 out of 32 (59.4%) of patients who relapsed, and in 18 out of 27 (66.7%) of patients who died.
Table 3 indicates that the incidence of relapses was statistically different between patients with methylated (19 out of 22, 86.3%) and patients with non-methylated BRMS1 promoter (13 out of 22, 59.1%) (P=0.044). The incidence of deaths was also statistically different between patients with methylated (18 out of 22, 82.0%) and patients with non-methylated BRMS1 promoter (9 out of 22, 40.9%) (P=0.006).
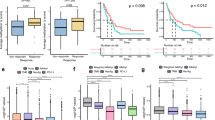
Subsequently, the prognostic significance of BRMS1 promoter methylation in these cfDNA samples was analysed. Kaplan–Meier estimates of the cumulative DFI and OS for NSCLC patients with methylated and non-methylated BRMS1 promoter methylation in cfDNA were significantly different in favour of patients with non-methylated BRMS1 promoter (P=0.048 and P=0.007, log-rank test, Figure 2A and B, respectively).
Prognostic significance of BRMS1 promoter methylation in cell-free DNA circulating in plasma: (A) training set (n=48): Kaplan–Meier estimates for DFI (P=0.048), (B) training set (n=48): Kaplan–Meier estimates for OS (P=0.007) and (C) independent cohort (n=74): Kaplan–Meier estimates for OS (P=0.003).
Multivariate analysis confirmed that only BRMS1 promoter methylation has a statistical significant influence on patients’ DFI (Table 4). Patients who present BRMS1 promoter methylation have a significant lower DFI than those who do not (HR: 2.158, 95% CI: 1.030–4.517, P=0.041). Multivariate analysis revealed that only BRMS1 promoter methylation has a statistical significant influence on patients’ OS time. Patients who present BRMS1 promoter methylation have a significantly lower OS time than those who do not (HR: 3.008, 95% CI: 1.295–6.989, P=0.010).
Evaluation of the prognostic significance of BRMS1 promoter methylation in cfDNA in an independent validation cohort of advanced NSCLC patients
To further verify the prognostic significance of BRMS1 promoter methylation in cfDNA circulating in plasma, we conducted a validation step in an independent cohort of 74 patients with advanced (stage IV) NSCLC.
The methylation status of BRMS1 promoter in cfDNA of these patients revealed that it was methylated in 47 out of 74 (63.5%) cases. After a median follow-up period of 43 months (range 1–84 months), 73 out of 74 (98.6%) patients relapsed and 72 out of 74 (97.3%) died due to disease progression. Methylation of BRMS1 was detected in plasma of all patients who relapsed and died.
Kaplan–Meier estimates of the cumulative PFS for patients with methylated and non-methylated BRMS1 promoter had marginal significance (P=0.059, log-rank test) whereas that of the cumulative OS was significantly different among the two groups (P=0.003, log-rank test) (Figure 2C).
Multivariate analysis confirmed that only BRMS1 promoter methylation has a statistical significant influence on patients’ PFS time (Table 5). Patients who present BRMS1 promoter methylation have a significant lower PFS time than those who do not (HR: 1.951, 95% CI: 1.175–3.238, P=0.010). Moreover, multivariate analysis revealed that BRMS1 promoter methylation has a statistical significant influence on patients’ OS time, while performance status has a marked but not significant trend (HR: 1.861, 95% CI: 0.989–3.500, P=0.054). Patients who present with BRMS1 promoter methylation have a significant lower OS than those who do not (HR: 2.057, 95% CI: 1.247–3.386, P=0.005) (Table 5).
Discussion
There is now an urgent need for blood-based, non-invasive molecular tests to assist in the detection, diagnosis and prognosis of cancers in a non-invasive and cost-effective manner especially at an early stage, when curative interventions are still possible. Cell-free DNA is released from cancer cells into plasma, and is representing a non-invasive liquid biopsy approach that can now give important information as a blood-based tumour biomarker. Especially information on the methylation status of tumour suppressor genes in cfDNA is a very promising approach, since it can offer a useful tool for lung cancer diagnostics, evaluation of cancer treatment efficiency and post-treatment monitoring.
In this study, we evaluated for the first time the prognostic significance of BRMS1 promoter methylation in cfDNA circulating in plasma of NSCLC patients. Our results clearly indicate that BRMS1 promoter methylation is highly methylated in NSCLC tissues, and that detection of BRMS1 promoter methylation in cfDNA isolated from plasma is highly specific and provides important prognostic information. The observed association between methylated BRMS1 and reduced OS may be relevant to the known role of BRMS1 as a tumour suppressor. Methylation of BRMS1 promoter, and subsequent silencing of this gene, may be indicative of a more aggressive tumour phenotype and, thus, the association with a poorer outcome. In the contrary, non-methylated BRMS1 may be indicative of a slower progressing tumour as the gene may partially maintain its tumour suppressing capacity.
It was recently reported that BRMS1 expression is diminished in NSCLC compared with non-cancerous lung tissues and it was also lower in squamous cell carcinoma compared with AD (Smith et al, 2009). Given these observations, the same group hypothesised that BRMS1 transcription is decreased in NSCLC through increased BRMS1 promoter methylation, and it was confirmed that BRMS1 promoter-associated CpG island was hypermethylated in both NSCLC cells and human NSCLC specimens (Nagji et al, 2010). For that specific study cohort, BRMS1 promoter methylation was significantly more robust in squamous cell carcinoma compared with AD histologies. Another recent study confirmed the above results. Yang et al (2011) have recently reported that BRMS1 promoter-associated CpG island is aberrantly methylated in NSCLC and that patients with a high level of BRMS1 mRNA expression had significantly better OS than those with low expression and that promoter methylation of BRMS1 was a significantly unfavourable prognostic factor.
The mechanism through which BRMS1 suppresses metastasis is not clearly understood. A recent study revealed that ubiquitous BRMS1 expression suppresses pulmonary metastasis and promotes apoptosis of tumour cells located in the lung but not in the mammary glands, suggesting that cell-location specific over expression of BRMS1 is important of BRMS1-mediated metastasis suppression (Cook et al, 2012).
Other studies focus on the strong correlation between loss of BRMS1 protein expression and reduced disease-free survival in subsets of breast cancer patients (Hang et al, 2006; Hicks et al, 2006). Furthermore, the loss of BRMS1 is associated with a decreased survival in patients with NSCLC (Nagji et al, 2010; Yang et al, 2011). In addition, aberrant methylation of BRMS1 is responsible for its loss of expression in breast cancer and in NSCLC. The BRMS1 low expression is also correlated with poor patient survival in nasopharyngeal carcinoma (Cui et al, 2012). These results suggest that downregulation caused by BRMS1 promoter methylation has an important role in tumorigenesis in many different types of cancer.
Our results clearly indicate that aberrant methylation of the BRMS1 gene promoter is a common event in operable NSCLC tissues. Our primers are designed to recognise BRMS1 promoter methylation in specific CpG sites, since we have already shown that methylation of these regions is of clinical importance in breast cancer (Chimonidou et al, 2013). However, the prognostic significance of BRMS1 promoter methylation in operable NSCLC tissues was not evident in our patients’ cohort. On the contrary, we report here for the first time that detection of aberrant methylation of the BRMS1 gene promoter in cfDNA circulating in plasma of these NSCLC patients provides prognostic information, both for DFI and for OS. Our results were further confirmed in an independent validation cohort of advanced NSCLC patients. In both groups of patients, training and independent cohort, the frequency observed for BRMS1 methylation was high. This may suggest that loss of BRMS1 expression is an early event in NSCLC tumorigenesis and remains in high levels in advanced stages of the disease. In operable NSCLC, Kaplan–Meier analysis has shown a strong correlation of BRMS1 promoter methylation in plasma and poor DFI and OS, and these results were further verified by multivariate analysis. In advanced NSCLC, Kaplan–Meier analysis has shown a strong correlation between BRMS1 promoter methylation in plasma and poor OS, while multivariate analysis has shown that BRMS1 promoter methylation has a statistical significant influence on both patients’ PFS and OS.
In conclusion, our data indicate for the first time that detection of BRMS1 promoter methylation in cfDNA circulating in plasma provides important prognostic information for NSCLC patients. We believe that BRMS1 promoter methylation in cfDNA should be further evaluated and validated as a non-invasive circulating tumour biomarker in a larger cohort of patients.
Change history
15 April 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Anglim PP, Alonzo TA, Laird-Offringa IA (2008) DNA methylation-based biomarkers for early detection of non-small cell lung cancer: an update. Mol Cancer 7: 81.
Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, Glöckner S, Piantadosi S, Gabrielson E, Pridham G, Pelosky K, Belinsky SA, Yang SC, Baylin SB, Herman JG (2008) DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 358: 1118–1128.
Chimonidou M, Kallergi G, Georgoulias V, Welch DR, Lianidou ES (2013) BRMS1 promoter methylation provides prognostic information in primary breast tumors. Mol Cancer Res 11: 1248–1257.
Chimonidou M, Strati A, Tzitzira A, Sotiropoulou G, Malamos N, Georgoulias V, Lianidou ES (2011) DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin Chem 57: 1169–1179.
Cook LM, Cao X, Dowell AE, Debies MT, Edmonds MD, Beck BH, Kesterson RA, Desmond RA, Frost AR, Hurst DR, Welch DR (2012) Ubiquitous Brms1 expression is critical for mammary carcinoma metastasis suppression via promotion of apoptosis. Clin Exp Metastasis 29 (4): 315–325.
Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A (2013) Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 10: 472–484.
Cui RX, Liu N, He QM, Li WF, Huang BJ, Sun Y, Tang LL, Chen M, Jiang N, Chen L, Yun JP, Zeng J, Guo Y, Wang HY, Ma J (2012) Low BRMS1 expression promotes nasopharyngeal carcinoma metastasis in vitro and in vivo and is associated with poor patient survival. BMC Cancer 29 (12): 376.
Diamandis EP, Bast RC, Lopez-Otín C (2013) Conquering cancer in our lifetime: new diagnostic and therapeutic trends. Clin Chem 59: 1–3.
Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG (1999) Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res 59: 67–70.
Fujiwara K, Fujimoto N, Tabata M, Nishi K, Matsuo K, Hotta K, Kozuki T, Aoe M, Kiura K, Ueoka H, Tanimoto M (2005) Identification of epigenetic aberrant methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res 11: 1219–1225.
Hanash SM, Baik CS, Kallioniemi O (2011) Emerging molecular biomarkers—blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol 8: 142–150.
Hang Z, Yamashita H, Toyama T, Yamamoto Y, Kawasoe T, Iwase H (2006) Reduced expression of the breast cancer metastasis suppressor 1 mRNA is correlated with poor progress in breast cancer. Clin Cancer Res 12: 6410–6414.
Heyn H, Esteller M (2012) DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 13: 679–692.
Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M, Choueiri TK, Tubbs RR, Gaile D, Nowak N, Accavitti-Loper MA, Frost AR, Welch DR, Casey G (2006) Loss of breast cancer metastasis suppressor 1 protein expression predicts reduced disease free survival in subsets of breast cancer patients. Clin Cancer Res 12: 6702–6708.
Hoque MO, Feng Q, Toure P, Dem A, Critchlow CW, Hawes SE, Wood T, Jeronimo C, Rosenbaum E, Stern J, Yu M, Trink B, Kiviat NB, Sidransky D (2006) Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol 24 (26): 4262–4269.
Hsu HS, Chen TP, Hung CH, Wen CK, Lin RK, Lee HC, Wang YC (2007) Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer 110 (9): 2019–2026.
Laird PW (2003) The power and promise of DNA methylation markers. Nat Rev Cancer 3: 253–266.
Marzese DM, Hirose H, Hoon DS (2013) Diagnostic and prognostic value of circulating tumor-related DNA in cancer patients. Expert Rev Mol Diagn 13 (8): 827–844.
Metge BJ, Frost AR, King JA, Dyyess DL, Welch DR, Samant RS, Shevde LA (2008) Epigenetic silencing contributes to the loss of BRMS1 expression in breast cancer. Clin Exp Metastasis 25: 753–763.
Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, Marass F, Humphray S, Hadfield J, Bentley D, Chin TM, Brenton JD, Caldas C, Rosenfeld N (2013) Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497: 108–112.
Nagji AS, Liu Yuan., Stelow EB, Stukenborg GJ, Jones DR (2010) BRMS1 transcriptional repression correlates with CpG island methylation and advanced pathological stage in non-small cell lung cancer. J Pathol 221 (2): 229–237.
Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA (2000) Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res 60: 5954–5958.
Parkin DM, Bray F, Ferlay M, Pisani P (2005) Global cancer statistics 2002. CA Cancer J Clin 55: 74–108.
Pavlou MP, Diamandis EP, Blasutig IM (2013) The long journey of cancer biomarkers from the bench to the clinic. Clin Chem 59: 147–157.
Schwarzenbach H, Hoon DS, Pantel K (2011) Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11: 426–437.
Smith PW, Liu Y, Siefert SA, Moskaluk CA, Petroni GR, Jones DR (2009) Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis and correlates with improved patient survival in non-small cell lung cancer. Cancer Lett 276: 196–203.
Usadel H, Brabender J, Danenberg KD, Jerónimo C, Harden S, Engles J, Danenberg PV, Yang S, Sidransky D (2002) Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum and plasma DNA of patients with lung cancer. Cancer Res 62: 371–375.
Vaidya KS, Welch DR (2007) Metastasis suppressors and their roles in breast carcinoma. J Mammary Gland Biol Neoplasia 12: 175–190.
Wang T, Nelson RA, Bogardus A, Grannis FW Jr (2010) Five-year lung cancer survival: which advanced stage non-small cell lung cancer patients attain long term survival? Cancer 116: 1518–1525.
Yang J, Shen Y, Liu B, Tong Y (2011) Promoter methylation of BRMS1 correlates with smoking history and poor survival in non-small cell lung cancer patients. Lung Cancer 74 (2): 305–309.
Zöchbauer-Müller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD (2001) Aberrant promoter methylation of multiple genes in non small cell lung cancer. Cancer Res 61: 249–255.
Acknowledgements
Ioanna Balkouranidou is a recipient of a PhD studentship from the Hellenic Oncology Research Group (HORG). We would like to thank Dr Dora Chatzidaki for her significant help in the statistical analysis of our results. Dr Welch is Hall Family Foundation Professor of Molecular Medicine and Kansas Bioscience Authority Eminent Scholar and this work was partly supported by the National Foundation for Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Balgkouranidou, I., Chimonidou, M., Milaki, G. et al. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br J Cancer 110, 2054–2062 (2014). https://doi.org/10.1038/bjc.2014.104
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.104
Keywords
This article is cited by
-
BRMS1: a multifunctional signaling molecule in metastasis
Cancer and Metastasis Reviews (2020)
-
Methylated DNA/RNA in Body Fluids as Biomarkers for Lung Cancer
Biological Procedures Online (2017)
-
Analysen epigenetischer Marker aus Liquid Biopsies: Informationen von jenseits des Genoms
Medizinische Genetik (2016)
-
Breast carcinoma metastasis suppressor gene 1 (BRMS1): update on its role as the suppressor of cancer metastases
Cancer and Metastasis Reviews (2015)
-
BRMS1L suppresses breast cancer metastasis by inducing epigenetic silence of FZD10
Nature Communications (2014)