Abstract
The phylogeography of the freshwater snail Biomphalaria glabrata remains poorly known, although this species is the major vector of schistosomiasis in the New World. It was here investigated in South America and the Lesser Antilles, based on partial mitochondrial large ribosomal subunit (16S rDNA) and nuclear internal transcribed spacer-2 (ITS-2) gene sequences. Sampling included 17 populations from a large part of the current geographic range of the species (Brazil, Venezuela and Lesser Antilles). Substantial variability was detected, as well as a high amount of phylogenetically informative signal. The molecular phylogeny inferred splits B. glabrata into Northern and Southern clades separated by the Amazon river, and may even suggest a supra-specific status for B. glabrata. Brazilian populations were the most diverse and appeared basal to the other populations. Venezuelan haplotypes formed a single clade, albeit not strongly supported. Two Venezuelan haplotypes appear rather similar to Brazilian haplotypes. Similarly, Lesser Antilles haplotypes clustered in the same monophyletic clade, which suggests that the recent colonisation of the Antilles has a northern South American origin. However, the estimated divergence time between Antilles and Venezuelan sequences is extremely large (conservatively higher than 105 years). These results are discussed in the light of (i) phylogeographic patterns at South American scale, and (ii) recurrent introduction of molluscs, especially in the Antilles, as a consequence of human activities.
Similar content being viewed by others
Introduction
The distribution of genetic diversity within species depends not only on currently acting forces (eg, migration), but also on species history. This is at the heart of the phylogeographic approach (Avise, 2000; Hewitt, 2001). In the New World, most phylogeographic studies have focused on North American species (Avise, 2000), but South American species are attracting increasing interest (Dutech et al, 2000; Moritz et al, 2000). In this context, there has been much interest in the so-called refuge hypothesis, that postulates the existence of forest refuges in the Amazon basin during the glaciated period of the Pleistocene (Haffer, 1997). This should have had an influence on population isolation and genetic divergence. However, the hypothesis holds only for forest-dwelling species, while species occupying the forest margins, whether terrestrial or freshwater, should have a different history: more habitats were available when the forest retreated, while inter-glaciation periods corresponded to decreased, potentially more patchy, distributions. In this geographic area, the Amazon river also constitutes an important barrier to gene flow for a large number of freshwater species (eg, snails). Human activities are a further aspect to consider when analysing genetic diversity at large geographic scales, since they deeply influence the current distribution of species (review in Williamson, 1997; eg, Meunier et al, 2001).
We focus here on the phylogeography of an invertebrate species with limited dispersal abilities and confined to disjunct freshwater bodies, the snail Biomphalaria glabrata. Besides adding to the literature on phylogeographic patterns in invertebrate freshwater species (eg, cladocerans, Crease et al, 1990; bryozoans, Freeland et al, 2000; snails, Viard et al, 1997), it allows a test for the role of the factors mentioned above, that is the extension of the equatorial forest, the Amazon, and human activities on phylogeographic patterns in the neotropics. Moreover, B. glabrata usually occupies fragmented habitats, such as ponds or small rivers, which can be submitted to temporal variations in water availability and even complete dryness or catastrophic floodings. These events can lead to demographic bottlenecks and even local population extinction (see for example, Pointier et al, 1993). Both habitat discontinuity and demographic instability may result in high genetic differentiation among populations (see McCauley, 1991). This is what previous studies have indicated in B. glabrata, whether analysing morphological (Malek, 1985) or genetic variation (eg, Mulvey et al, 1988; Vidigal et al, 1994; DeJong et al, 2001; Mavárez et al, 2002a,b). A further important point is that, although this species is the major intermediate host of the parasite Schistosoma mansoni in the neotropics, its phylogeography at large geographic scales has not been studied.
Our work is based on sequence data (one mitochondrial and one nuclear gene) obtained from populations sampled at the scale of the current geographic distribution of the species (South America and the Caribbean islands), and aims at reconstructing the phylogeography of B. glabrata. Note that a similar analysis has been performed using much more variable microsatellite markers, and results are presented in a companion paper (Mavárez et al, 2002b). Based on the standing variation within regions (Brazil, Venezuela and Lesser Antilles) and the genetic divergence among populations (haplotypes), we were interested in evaluating the following aspects or processes. (i) It has been suggested that what is known as B. glabrata might indeed constitute a species complex (Bandoni et al, 1995; Woodruff and Mulvey, 1997). DNA data might help in addressing this question. (ii) We mentioned above that the equatorial forest of the Amazon basin and the Amazon river itself constitute barriers to gene flow. The expectation is a phylogenetic split between groups of populations north and south of the Amazon. The historical variation in the extension of the Amazon, as well as the patchy distribution of population and extinction-colonisation cycles, should also limit the genetic variation. (iii) Given the limited dispersal abilities of freshwater snails, a pattern of isolation-by-distance is expected. However this should be counterbalanced by introductions through human activities, especially in the Antilles. (iv) The simpler explanation for the colonisation of the Lesser Antilles is that source populations are (or were) located in northern South America. We tried to locate these source populations, and more generally to reconstruct the colonisation history.
Materials and methods
Sample collection, DNA extraction, amplification and sequencing
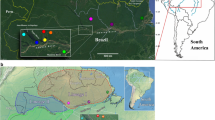
One individual was analysed from each of three B. glabrata populations from Brazil, seven from Venezuela and five from the Lesser Antilles (one from Martinique, three from the Guadeloupe archipelago and one from Saint-Lucia; Figure 1 and Table 1). However, both the VE13 and VE6 localities in Venezuela seem to be composed of two groups of widely different multilocus microsatellite genotypes (Mavarez et al, 2002b). Thus, two individuals (one from each group) were analysed from both localities. Individuals were collected by hand or with a lending net, killed by immersion (30 sec) in 80°C water and preserved in 96% ethanol until genetic analysis. The formula DAB = R . Arccos {sin(π/2 − LA) . sin(π/2 − LB) . cos(lA − lB) + cos(π/2 − LA) . cos(π/2 − LB)}, where R is the radius of the earth, LA and LB the latitudes and lA and lB the longitudes of localities A and B in radians, was used to calculate the geographic distances between both localities.
Map of South America and the Lesser Antilles showing the localities sampled. The current geographic distribution of Biomphalaria glabrata is shown in grey. The tree represents ML (right bootstrap values) and MP (left bootstrap values) analysis of concatenated 16S and ITS-2 genes (943 nt). Only bootstrap values higher than 50 are shown.
Total genomic DNA was extracted from foot muscle using the QIAmp tissue kit (Qiagen). The polymerase chain reaction (PCR) was used to amplify the 5′ fragment of the mitochondrial 16S rRNA gene (422 bp) and the complete sequence of the nuclear ITS-2 rRNA (521 bp) using the conserved primers described by Palumbi (1996). Both strands of each gene were sequenced using an automatic sequencer (ABI 310 Perkin Elmer) with big dye terminator cycle sequencing (Applied Biosystems).
Sequence analysis
Sequence alignment was performed using the MUST 2000 software (Philippe, 1993). The total number of substitutions (transitions and transvertions) and pairwise Kimura 2-parameter distance (K2p; Kimura, 1980) were calculated for single genes and for concatenated sequences in MUST 2000 (Philippe, 1993). The relationship between genetic and geographical distances was evaluated with a Mantel-like procedure (1000 permutations) using the GENETIX 4.02 package (Belkhir et al, 2000).
The phylogenetic relationships among taxa were analysed though maximum parsimony (MP) and distance (K2p) methods using PAUP version 4.0b8 (Swofford, 1998). Indels were not considered in the phylogenetic analysis. MP trees were constructed using heuristic searches with 100 random orders of taxa with tree bisection-reconnection (TBR) branch swapping. Distance trees (gamma corrected) were calculated from the general time-reversible model (GTR) using the neighbour-joining algorithm. The shape parameter of the gamma distribution, α, was estimated using maximum likelihood methods (Kishino and Hasegawa, 1989). When multiple minimum-length trees were found, both strict and 50% majority-rule consensus trees were obtained. The strength of internal nodes was estimated by bootstrap analysis, with 500 replicates (Felsenstein, 1985). ITS-2 and 16S gene sequences from an individual B. tenagophila from Contagem (Brazil, GenBank AF449614, AF449615) and available sequences from B. pfeifferi were used as outgroups.
16S and ITS-2 gene sequences were also analysed simultaneously (ie, a single concatenated sequence for each individual) using MP and maximum likelihood methods (ML) in order to increase the number of parsimony-informative sites and the power of phylogenetic reconstructions. ML analyses were conducted in two steps in order to accelerate computation times. First, ML parameters were estimated using a heuristic search with nearest-neighbour interchange (NNI) branch swapping on a neighbour-joining starting tree. Second, a new search was conducted with random addition of sequences and TBR branch swapping using the previously estimated parameters. Substitution rates were described from GTR models of sequence evolution for nucleotides. The heterogeneity in substitution rate among DNA sites was described with a gamma distribution including eight categories (Yang, 1996). The strength of internal nodes was estimated by bootstrap analysis, with 200 replicates.
Results
Sequence variation
Table 2 shows a summary of molecular diversity when the two genes were concatenated (943 bp). Forty-four sites were variable and 14 parsimony-informative (GenBank AF449580-AF449613). The pairwise number of substitutions was large and ranged from 0 to 2.84% in the 16S gene and from 0 to 3.84% in the ITS-2 gene. All individuals from the Valencia lake basin in Venezuela (VE2, VE4, VE8 and VE10) and some from the Lesser Antilles (MA1, GU1 and GU3) shared the same 16S haplotype, respectively (MA1 and GU1 are in fact identical for both genes). Within regions, mean pairwise number of substitutions were higher in Brazil (16S: 1.26%, ITS-2: 1.28%) than in Venezuela (16S: 0.55%, ITS-2: 1.21%) and the Antilles (16S: 0.52%, ITS-2: 0.52%) (Table 2a), the difference being statistically significant (Kruskal-Wallis test on concatenated variation, n = 49, P = 0.017). However, the higher values within Brazil were largely due to the BR2 haplotype (Colibri), which seems relatively divergent, and much lower values were obtained when this haplotype was discarded (16S: 0.47%, ITS-2: 0.77%). The mean pairwise number of substitutions between regions were much higher for Brazil/Venezuela (16S: 2.06%, ITS-2: 1.86%) and Brazil/Antilles (16S: 2.26%, ITS-2: 1.00%) than for Venezuela/Antilles (16S: 0.79%, ITS-2: 1.64%) (Table 2b), the difference being statistically significant (Kruskal-Wallis test on concatenated variation, n = 89, P < 0.001). Pairwise K2p distances showed very similar patterns (data not shown). In agreement with these results, a correlation was detected between pairwise genetic and geographical distances (P < 0.001 for both K2p and number of substitutions).
Phylogenetic relationships among sequences
The MP reconstruction using either the 16S or ITS-2 sequences gave essentially the same results. The phylogenetic signal seems to be additive, as the analysis of each data set shows that there were few strongly supported conflicts between them. Results from the two genes were combined as it generally improved phylogenetic resolution (ie, higher consistency index and bootstrap values).
A ML tree (-ln likelihood = 2114.17716) is given in Figure 1. Three major lineages were revealed (A, B and C; Figure 1). Lineages A and B include only Brazilian haplotypes, which appear ancestral and paraphyletic in our trees. Lineage C included all the Venezuela and Lesser Antilles haplotypes, suggesting a common origin for both regions. The introduction of B. glabrata in Saint-Lucia seems to be older than in the French Antilles. The MP analysis tree (522 steps, consistency index = 0.9272) provided exactly the same results, with only minor differences concerning the robustness of tree nodes (Figure 1).
Our phylogenetic analyses showed that localities VE6 and VE13 both harboured two highly divergent haplotypes (16S: 1.18% and 0.78%, ITS-2: 1.15% and 1.54%, respectively). ITS-2 haplotypes VE6b and VE13a appear more similar to Brazilian haplotypes than to the other Venezuelan ones (Figure 2). For example, they share a six-nucleotide insertion with haplotype BR1, which is absent from other haplotypes in Venezuela and the Antilles. In addition, the other Venezuelan haplotypes share a characteristic three-nucleotide insertion absent in the VE6b and VE13a haplotypes.
The phylogenetic analyses based on distances (single and concatenated sequences) gave completely different and somewhat incoherent results (trees not shown). This is most probably due to the low level of variability in our dataset.
Discussion
Our study is the first investigation of intra-specific phylogenetic relationships in B. glabrata, and one of the first in freshwater snails based on DNA sequence variation (see Angers et al, submitted). The results showed that the phylogenetic relationships among haplotypes (16S and ITS-2) are consistent with inferences based on biogeography and the present distribution of the species. The discussion below focus more specifically on the questions outlined in the Introduction.
The systematic status of B. glabrata
Four interesting results can be retained here: (i) the estimated level of molecular divergence for nuclear and mitochondrial genes is relatively high as compared with other Biomphalaria species, especially when considering Brazilian haplotypes (see DeJong et al, 2001 for comparison). Although no previous study made use of DNA sequence variation, this is consistent with results based on other markers which suggest that levels of genetic variation in B. glabrata are particularly high (Bandoni et al, 1995; Woodruff and Mulvey, 1997; Mavárez et al, 2002a,b). (ii) The Brazilian group is paraphyletic and the BR2 haplotype (Colibri) appears extremely divergent from the rest. The mean number of substitutions between BR2 and other haplotypes was 2.16% (16S) and 1.96% (ITS-2), and 1.13% (16S) and 1.38% (ITS-2) among other haplotypes. In addition, haplotype BR2 appears basal to the other B. glabrata haplotypes (see Figure 1). (iii) The phylogenetic reconstructions show the existence of three major clades. Brazilian haplotypes appear ancestral and harbour a large amount of the current molecular diversity of B. glabrata. (iv) The degree of pairwise differentiation was related to the distance among localities, which confirm our previous analyses using microsatellite markers. Thus, isolation by distance seems to be significant at both wide and restricted geographic scales (Mavárez et al, 2002a,b).
These results suggest that B. glabrata might constitute a species complex, as already suggested on the basis of allozymic data (Bandoni et al, 1995; Woodruff and Mulvey, 1997), and on the basis of reproductive system (Mavárez et al, 2002b; see below). B. glabrata indeed displays a wide range of anatomical and morphological (shell) variation, and some variants have even been described as nominal species (see Paraense, 1961). However, the presence of a renal ridge has been considered as a sufficient diagnostic character of B. glabrata (Paraense and Deslandes, 1959). On the other hand, DeJong et al (2001) suggested that B. glabrata is a single species on the basis of 16S, ITS1 and ITS-2 sequence variation. However, these authors studied only four individuals, including only one from Brazil, and they certainly failed to detect most of the variation present in this country. For example, when the BR2 haplotype is excluded from our analysis, the molecular divergence is only slightly higher than that reported by DeJong et al (2001). This haplotype certainly deserves further attention, and the taxonomical status of B. glabrata might have to be reconsidered.
Biomphalaria glabrata is currently distributed in South America on both sides of the Amazon river. Our results suggest that this has probably been so for some time, and that the Amazon is a barrier to gene flow. Moreover the equatorial forest of the Amazon basin does not provide appropriate habitats for B. glabrata, which constitute a further barrier to gene flow (Paraense, 1983). It is therefore likely that the repeated cycles of forest fragmentation and expansion in the Amazon basin due to Pleistocene glaciations strongly affected the distribution of this species, and hence its population differentiation. Periods of population differentiation might have coincided with times of large forest coverage (ie, relatively humid periods between glaciations), while drier periods might have offered more favourable habitats. This is somewhat in opposition with the common ‘refuge-based’ view of speciation and differentiation in the Amazon basin (Moritz et al, 2000). A similar scenario has been previously proposed for other non-forest dwelling organisms, such as teiid lizards (Cole and Dessauer, 1993).
The northern clade
Our study revealed the existence of a Venezuela + Lesser Antilles clade showing lower levels of sequence divergence than the Brazilian clades. As expected, the Venezuelan haplotypes show higher values of sequence divergence than the Lesser Antilles ones. Moreover, the Venezuelan haplotypes appear clearly divided into two well-differentiated groups of: (a) relatively variable haplotypes (VE1, 2, 4, 6a, 8, 10 and 13b; Figure 1) that also exhibit a highly diverse array of multilocus microsatellite genotypes, and (b) two almost identical haplotypes (VE6b and 13a; Figure 1), exhibiting the same multilocus microsatellite genotype (Mavárez et al, 2002b). These two haplotypes are more closely related to Brazilian than to Venezuelan ITS-2 haplotypes, while the reverse holds for the 16S gene. This constitutes the most evident case of conflict between the two genes in our dataset. Note that VE6b and VE13a are also genetically closer to BR1 individuals than to other Venezuelan individuals, when using microsatellite (nuclear) markers (Mavarez et al, 2002b). A possible explanation to such a nuclear-mitochondrial conflict involves interspecific (or inter-clade) mitochondrial gene flow via hybridisation, which is common in animals (Barton and Hewitt, 1985), and has been documented in Biomphalaria (Mello-Silva et al, 1998; Kristensen et al, 1999). However it is also possible that the 16S gene tree is poorly resolved, due to a lower evolutionary rate.
The occurrence of these two particular haplotypes in Venezuela is somewhat amazing, because they presently occur at two sites separated by about 250 km. A first possibility is that these haplotypes have been recently introduced in Venezuela. There are well documented reports for the recent arrival of B. glabrata into Caripe, eastern Venezuela (Chrosciechowski and Arcas, 1986) and Haiti (Raccurt et al, 1985). However, the VE6 and VE13 sites are artificial ponds located in rural and mountainous areas with difficult access. Another possibility is that these haplotypes belong to a lineage that was already present in Venezuela, and more widespread, before the arrival of other haplotypes which are now distributed in the whole country.
The Lesser Antilles
The Antilles sequences form a monophyletic clade with low divergence. Haplotypes GU1, GU3 and MA1 show no variation at all (16S) or just one substitution (ITS-2). This suggests a very recent common origin for these populations: the colonisation of these islands might have occurred from the same source population, or colonisation took place from island to island. The analyses also suggest an older status for the Saint-Lucia haplotype. A difference in colonisation history of Saint-Lucia and the French Antilles by B. glabrata (through human activities) is not unexpected given that these islands were respectively colonised by Great Britain and France. However, the mean molecular divergences between the Lesser Antilles and Venezuela (the most probable source population) are 0.76 (16S) and 1.64 (ITS-2). Assuming a standard value of 2–4% animal mtDNA divergence per My, the haplotypes from the Lesser Antilles and Venezuela are separated by 1.9 − 3.8×105 years, which greatly antedates the occupation of the New World by human populations. Even calibrating with the exceptionally high rates reported for some mitochondrial DNA genes in molluscs (Hoeh et al, 1996; 10% per My, Chiba, 1999) provides a lineage split tens of thousands years ago. Two scenarios might then be considered for explaining the colonisation of these islands: (i) an ancient introduction, probably involving aquatic birds (see Madsen and Frandsen, 1989) or (ii) a recent introduction of already divergent lineages.
The colonisation of the Lesser Antilles by man is relatively recent (6000 years BP) and the deforestation of these islands, linked to the introduction of the sugar cane and banana cultures, is more recent (400–500 years BP). These agricultural practices were responsible for the creation of numerous artificial habitats such as sugar cane canals, irrigation ponds or banana drains which are highly favourable sites for B. glabrata (Golvan et al, 1981; Jordan, 1985). Before the economic development of the islands, the aquatic habitats were mainly represented by forest streams and rivers which are largely unsuitable biotopes for B. glabrata (Golvan et al, 1981). This makes scenario (i) rather unlikely. On the other hand, numerous well-documented introductions of freshwater snails into the Caribbean area have been reported over the last few decades, showing that the invasion process are not only underway, but also strongly increasing (see review in Pointier, 1999). This gives some credence to scenario (ii). Moreover, the recent introduction of an already divergent haplotype in Caripe (see Table 1; JPP, unpublished data) goes in the same direction. However, our sample does not include the Venezuelan haplotypes that were the source (or are closely related to) of colonisation of the Antilles. There are two possible explanations: either we failed to sample this source population (eg, no samples from the Guiana region were available), or it does not exist anymore in Venezuela. More analyses will be necessary before we can chose between these alternatives.
References
Angers, B, Charbonnel, N, Galtier, N, Jarne, P . Selective sweep and demography affect molecular diversity in the freshwater snail Biomphalaria pfeifferi. Genet Res (submitted).
Avise, JC (2000). Phylogeography The history and formation of species. Harvard University Press: Cambridge, Massachusetts.
Bandoni, SM, Mulvey, M, Loker, E (1995). Phylogenetic analysis of eleven species of Biomphalaria Preston, 1910 (Gastropoda: Planorbidae) based on comparisons of allozymes. Biol J Linn Soc 54: 1–27.
Barton, NH, Hewitt, GM (1985). Analysis of hybrid zones. Ann Rev Ecol Syst 16: 113–148.
Belkhir, K, Borsa, P, Chikhi, L, Raufaste, N, Bonhomme, F (2000). GENETIX 402, logiciel sous Windows TM pour la génétique des populations. Université de Montpellier II, Montpellier (France).
Chiba, S (1999). Accelerated evolution of land snail Mandarina in the oceanic Bonin islands: evidence from mitochondrial DNA sequences. Evolution 53: 460–471.
Chrosciechowski, P, Arcas, H (1986). Freshwater snails of the Caripe Valley, Monagas, Venezuela. Bol Min San Asist Soc Venez. 29pp.
Cole, C, Dessauer, H (1993). Unisexual and bisexual whiptail lizards of the Cnemidophorus lemniscatus complex (Squamata: Teiidae) of the Guiana region, South America, with descriptions of new species. Am Mus Nov 3081: 30pp
Crease, TJ, Lynch, M, Spitze, KS (1990). Hierarchical analysis of population genetic variation in mitochondrial and nuclear genes of Daphnia pulex. Mol Biol Evol 7: 444–458.
Dejong, RJ, Morgan, JAT, Paraense, WL, Pointier, J-P, Amarista, M, Ayeh-Kumi, PFK et al. (2001). Evolutionary relationships and biogeography of Biomphalaria (Gastropoda; Planorbidae) with implications regarding its role as host of the human bloodfluke, Schistosoma mansoni. Mol Biol Evol 18: 2225–2239.
Dutech, C, Maggia, L, Joly, HI (2000). Chloroplast diversity in Vouacapoua americana (caesalpiniaceae), a neotropical forest tree. Mol Ecol 9: 1427–1432.
Felsenstein, J (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791.
Freeland, JR, Noble, L, Okamura, B (2000). Genetic diversity of North American populations of Cristatella mucedo, inferred from microsatellite and mitochondrial DNA. Mol Ecol 9: 1375–1389.
Golvan, YJ, Combes, C, Euzeby, J, Salvat, B (1981). Enquête d’épidémiologie écologique sur la schistosomose à Schistosoma mansoni en Guadeloupe (Antilles franaises). Mém Mus Nat Hist Nat Paris 119: 1–229.
Haffer, J (1997). Alternative models of vertebrate speciation in Amazonia: an overview. Biodiversity and Conservation 6: 451–476.
Hewitt, GM (2001). Speciation, hybrid zones and phylogeography -seeing genes in space and time. Mol Ecol 10: 537–549.
Hoeh, WR, Stewart, DT, Sutherland, BW, Zouros, E (1996). Cytochrome c oxidase sequence comparisons suggest an unusually high rate of mitochondrial DNA evolution in Mytilus (Mollusca: Bivalvia). Mol Biol Evol 13: 418–421.
Jordan, P (1985). Schistosomiasis The St Lucia Project. Cambridge University Press: Cambridge.
Kimura, M (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120.
Kishino, H, Hasegawa, M (1989). Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, the branching order in Hominoidea. J Mol Evol 29: 170–179.
Kristensen, TK, Yousif, F, Raahauge, P (1999). Molecular characterization of Biomphalaria spp in Egypt. J Moll Stud 65: 133–136.
Madsen, H, Frandsen, F (1989). The spread of freshwater snails including those of medical and veterinary importance. Acta Trop 46: 139–146.
Malek, EA (1985). Snail Hosts of Schistosomiasis and Other Snail-Transmitted Diseases in Tropical America: A Manual. Burgess Publishing Company: Minneapolis.
Mavárez, J, Amarista, M, Pointier, J-P, Jarne, P (2002). Fine-scale population structure and dispersal in Biomphalaria glabrata, the intermediate snail host of Schistosoma mansoni, in Venezuela. Mol Ecol 11: 879–889.
Mavárez, J, David, P, Pointier, J-P, Jarne, P Genetic differentiation, dispersal and mating system in the Schistosome-transmitting freshwater snail Biomphalaria glabrata. Heredity 89: 258–265.
McCauley, DE (1991). Genetic consequences of local population extinction and recolonization. Trends Ecol Evol 6: 5–8.
Mello-Silva, CC, Grault, CE, Da Costa, VA, Barbosa, FS (1998). Possible hybridisation of Brazilian planorbid snails and its importance in population dynamics. Mem Inst Osw Cruz 93: 227–232.
Meunier, C, Tirard, C, Hurtrez-Boussès, S, Durand, P, Bargues, MD, Mas-Coma, S et al. (2001). Lack of molluscan host diversity and the transmission of an emerging parasitic disease in Bolivia. Mol Ecol 10: 1333–1340.
Moritz, C, Patton, JL, Schneider, CJ, Smith, TB (2000). Diversification of rainforest faunas: an integrated molecular approach. Ann Rev Ecol Syst 31: 533–563.
Mulvey, M, Newman, MC, Woodruff, DS (1988). Genetic differentiation among West Indian populations of the schistosome-transmitting snail Biomphalaria glabrata. Malacologia 29: 309–317.
Palumbi, SR (1996). Nucleic acids II: the polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK (eds) Molecular Systematics Sinauer Associates, Inc: Sunderland. pp 205–247.
Paraense, WL (1961). The nomenclature of Brazilian planorbids. I. “Australorbis glabratus” (Say, 1818). Rev Bras Biol 21: 287–296.
Paraense, WL (1983). A survey of planorbid molluscs in the Amazonian region of Brazil. Mem Inst Oswaldo Cruz 78: 343–361.
Paraense, WL, Deslandes, N (1959). The renal ridge as a reliable character for separating Taphius glabratus from Taphius tenagophilus. Am J Trop Med Hyg 8: 456–472.
Philippe, H (1993). MUST: a computer package of management utilities for sequences and trees. Nucl Acid Res 21: 5264–5272.
Pointier, J-P (1999). Invading freshwater gastropods: some conflicting aspects for public health. Malacologia 41: 403–411.
Pointier, J-P, Théron, A, Borel, G (1993). Ecology of the introduced snail Melanoides tuberculata (Gastropoda: Thiaridae) in relation to Biomphalaria glabatra in the marshy forest zone of Guadeloupe French West Indies. J Moll Stud 59: 421–428.
Raccurt, CP, Sodeman, WA, Rodrick, GL, Boyd, WP (1985). Biomphalaria glabrata in Haiti. Trans Roy Soc Trop Med Hyg 79: 455–477.
Swofford, DL (1998). PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods). Sinauer Associates, Inc: Sunderland, Massachusetts.
Viard, F, Justy, F, Jarne, P (1997). The influence of self-fertilization and bottlenecks on the genetic structure of subdivided populations: a case study using microsatellite markers in the freshwater snail Bulinus truncatus. Evolution 51: 1518–1528.
Vidigal, THDA, Neto, ED, Carvalho, ODS, Simpson, AJG (1994). Biomphalaria glabrata: extensive genetic variation in Brazilian isolates revealed by Random Amplified Polymorphic DNA analysis. Exp Parasit 79: 187–194.
Williamson, M (1997). Biological Invasions. Chapman and Hall: London.
Woodruff, DS, Mulvey, M (1997). Neotropical schistosomiasis: African affinities of the host snail Biomphalaria glabrata (Gastropoda: Planorbidae). Biol J Linn Soc 60: 505–516.
Yang, Z (1996). Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol Evol 11: 367–372.
Acknowledgements
The authors thank M Amarista, P David and T Lenormand for help in collecting snails, S Loker for providing snails from Brazil, and B Delay for help in the early stages of the project. This work was supported by funds from the Programme National Dynamique de la Biodiversité et Environnement, CNRS (Programme Jeune Equipe) and the French Ministry of Foreign Affairs (Ecos-Nord Committee V99A02) to PJ. JM benefited from a fellowship from the Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICIT), Venezuela.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mavárez, J., Steiner, C., Pointier, JP. et al. Evolutionary history and phylogeography of the schistosome-vector freshwater snail Biomphalaria glabrata based on nuclear and mitochondrial DNA sequences. Heredity 89, 266–272 (2002). https://doi.org/10.1038/sj.hdy.6800128
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800128
Keywords
This article is cited by
-
Population genetics and geometric morphometrics of the freshwater snail Segmentina nitida reveal cryptic sympatric species of conservation value in Europe
Conservation Genetics (2021)
-
Neither variation loss, nor change in selfing rate is associated with the worldwide invasion of Physa acuta from its native North America
Biological Invasions (2014)
-
Genetic differentiation, dispersal and mating system in the schistosome-transmitting freshwater snail Biomphalaria glabrata
Heredity (2002)