Abstract
Background:
The ING family of type II tumour suppressors serve as both epigenetic ‘readers’ and target histone acetyl transferase (HAT) and histone deacetylase (HDAC) ‘writers’ of the epigenetic histone code. The ING1 protein has also been implicated in regulating microRNA (miRNA) levels. In this study, we identify a link between ING1b and the miRNA epigenetic network.
Methods:
Primary fibroblasts infected with adenoviruses expressing GFP control or GFP plus ING1b were examined for alterations in miRNA profiles using a miRNA PCR array. Additional experiments confirmed specificity and consequences of altered miRNA expression.
Results:
MicroRNAs miR-203, miR-375, miR-449b and miR-200c were increased by ING1b overexpression. Ectopic expression of miR-203 inhibited U2OS and MDA-MB-231 cancer cell growth, and induced G1 cell cycle arrest in U2OS cells as estimated by flow cytometry. Transfection with miR-203 inhibitor reversed the proliferation inhibition induced by ING1b in U2OS cells. CHIP assays showed that ING1b bound to the promoter of miR-203. Western blot analyses showed that CDK6, c-Abl and Src were downregulated by the transfection of miR-203.
Conclusion:
These results indicate that ING1b epigenetically regulates several miRNAs including miR-203. The several-fold increase in miR-203 by ING1b might inhibit cancer cell proliferation through coordinate downregulation of CDK6, c-Abl and Src.
Similar content being viewed by others
Main
The inhibitor of growth (ING) family of type II tumour suppressors contributes to the neoplastic growth of various tumours. This family is well conserved and includes five genes. Among them, ing1 is the founding member and encodes four protein isoforms. P33ING1b (ING1b) is the most highly expressed isoform in human cells and contributes to most recognised functions of ing1. Overexpression of ING1b caused cell cycle arrest at G1 phase with ensuing apoptosis, whereas suppression of its expression increased colony focus formation and growth in vitro and tumour formation in vivo (Garkavtsev et al, 1996). Deletion of ing1 in mice, and in particular, the orthologue of the human exon encoding ING1b, resulted in an earlier onset and higher incidence of B-cell lymphomas (Coles et al, 2007). However, the detailed molecular mechanisms by which ING1b acts as a tumour suppressor remain incompletely defined.
One mechanism by which INGs exert their effects appears to be by regulating the expression of a broad variety of genes (Feng et al, 2006). ING1b regulates gene expression through regulating histone acetylation (Loewith et al, 2000), acting as a stoichiometric component of histone deacetylase-1 and -2 complexes (Doyon et al, 2006). ING2, which is closely related to ING1b evolutionarily (He et al, 2005) is the major target of the HDAC inhibitor SAHA and thus ING1b and ING2 appear to regulate chromatin compaction and subsequently, the expression of subsets of genes through, primarily, epigenetic mechanisms (Russell et al, 2006; Soliman and Riabowol, 2007). The plant homeodomain (PHD) form of zinc finger is highly conserved amongst ING family members (He et al, 2005). It enables ING family members to specifically recognise the trimethylated form of lysine 4 on histone H3 (H3K4Me3) epigenetic mark that is associated with active gene transcription (Martin et al, 2006; Peña et al, 2006; Shi et al, 2006), allowing INGs to subsequently target histone HDAC (for ING1 and ING2) or HAT complexes (for ING3-5) to this mark, thus serving as agents to target ‘writers’ of the histone code (Vieyra; Doyon et al, 2006; Soliman and Riabowol, 2007).
MicroRNAs (miRNAs) are small, endogenous and non-coding RNA molecules, which regulate gene expression by directly inhibiting translation or by decreasing target mRNA levels. The discovery of miRNAs has shed new insights regarding the regulation of cell proliferation, apoptosis and senescence and has also contributed to clinical medicine in the areas of diagnosis and therapy. For example, antagonizing miR-122 by locked-nucleic-acid-modified oligonucleotides in primates showed a dose-dependent and long-lasting decrease in plasma cholesterol without any acute or subchronic toxicity (Elmén). Many miRNAs have been reported to be regulated by epigenetic mechanisms, such as histone acetylation (Sato. et al, 2011). Therefore, we asked whether miRNAs, which are regulated by ING1b, might have critical roles in the regulation of cell proliferation. In the current study, we show that the expression of miR-203 is epigenetically regulated by ING1b and that miR-203 mediates a significant proportion of the inhibitory effects of ING1b on cell proliferation. This study has identified a novel mechanism underlying ING1b-induced inhibition of cell proliferation, further establishing links between miRNAs and the ING1b tumour suppressor.
Materials and methods
Cell culture and microarray materials
U2OS (osteosarcoma) and MDA-MB-231 (breast cancer) cell lines were obtained from ATCC and were grown in high-glucose DMEM media supplemented with 10% foetal bovine serum, and 1% penicillin and streptomycin. Alamar Blue reagent, Lipofectamine 2000, the mirVana miRNA Isolation Kit, and Propidium iodide (PI) were purchased from Invitrogen Canada (Burlington, ON, Canada). The TaqMan Array Human MicroRNA Card Set v3.0, TaqMan MicroRNA Reverse Transcription Kit, the primers for miRNAs and U6B and universal PCR Master Mix were purchased from Applied Biosystems (Foster City, CA, USA). MicroRNA mimics and MicroRNA mimic controls were purchased from Dharmacon (Lafayette, CO, USA). AKT-2, CDK6, c-Abl, Src and β-actin antibodies were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA).
MiRNA and mRNA profiling and miRNA target prediction
It was previously established that overexpression of ING1b inhibits cell proliferation (Garkavtsev et al, 1996) and promotes apoptosis in established cancer cell lines (Helbing et al, 1997) and in Hs68 human primary fibroblasts (Feng et al, 2006). To ask whether miRNAs might be involved with these phenotypes, Hs68 cells were infected with GFP alone or with GFP-ING1b adenovirus at titres known to arrest cell growth (Feng et al, 2006; Thalappilly et al, 2011) for 48 h, followed by isolation of total RNA. The isolated total RNA was used to probe a TaqMan Array Human MicroRNA Card Set (v3.0), and hits were confirmed using an Applied Biosystems 7900HT Fast Real-Time PCR System, following the manufacturer’s manual (n=1). For mRNA profiling, three biological replicates were used in different groups (n=3). For miRNA target prediction, the miRecords programme was used for identifying the target gene. This programme includes 11 miRNA target prediction algorithms. Target genes that have been selected by more than one algorithm were regarded as potential true targets for further analysis.
Quantification of PCR analyses
Quantification of miRNAs level was performed as described previously (Chen et al, 2011). Briefly, miRNAs from Hs68 cells and U2OS were isolated with mirVana miRNA Isolation Kits and miRNAs were measured following the Applied Biosystems TaqMan MicroRNA assay protocol. Real-time PCR was performed using universal PCR Master Mix in the Applied Biosystems 7900HT Fast Real-Time PCR System. U6B expression was used to normalise the expression of miRNAs. The delta-Ct method was used to evaluate the analyses of real-time PCR data.
Transfection of MiR-203 and cell proliferation assays
For cell-proliferation assays, U2OS cells (4000 per well) were plated in 96-well plates. After overnight incubation, U2OS cells were transfected with 10 nM, 100 nM and 1000 nM of miR-203 mimic or miRNA mimic control using Lipofectamine 2000. After 48 h, Alamar Blue (10% v/v) was added and fluorescence was measured 3 h later with a fluorescence plate reader with excitation and emission at 560 and 590 nm, respectively.
For assessing the effect of miR-203 inhibitor on the ability of ING1b to inhibit cell proliferation, U2OS cells were simultaneously transfected with either miR-203 inhibitor or miRNA inhibitor negative control and empty vector or ING1b plasmids. Cell proliferation was then evaluated 48 h later using the Alamar Blue method. A similar experiment was repeated in Hs68 cells. Hs68 cells were transfected with miR-203 inhibitor or miRNA inhibitor negative control for 6 h, followed by infecting with GFP alone or with GFP-ING1b adenovirus. Cell proliferation was evaluated by the Alamar Blue method 48 h later. Lysates were harvested for western blot analysis.
Chromatin immunoprecipitation assay (ChIP)
ING1b binding to the promoter of miRNA203 was tested using ChIP analysis as described previously (Yan et al, 2004). Briefly, about 3 × 106 Hs68 fibroblasts, infected with either Ad-GFP or Ad-GFP-ING1b adenoviruses were crosslinked using 1% formaldehyde (Sigma, St Louis, MO, USA) for 15 min at 37 °C. Cells were harvested after quenching with 0.125 M glycine and lysed in ChIP lysis buffer (150 mM NaCl, 50 mM Tris (pH 8.0), 1% Triton X-100, 0.1% deoxycholate, 1 mM EDTA, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml pepstatin and 1 μg/ml leupeptin). Extracts were sonicated eight times for 10 s each and lysates were clarified by centrifugation at 13 000 rpm for 15 min at 4 °C. Of this sample, 100 μl was used as input. The clarified supernatants were immunoprecipiated with either α-ING1 or with mouse IgG (negative control) at 4 °C for 3 h, followed by protein G Sepharose (GE healthcare, New Haven, CT, USA) for 1 h at 4 °C. The immunoprecipitates were sequentially washed with 1 ml of ChIP lysis buffer twice, ChIP lysis buffer with 500 mM NaCl twice and with LiCl/detergent solution (10 mM Tris-HCl, pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA) twice and finally with TE buffer (10 mM Tris and 1 mM EDTA, pH 8.0). The beads were eluted using 1% SDS and 0.1 M sodium bicarbonate solution. The eluent and the input samples were reverse-crosslinked using NaCl for 6 h at 65 °C. The DNA from the samples was isolated by phenol–chloroform, followed by ethanol precipitation. Promoter binding was tested by PCRpolymerase chain reaction using primers spanning the upstream regions of miRNA203 and miRNA34a start sites, sequences retrieved from UCSC genome browser (primer sequences available upon request).
Western blot analysis
U2OS cells were harvested 48 h after transfection with expression constructs or miRNAs using Lipofectamine 2000, followed by protein sample preparation by boiling in Laemmli sample buffer. Equal amounts of protein from each sample (20 μg) were separated by 10% SDS–PAGE and transferred to nitrocellulose membranes. Different primary antibodies were used to detect their respective proteins. The signals were developed using an ECL detection kit (Amersham Canada, Oakville, ON, Canada) after incubation with appropriate horseradish peroxidase-coupled secondary antibodies.
Data analysis
The mean±standard error of mean (s.e.m.) were used for data presentation. Differences were evaluated by Student’s t-test to estimate statistical significance when two groups were compared, and by ANOVA when three or more groups were compared. A value of P<0.05 was considered to be significant.
Results
ING1b affects the levels of a subset of miRNAs
To identify miRNAs regulated by ING1b, Hs68 fibroblasts were infected with adenovirus expressing GFP alone or with adenovirus expressing both GFP and ING1b under separate promoters. The expression level of ING1b is shown in Supplementary Figure 1A. MicroRNAs were isolated after 48 h of virus treatment and used to probe a TaqMan Array Human MicroRNA Card Set v3.0, enabling quantitation of 754 human mature microRNAs. We found that 65 miRNAs reproducibly increased more than two-fold and 43 miRNAs decreased more than two-fold compared with Hs68 cells infected with GFP control (Supplementary Table 1). Several miRNAs chosen on the basis of previously determined functions related to cell proliferation were further examined by real-time PCR. Of these, miR-203, miR-375, miR-449b and miR-200c were confirmed to increase significantly and reproducibly in response to ING1b (Figure 1).
ING1b regulates miR-203 expression
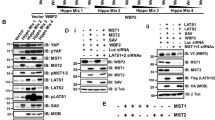
MiR-203 was chosen for further study based upon several criteria. The mRNAs regulated by ING1b were determined using microarray after overexpression of ING1b (Tran et al, manuscript in preparation). When comparing this list with the list of miRNAs predicted to be targets of miR-203 using several miRNA target prediction software packages, we found that ∼25% of the mRNAs downregulated by ING1b were predicted to be target genes of miR-203 (301 of 1199 genes; Figure 2A). As miRNAs have repressive effects on gene expression, only the mRNAs downregulated by ING1b were used for the comparison. The identities of these 301 genes are shown in Supplementary Table 2 and they are described in more detail in the functional annotations shown in Supplementary Table 3. In addition, as shown in Figure 2B, suberoylanilide hydroxamic acid (SAHA), a histone deacetylase (HDAC) inhibitor, also increased miR-203 expression significantly in normal primary Hs68 fibroblasts, which is consistent with epigenetic regulation by ING1b as part of HDAC1 and HDAC2 complexes. ChIP analysis of ING1b showed that ING1b bound to the promoter area of miR203 (Figure 2C), indicating that ING1b directly regulated miR203. Thus, we focused on miR-203 in subsequent investigations.
ING1b and miR-203 gene targets. (A) Combining transcriptional repression targets of ING1b from microarray data with predicted mRNA targets of miR-203 showed that ∼25% of ING1b target genes were also predicted to be targets of miR-203. (B) miR-203 level was determined using real-time RT-PCR in Hs68 cells in response to treatment with the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA). Bars represent the mean±s.e.m (*P<0.05, n=4). (C) Cells infected with control GFP adenovirus or with the same virus also encoding ING1b were harvested 24 h later and prepared for ChIP analysis using control or ING1 antibodies. PCR of immunoprecipitation products showed that ING1 bound to the upstream region of mir-203, but not to miR-34a.
MicroRNA 203 inhibits the proliferation of different cancer cell lines
Overexpression of ING1b inhibits cell proliferation and causes G1/S arrest (Garkavtsev et al, 1996), but the molecular mechanism(s) responsible has not yet been fully elucidated. To investigate whether miR-203 might contribute to the growth inhibition by ING1b, the effect of miR-203 on U2OS and MDA-MB-231 cell proliferation was evaluated. Compared with cells treated with control miRNA, cells treated with miR-203 showed significantly reduced cell growth (Figure 3A and B). Expression levels of miR-203 are shown in Supplementary Figure 1B. To further investigate the mechanism by which miR-203 inhibited cell proliferation, we performed cell cycle analysis using flow cytometry. As shown in Figure 3C, a larger proportion of U2OS cells transfected with mir-203 were arrested in G1 phase, compared with cells transfected with control miRNA. Graphs representative of each condition are shown in Supplementary Figures 2A and 2B. Transfection with miR-203 inhibitor, but not with the miR inhibitor control, significantly reversed the inhibition of proliferation induced by ING1b (Figure 3D), suggesting that miR-203 functions downstream of ING1b to regulate cell proliferation in U2OS cells. A similar result was found in HS68 cells (Supplementary Figure 3A). In addition, ING1b overexpression reduced c-Abl protein level, whereas miR-203 inhibitor could partially reverse the reduction in c-Abl protein level (Supplementary Figure 3B).
Effects of miR-203 on cell proliferation. (A) U2OS and (B) MDA-MB-231 cells were transfected with miR-203 or miRNA negative control and cell proliferation was evaluated using the Alamar Blue method 48 h after transfection (*P<0.05; **P<0.01, n=3). (C) Overexpression of miR-203 increases the proportion of cells in G1 as estimated by using flow cytometry (**P<0.01, n=3). (D) U2OS cells were simultaneously transfected with either miR-203 inhibitor or miRNA inhibitor negative control and empty vector or ING1b plasmids. Cell proliferation was then evaluated 48 h later using the Alamar Blue method (** indicates P<0.01, n=3).
Cell cycle regulators are target genes of miR-203 in U2OS cells
To investigate the molecular mechanism by which miR-203 inhibited cell proliferation, target genes of miR-203 were further examined. Among validated target genes of miR-203 that had the potential to affect cell proliferation, we examined CDK6, c-Abl, Src and AKT2 protein levels. Western blot analysis showed that endogenous levels of CDK6, c-Abl and Src were decreased significantly and reproducibly by miR-203 in U2OS cells (Figure 4A–C), whereas the expression of AKT2 was largely unaffected (Figure 4D). These results suggested that the inhibitory effects of miR-203 on cell proliferation might be through regulating the expression of CDK6, c-Abl and Src, which can act to inhibit growth at the G1/S phase of the cell cycle.
Gene expression in response to miR-203. Relative expression of (A) CDK6, (B) Src, (C) c-Abl and (D) Akt2 proteins in response to miR-203 in U2OS cells. Lysates harvested 48 h after transfection of miR-203 or control miRNA were boiled in Laemmli buffer, electrophoresed and blotted with the antibodies indicated. Actin was used as a loading and blotting control.
Discussion
In this study, we have shown that ING1b affects the expression of a significant number of miRNAs. Approximately 25% of the mRNAs downregulated by ING1b are also predicted to be target genes of miR-203 using several prediction programs. This microRNA has been recently reported to repress the proliferation of several different cancer cell lines in independent studies (Li et al, 2011; Saini et al, 2011).
Previous studies have shown that overexpression of ING1b can result in increased global levels of acetylated histones H3 and H4 (Vieyra et al, 2002), which are chromatin marks associated with active gene transcription. This study showed that consistent with an increase in acetylated histone marks, ING1b overexpression led to miR-203 induction. Furthermore, we demonstrated that the inhibition of histone deacetylases using SAHA also significantly increased miR-203 expression (Figure 2B), whereas ChIP assays showed that ING1b bound to the promoter of miR-203 (Figure 2C). Recently, several reports have indicated that the expression of many miRNAs can be regulated by histone acetylation and DNA methylation (Sato. et al, 2011). Therefore, miRNAs might be epigenetic targets of ING1b, and thus, they could be mediating ING1b’s downstream functions. In our miRNA PCR array, we found that overexpression of ING1b caused 65 miRNAs to increase and 43 miRNAs to decrease when compared with control cells. The expression of a subset of these miRNAs was validated using quantitative real-time RT-PCR. Previously, an independent study looking at ING1b-regulated miRNAs using a miRNA microarray approach found that only seven miRNAs were differentially regulated in mouse embryonic fibroblasts (Gómez-Cabello et al, 2010). In this study, ING1 knockout cells were compared with those expressing physiological levels of ING1. In contrast, we have chosen to amplify the signals regulated by ING1 and to use a more sensitive miRNA PCR array approach to determine what miRNAs might be able to transduce the apoptotic signal initiated upon ING1 overexpression (Helbing et al, 1997). Thus, the discrepancy between their result and ours could be attributed to a variety of factors, including the differences in our chosen methods and experimental conditions. For example, compared with the common miRNA microarray, miRNA PCR array provides a larger dynamic range of microRNA gene expression, typically from three to seven logs. Perhaps more importantly, miRNA PCR array detects only mature miRNAs that directly perform the functions of miRNAs, instead of both precursors and mature miRNAs as in the miRNA microarray method (Chen et al, 2009). It is also possible that although ING1b directly binds upstream of miR-203, the effects upon miRNA expression may be partially mediated through p53. As ING1b can stabilise p53 through prevention of its proteosomal-mediated degradation (Thalappilly et al, 2011), overexpression of ING1b would be expected to stabilise p53, resulting in a possible synergistic effect of these tumour suppressors upon miRNA expression.
Both miR-203 and ING1b are evolutionally conserved across several species and can have essential functions in various cancer types (Soliman Riabowol, 2007; Craig et al, 2011; and Li et al, 2011). We found that there was a significant overlap in the number of mRNAs regulated by ING1b and those that are predicted to be targets of miR-203. This finding suggests that certain biological functions of ING1b may be mediated by miR-203. We and others (Li et al, 2011; Saini et al, 2011) have found that miR-203 inhibits the proliferation of various cancer cell lines including U2OS, MDA-MB-231 and bladder cancer cell lines (Saini et al, 2011). In bladder cancer cell lines, miR-203 decreased proliferation and increased apoptosis by targeting Akt2 and Src (Saini et al, 2011). In addition, miR-203 was reported to inhibit growth in human colon (Li et al, 2011) and lymphoma (Craig et al, 2011) cell lines. These observations are consistent with ING1b being downregulated in several types of cancer cells (Soliman and Riabowol, 2007). Finally, as shown in Supplementary Table 3, among the 301 genes which were downregulated by ING1b and also predicted to be targets genes of miR-203, the most relevant gene pathway was cancer, which further strengthens links between ING1b and miR-203.
Our understanding of the biological functions of microRNA relies on the identification of relevant miRNA target genes. However, the prediction of mammalian miRNA targets is still a challenging research area. In mammalian cells, the majority of the miRNA-target sequence matching is based on imperfect complementarity of the miRNA with the 3′-UTRs of target mRNAs. The selection of mRNA targets is largely determined by the seed sequence of the miRNA, which consists of nucleotides two to eight at the 5′-end of the microRNA. However, the requirement for complementarity of only seven or eight nucleotides could result in hundreds of possible targets (Lim et al, 2005). Thus, further experimentation is required to validate the individual microRNA targets. To date, only a few target genes of miR-203 have been confirmed in different biological systems. In this study, we examined four proliferation-related genes previously reported to be direct targets of miR-203 including AKT-2, c-Abl, CDK6 and Src (Bueno et al, 2008; Furuta et al, 2010; Saini et al, 2011). CDK6, c-Abl and Src were found to be downregulated by miR-203 in U2OS cells, an osteosarcoma cell line. This is consistent with the concept that target genes of miRNAs might have tissue specificity (Chen et al, 2011). CDK6 and Src have key roles in the G1/S phase of the cell cycle. As expected, overexpression of miR-203, which can inhibit CDK6 and Src gene expression, caused G1 arrest. Thus, a potential mechanism for ING1b-mediated G1 cell cycle arrest could involve the induction of miR-203 and subsequent silencing of CDK6 and Src.
In summary, we report the novel observation that miR-203 is epigenetically regulated by ING1b. Our data shows that miR-203 contributes to the inhibition of cell proliferation through downregulation of CDK6, c-Abl and Src. MiR-203 might also mediate the tumour-suppressive functions of ING1b, and thus have potential therapeutic value in the future treatment of osteosarcomas and other cancers.
Change history
19 March 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernández-Piqueras J, Malumbres M (2008) Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 13: 496–506
Chen J, Yin H, Jiang Y, Radhakrishnan SK, Huang ZP, Li J, Shi Z, Kilsdonk EP, Gui Y, Wang DZ, Zheng XL (2011) Induction of microRNA-1 by myocardin in smooth muscle cells inhibits cell proliferation. Arterioscler Thromb Vasc Biol 31: 368–375
Chen Y, Gelfond JA, McManus LM, Shireman PK (2009) Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics 10: 407
Chen HY, Han ZB, Fan JW, Xia J, Wu JY, Qiu GQ, Tang HM, Peng ZH (2011) miR-203 expression predicts outcome after liver transplantation for hepatocellular carcinoma in cirrhotic liver. Med Oncol 29 (3): 1859–1865
Craig VJ, Cogliatti SB, Rehrauer H, Wündisch T, Müller A (2011) Epigenetic silencing of microRNA-203 dysregulates ABL1 expression and drives Helicobacter-associated gastric lymphomagenesis. Cancer Res 71: 3616–3624
Coles AH, Liang H, Zhu Z, Marfella CG, Kang J, Imbalzano AN, Jones SN (2007) Deletion of p37Ing1 in mice reveals a p53-independent role for Ing1 in the suppression of cell proliferation, apoptosis, and tumorigenesis. Cancer Res 67: 2054–2061
Doyon Y, Cayrou C, Ullah M, Landry AJ, Côté V, Selleck W, Lane WS, Tan S, Yang XJ, Côté J (2006) ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell 21: 51–64
Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S (2008) LNA-mediated microRNA silencing in non-human primates. Nature 452: 896–899
Feng X, Bonnie S, Riabowol K (2006) Induction of HSP70 by ING Proteins Sensitizes Cells to TNFa-mediated Apoptosis. Mol Cell Biol 26: 9244–9255
Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J (2010) miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 31: 766–776
Garkavtsev I, Kazarov A, Gudkov A, Riabowol K (1996) Suppression of novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet 14: 415–420
Gómez-Cabello D, Callejas S, Benguría A, Moreno A, Alonso J, Palmero I (2010) Regulation of the microRNA processor DGCR8 by the tumor suppressor ING1. Cancer Res 70: 1866–1874
Helbing CC, Veillette C, Riabowol K, Johnston RN, Garkavtsev I (1997) A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res 57: 1255–1258
He GH, Helbing CC, Wagner MJ, Sensen CW, Riabowol K (2005) Phylogenetic analysis of the ING family of PHD finger proteins. Mol. Biol. Evol. 22: 104–116
Li J, Chen Y, Zhao J, Kong F, Zhang Y (2011) miR-203 reverses chemoresistance in p53-mutated colon cancer cells through downregulation of Akt2 expression. Cancer Lett 304: 52–59
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773
Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D (2000) Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Biol 20: 3807–3816
Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L (2006) The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol 26: 7871–7879
Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG (2006) Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442: 100–103
Russell M, Berardi P, Gong W, Riabowol K (2006) Grow-ING, Age-ING and Die-ING: ING proteins link cancer, senescence and aging. Exp Cell Res 312: 951–961
Saini S, Arora S, Majid S, Shahryari V, Chen Y, Deng G, Yamamura S, Ueno K, Dahiya R (2011) Curcumin modulates microRNA-203-mediated regulation of the Src-Akt axis in bladder cancer. Cancer Prev Res (Phila) 4: 1698–1709
Sato F, Tsuchiya S, Meltzer SJ, Shimizu K (2011) MicroRNAs and epigenetics. FEBS J 278: 1598–1609
Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Côté J, Chua KF, Gozani O (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99
Soliman MA, Riabowol K (2007) After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci 32: 509–519
Thalappilly S, Feng X, Pastyryeva S, Suzuki K, Muruve D, Larocque D, Richard S, Truss M, von Deimling A, Riabowol K, Tallen G (2011) The p53 tumor suppressor is stabilized by inhibitor of growth 1 (ING1) by blocking polyubiquitination. PLoS One 6: e21065
Yan Y, Chen H, Costa M (2004) Chromatin immunoprecipitation assays. Methods Mol Biol 287: 9–19
Vieyra D, Loewith R, Scott M, Bonnefin P, Boisvert FM, Cheema P, Pastyryeva S, Meijer M, Johnston RN, Bazett-Jones DP, McMahon S, Cole MD, Young D, Riabowol K (2002) Human ING1 proteins differentially regulate histone acetylation. J Biol Chem 277: 29832–29839
Acknowledgements
This work was supported by grants to KR from the Canadian Institutes of Health Research and Alberta Innovates-Health Solutions. KR and JC are recipients of AHFMR Scientist and Fellowship awards, respectively and UT holds an Alberta Cancer Foundation Studentship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies the paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Chen, J., Tran, U., Rajarajacholan, U. et al. ING1b-inducible microRNA203 inhibits cell proliferation. Br J Cancer 108, 1143–1148 (2013). https://doi.org/10.1038/bjc.2013.50
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.50
Keywords
This article is cited by
-
INGs are potential drug targets for cancer
Journal of Cancer Research and Clinical Oncology (2017)