Abstract
Background:
Inhibitors of DNA-binding proteins (Id1-4), lacking the basic DNA-binding domain, function as dominant inhibitors of cell-cycle regulators. Overexpression of Id proteins promotes cancer cell proliferation and resistance against apoptosis. Level of Id protein expression, especially of Id1, correlates with poor differentiation, enhanced malignant potential and more aggressive clinical behaviour of ovarian tumours. Although overexpression of Ids has been found and shown to correlate with poor clinical outcome, their inhibition at protein level has never been studied.
Methods:
A peptide aptamer, Id1/3-PA7, targeting Id1 and Id3, was isolated from a randomised combinatorial expression library using yeast and mammalian two-hybrid systems. Id1/3-PA7 was fused, expressed and purified with a cell-penetrating protein transduction domain.
Results:
Intracellular-delivered Id1/3-PA7 colocalised to Id1 and Id3. It induced cell-cycle arrest and apoptosis in ovarian cancer cells ES-2 and PA-1. It activated the E-box promoter and increased the expression level of cyclin-dependent kinase inhibitor (CDKN2A) in a dose-dependent manner that is paralleled by the cleavage of poly-ADP ribose polymerase. These effects were counteracted by ectopically overexpressed Id1 and Id3.
Conclusion:
Id1/3-PA7 could represent an exogenous anti-tumour agent that can significantly trigger cell-cycle arrest and apoptosis in ovarian cancer.
Similar content being viewed by others
Main
Inhibitors of DNA-binding or differentiation proteins (Id) belong to the helix–loop–helix (HLH) family of transcription factors. There are four members of the Id family, Id1 to Id4. Id proteins, lacking the basic DNA-binding domain, associate with members of the basic HLH (bHLH) transcription factors and form transcriptionally inactive heterodimers. Thus, they function as dominant-negative regulators of bHLH transcription factors (Benezra et al, 1990). Furthermore, Id proteins regulate cellular growth and senescence through direct sequestration of Ets and Rb proteins (Iavarone et al, 1994; Ohtani et al, 2001; Ruzinova and Benezra, 2003). Id proteins are essential for cell differentiation, proliferation, cell-cycle progression, migration and angiogenesis (Norton and Atherton, 1998; Israel et al, 1999; Norton, 2000). Although barely expressed in most normal tissues, deregulated expression of Id proteins has been demonstrated in a variety of human tumours (Lin et al, 2000; Schindl et al, 2001; Wang et al, 2004). Overexpression of Id proteins promotes cancer cell proliferation and resistance against apoptosis (Ouyang et al, 2002a, 2002b; Ling et al, 2003). Id overexpression in different tumours is significantly associated with poor clinical outcome (Schoppmann et al, 2003; Han et al, 2004). Level of Id protein expression, especially of Id1, correlates with poor differentiation, enhanced malignant potential and more aggressive clinical behaviour of epithelial ovarian tumours (Schindl et al, 2003). Recently, it has been shown that ectopic Id1 expression stimulates ovarian cancer cell proliferation, and this process is mediated through upregulation of epidermal growth factor receptor (EGFR), suggesting that Id1 might serve as an upstream regulator of the EGFR pathway in promoting ovarian cancer cell growth (Zhang et al, 2004). In various forms of tumours, including ovarian cancer, Id1 and Id3 are overexpressed extensively and in an overlapping pattern (Fong et al, 2000; Coppe et al, 2003). They have been shown to inactivate Ets1 and Ets2, which leads to the inhibition of CDKN2A expression and consequently allows phosphorylation of retinoblastoma protein (pRb) (Ohtani et al, 2001). Furthermore, DNA-binding motif (E-box)-mediated repression of the CDKN2A promoter by Id1 has been reported (Alani et al, 2001). A significant positive association has been reported between CDKN2A expression and clinical outcome for epithelial ovarian cancer patients (Kusume et al, 1999).
A peptide-conjugated Id1 antisense oligonucleotide homed to tumour endothelium inhibited tumour growth and metastasis in two different murine models (Henke et al, 2008). Mice lacking Id1 and Id3 genes (Id1+/−Id3−/−) are resistant to xenotransplanted tumour grafts and show defects in tumour neoangiogenesis (Lyden et al, 1999; de Candia et al, 2003). Therefore, Id1 and Id3 are considered as potentially therapeutic targets.
So far, a joint inhibition of Id1 and Id3 at protein level has hardly been studied. To develop a new strategy for the inhibition of Id1 and Id3, we isolated a novel peptide aptamer Id1/3-PA7 that specifically interacts with Id1 and Id3 (Mern et al, 2010). Peptide aptamers represent short peptides of random sequences. On the basis of their in vivo binding affinity to their target protein, they can be selected from randomised combinatorial expression libraries using yeast and mammalian two-hybrid systems (Fields and Song, 1989; Chien et al, 1991; Bartel et al, 1993). In this study, we investigated the role of the peptide aptamer Id1/3-PA7 on the progression of cell cycle and apoptosis in Id1- and Id3-overexpressing ovarian cancer cells ES-2 and PA-1. For its delivery into ovarian cancer cells, Id1/3-PA7 was fused with a cell-penetrating protein transduction domain (PTD) (Elliott and O'Hare, 1997; Phelan et al, 1998; Narita et al, 2001), expressed and purified from bacteria. We demonstrated that intracellular-delivered Id1/3-PA7 colocalised to Id1 and Id3 and promoted increased expression of the tumour suppressor CDKN2A in a dose-dependent manner. In addition, Id1/3-PA7 induced cleavage of the apoptosis indicator poly-ADP ribose polymerase (PARP). These effects were counteracted by ectopically overexpressed Id1 and Id3. Peptide aptamer Id1/3-PA7 significantly inhibited proliferation and induced apoptosis in ovarian cancer cells, with deregulated expression of Id1 and Id3. Therefore, we suggest that Id1/3-PA7 could represent an exogenous anti-tumour agent that could find applications in targeted therapy.
Materials and methods
Cell lines and cell culture
ES-2 and PA-1 cell lines were purchased in October 2007 from the American Type Culture Collection (ATCC, Manassas, VA, USA). PA-1 cell lines were grown in EMEM (ATCC), and ES-2 cell lines in McCoy's 5a (ATCC) supplemented with 10% fetal bovine serum (ATCC), penicillin (50 U ml–1), streptomycin (50 mg ml–1) and 2 mM glutamine (Sigma, Deisenhofen, Germany). All cell lines were tested and authenticated in February 2008 by the Genetic Core Facility at the German Cancer Research Center (Heidelberg, Germany) using the method Multiplex cell Contamination Test (McCT) (http://www.dkfz.de/gpcf/contamination-control.html).
Transfections
For luciferase assays, ES-2 cells were transiently transfected using the calcium phosphate method (Chen and Okayama, 1987). For the analysis of CDKN2A expressions or PARP cleavage, ES-2 and PA-1 cells were transfected using FuGENE 6 Transfection Reagent (Roche Molecular Biochemicals, Indianapolis, IN, USA). Cells were selected with 1 mg ml–1 G418 (Sigma). Transfections were monitored by immunoblot detection of the expressed proteins.
Bacterial expression and purification of PTD-fused peptide aptamers
A PTD, truncated VP22 ORF, was used for the intracellular delivery of the peptide aptamer (Elliott and O'Hare, 1997; Phelan et al, 1998; Narita et al, 2001). PTD was fused to the C-terminus of peptide aptamers or TrxA, which were inserted into vectors pCR T7/VP22-1-TOPO and pCR T7/VP22/NES-2-TOPO (Invitrogen, Carlsbad, CA, USA). Bacterial expression of the peptide aptamer was induced with isopropyl-1-thio-β-D-galactopyranoside (1 mM) for 5 h at room temperature. The peptide aptamers were furnished with 6xHis-tag to facilitate their purification by nickel chelate chromatography. The peptide aptamer was purified under native conditions using the ProBond Purification System (Invitrogen). The purity of the peptide aptamer was investigated by gel electrophoresis and Coomassie staining. The concentration was determined using the BCA Protein Assay Kit (Pierce, Rockford, IL, USA).
Immunofluorescence and immunoblotting
For immunofluorescence analysis, cells were grown on coverslips, fixed and permeabilised in methanol for 20 min at −20°C and rehydrated with PBS. Before immunostaining with the indicated antibodies, cells were blocked in normal serum (1 : 10 in PBS containing 5% BSA). Nuclei were visualised by using Prolong Gold Antifade Reagent with DAPI (Invitrogen). Cells were analysed using a Carl Zeiss AxioVision 4 microscope equipped with a Carl Zeiss AxioCam digital camera and software version Carl Zeiss AxioVision Rel. 4.6.3 (Carl Zeiss Vision GmbH, Jena, Germany).
For immunoblot experiments, cell lysates prepared in EBC buffer (50 mM Tris–HCl, 120 mM NaCl, 1% (v/v) Nonidet P40, pH 8.0) supplemented with protease and phosphatase inhibitors were separated by SDS–PAGE and electrotransferred to polyvinylidene fluoride membranes (Millipore, Temecula, CA, USA). To detect antigen/antibody complexes, membranes were incubated with appropriate horseradish peroxidase-labelled secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and developed for enhanced chemiluminescence using the ECL WB Detection kit (Millipore).
Primary antibodies used were anti-Id1 (C-20), anti-Id3 (C-20) (Santa Cruz Biotechnology), anti-α-tubulin, anti-Trx (Sigma), anti-CDKN2A (Millipore) and anti-PARP (BD Biosciences, San Jose, CA, USA). Secondary antibodies used were goat anti-mouse or goat anti-rabbit Alexa Fluor 488, 660 and 680 for immunofluorescence experiments and horseradish peroxidase-conjugated goat anti-rabbit and goat anti-mouse (Santa Cruz Biotechnology) for immunoblotting.
Coimmunoprecipitation
ES-2 and PA-1 cells were treated with peptide aptamer Id1/3-PA7 (5 μg per 106 cells). After 1.5 h, cells were lysed in RIPA buffer (Pierce) containing protease and phosphatase inhibitors. Protein samples were purified by nickel chelate chromatography under native conditions using the ProBond Purification System (Invitrogen) and analysed by western blotting using antibodies against Trx, Id1 and Id3.
E-box promoter-reporter assays
pGL4.1-4Rtk-luc (E-box-dependent reporter construct), pcDNA3-E47, pCMV-Id1, pCMV-Id3, along with pCMV-LacZ as an internal standard, were used for transfection of ES-2 cells. After transfection, cells were treated by adding Id1/3-PA7 (5 μg per 106 cells) at 4 h intervals for 48 h. Cells were lysed in Triton buffer (1% Triton X-100, 25 mM glycylglycin (pH 7.8), 15 mM MgSO4, 4 mM EGTA and 1 mM DTT) for 10 min on ice. The lysates were clarified by centrifugation for 10 min at 16 000 g. Lysates (10 μl) were measured using a luminometer (Berthold, Vista, CA, USA) by injecting luciferin reagent (25 mM glycylglycin (pH 7.8), 5 mM ATP (pH 7.8) and 330 mM beetle luciferin). The samples were normalised for β-galactosidase activity, which was measured after incubating 3 μl lysate with 33 μl reaction buffer (100 mM Na2HPO4, 1 mM MgCl2 and 1 × Galacton (Tropix, Bedford, MA, USA)) for 30 min in the dark. Galactosidase activity was measured in a luminometer by injection of amplifier (10% Emerald (Tropix), 0.2 M NaOH).
RNA analysis
Cells were treated with different doses of Id1/3-PA7 (1–7.5 μg per 106 cells) at 4 h intervals for 48 h. Total RNA was purified using an RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) and inspected by agarose gel electrophoresis. cDNAs were synthesised using TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA, USA). The mRNAs of CDKN2A, CDKN1A, CDKN1B and β-actin as internal standard were measured by real-time PCR with TaqMan gene expression assays (Applied Biosystems), using the LightCycler 480 Real-Time PCR System and LightCycler 480 Probes Master (Roche Applied Science, Mannheim, Germany) under the following conditions: 15 min at 95°C activation, 40 cycles of 15 s at 95°C, followed by 30 s at 55°C and 1 s at 72°C, melting for 1 s at 55°C and cooling for 1 min at 40°C. For the relative quantification of mRNA levels, three independent amplifications were performed for each probe, with triplicate samples.
Flow cytometry cell-cycle analysis
Cells were treated with Id1/3-PA7 (5 μg per 106 cells) at 4 h intervals for 48 h. For quantitative cell-cycle analysis, the BD Pharmingen FITC bromodeoxyuridine (BrdU) Flow Kit (BD Biosciences) was used. Prolonged cell exposure (6 h) to BrdU allowed the identification and analysis of actively cycling, as opposed to non-cycling, cell fractions. Cell-incorporated BrdU (with FITC anti-BrdU) and total DNA content (with 7-AAD) in cells were measured using the BD FACSCalibur Flow Cytometer (BD Biosciences).
Results
Peptide aptamer Id1/3-PA7 colocalises to Id1 and Id3
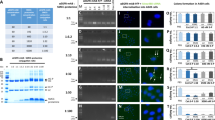
Using yeast and mammalian two-hybrid systems, we isolated from the randomised expression library a constrained peptide aptamer Id1/3-PA7 that interacts specifically with Id1 and Id3 (Mern et al, 2010). Here, we used Id1/3-PA7 to analyse its functional interference in Id1- and Id3-overexpressing ovarian cancer cells ES-2 and PA-1. For its delivery into ovarian cancer cells, Id1/3-PA7 was fused with cell-penetrating PTD (Elliott and O'Hare, 1997; Phelan et al, 1998; Narita et al, 2001), expressed in bacteria and purified under native conditions. Ovarian cancer cells ES-2 and PA-1 were treated with Id1/3-PA7 (5 μg per 106 cells). At 1.5 h after treatment, the internalisation of the peptide aptamer into cells and its colocalisation with Id1 and Id3 were verified by His-tag coimmunoprecipitation (Figure 1A and B). Cells treated with TrxA (5 μg per 106 cells) were used as negative control in coimmunoprecipitation (Figure 1C and D). In addition, the colocalisation was verified by coimmunofluorescence staining (Figure 1E–H).
Colocalisation of the intracellular-delivered peptide aptamer Id1/3-PA7 to Id1 and Id3. (A, B) His-tag coimmunoprecipitation of Id1/3-PA7 with Id1 and Id3 in ovarian cancer cells ES-2 and PA-1. (C, D) His-tag coimmunoprecipitation of TrxA as negative control. (E–H) Coimmunofluorescence staining of Id1/3-PA7 with Id1 and Id3 in ES-2 and PA-1 cells. DAPI, 4,6-diamidino-2-phenylindole.
Functional interference of Id1/3-PA7 with Id1 and Id3
E-proteins activate transcription by binding to promoter E-boxes. Formation of heterodimers between Id protein and E-protein prevents E-proteins from forming DNA-binding transcriptionally active complexes. To analyse the potency of Id1/3-PA7 with respect to Id1 and Id3 inhibition and restoration of E-box promoter activity, we used the E-box-dependent reporter gene 4Rtk-luc, which contains four tandem E-boxes from the MCK enhancer upstream of the thymidine kinase basal promoter (Weintraub et al, 1990). ES-2 cells were transfected with 4Rtk-luc, pcDNA3-E47, pCMV-Id1, pCMV-Id3 and pCMV-LacZ as an internal standard. After transfection, cells were treated with different doses of Id1/3-PA7 (1, 3 and 5 μg per 106 cells, half-life 2.5 h) at 4 h intervals for 48 h. The reporter gene was upregulated in the presence of the E-protein, E47. E47-dependent activation was reduced in the presence of Id1 or Id3, and, by addition of Id1/3-PA7, the activation was restored again (Figure 2A). Id1/3-PA7 restored E47-dependent activation in a dose-dependent manner (Figure 2B and Supplementary data 1). Treatment of cells with different doses of TrxA ((1, 3 and 5 μg per 106 cells) had no effect on E-box promoter activity (Figure 2A and Supplementary data 1).
(A) Peptide aptamer Id1/3-PA7 restores the E-box promoter activity. The transcription factor E47 upregulated the E-box promoter-dependent reporter gene (6), Id1 and Id3 reduced E47-dependent activation (13, 14) and addition of peptide aptamer Id1/3-PA7 restored E47-dependent activation (15, 16). The control with TrxA showed no effect on E-box promoter activity. (B) With Id1 and Id3, reduced E47-dependent activation is restored by the addition of Id1/3-PA7 in a dose-dependent manner (RLU/β-Gal: 250, 290, 720 and 1252 units respectively). Bars: black, Id1; white, Id3. Results are represented as mean values of three independent experiments with s.d. (P⩽0.05).
Id1/3-PA7 enhances the expression level of CDKN2A
E-box promoter-mediated repression of CDKN2A, CDKN1A and CDKN1B expression by Ids has been reported (Prabhu et al, 1997; Alani et al, 2001; Perk et al, 2005). Furthermore, it has been shown that Id1 and Id3 inactivate Ets1 and Ets2 and inhibit the transcriptional expression of CDKN2A (Ohtani et al, 2001). Therefore, we analysed the effect of Id1/3-PA7 on the expression level of CDKN2A, CDKN1A and CDKN1B, using real-time PCR and western blotting. Ovarian cancer cells ES-2 and PA-1 were treated with different doses of Id1/3-PA7 (1–7.5 μg per 106 cells) at 4 h intervals for 48 h. Intracellular delivery of Id1/3-PA7 caused a dose-dependent increase in the mRNA and protein levels of CDKN2A (Figure 3A and B). Untreated cells or cells treated with TrxA (5 μg per 106 cells) did not show any change in CDKN2A expression (Figure 3A and B). This increased expression level of CDKN2A is Id1 and Id3 specific, being counteracted by ectopically overexpressed Id1 and Id3 (Figure 3A). The expression levels of CDKN1A and CDKN1B were moderately increased after treatment with only high doses of Id1/3-PA7 (5 and 7.5 μg per 106 cells) (Supplementary data 2). However, these moderately increased expression levels of mRNA could not be confirmed at protein level. The treatment of PA-1 and ES-2 cells with lower doses of Id1/3-PA7 (1–4 μg per 106 cells) did not change the expression levels of CDKN1A and CDKN1B (Supplementary data 2).
Intracellular-delivered Id1/3-PA7 increases the expression level of CDKN2A. (A) Results of quantitative real-time PCR. Intracellular-delivered Id1/3-PA7 caused a dose-dependent increase in the relative mRNA level of CDKN2A compared with that in untreated or TrxA-treated (5 μg per 106 cells) cells. The increased expression level of CDKN2A was counteracted by ectopically overexpressed Id1 and Id3. Bars: hatched, ES-2 cells; black, PA-1 cells. Results are represented as mean values of three independent experiments with s.d. (P⩽0.05). (B, C) Western blotting showed increased CDKN2A protein levels in Id1/3-PA7-treated (5 μg per 106 cells) ES-2 and PA-1 cells, compared with that in untreated or TrxA-treated (5 μg per 106 cells) cells.
Id1/3-PA7 induces cell-cycle arrest and apoptosis in ovarian cancer cells
Tumour suppressor CDKN2A inhibits the cyclin-dependent kinases (Cdk4 and Cdk6) that initiate the phosphorylation of the pRb (Ruas and Peters, 1998; Sherr and Roberts, 1999). Therefore, CDKN2A has the capacity to arrest cells in the G1 phase of the cell cycle. Applying immunofluorescent staining of incorporated BrdU, an analogue of the DNA precursor thymidine, and flow cytometric analysis, we quantified individual cells that have incorporated BrdU into newly synthesised DNA as cells entering and progressing through the S (DNA synthesis) phase of the cell cycle. ES-2 and PA-1 cells were treated with TrxA or Id1/3-PA7 (5 μg per 106 cells) at 4 h intervals for 48 h. For the identification and analysis of actively cycling, as opposed to non-cycling, cell fractions, we exposed untreated and TrxA- or Id1/3-PA7-treated cells to BrdU for 6 h. The cell-cycle positions and active DNA synthetic activities of cells were determined by analysing the correlated expression of total DNA and incorporated BrdU levels as shown by the region gates applied to the 7-AAD vs BrdU dot plot (Figure 4A–F). Flow cytometric analysis of untreated cells vs Id1/3-PA7-treated cells demonstrated that the anti-proliferative and apoptotic effects of Id1/3-PA7 reduced the number of actively cycling cells in S from 93.1 to 54.8% for ES-2 cells (Figure 4G) and from 91.4 to 54.6% for PA-1 cells (Figure 4I). The number of apoptotic cells in sub-G0/G1- and in G0/G1- or G2/M-resided cells increased from 1.4 to 21.1%, from 3.2 to 13.2% and from 0.2 to 5.9%, respectively, for ES-2 cells (Figure 4H), and from 1.7 to 14,6%, from 3.9 to 16% and from 0.3 to 7.2%, respectively, for PA-1 cells (Figure 4J). There were no significant differences between untreated and TrxA-treated cells (Figure 4A–J).
Id1/3-PA7 induces cycle arrest and apoptosis in ovarian cancer cells. (A–J) Quantification of cell-incorporated bromodeoxyuridine (BrdU) (fluorescein isothiocyanate (FITC) anti-BrdU) and total DNA content (7-AAD) in untreated and TrxA-treated (5 μg per 106 cells) or Id1/3-PA7-treated (5 μg per 106 cells) ES-2 cells (A, C) and PA-1 cells (D–F). Flow cytometric analysis of untreated cells vs Id1/3-PA7-treated cells showed S-phase reduction of actively cycling ES-2 cells (G) and PA-1 cells (I), increasing apoptotic cells in sub-G0/G1- and G2/M-resided ES-2 cells (H) and PA-1 cells (J). There were no significant differences between untreated and TrxA-treated cells (A–J). Bars: black, untreated; white dotted, TrxA treated; hatched, Id1/3-PA7 treated. Results are represented as mean values of three independent experiments with s.d. (P⩽0.05).
Apoptotic PARP cleavage in response to Id1/3-PA7
It has been reported that wild-type CDKN2A expression from an adenovirus vector (Adv/p16) in non-small-cell lung cancer cell line A549, which carries the wild-type p53 gene, results in activation of caspase-3, accompanied by the cleavage of its substrate PARP (Koh et al, 2002). Furthermore, it has been shown that ectopically overexpressed Id1 is able to suppress PARP cleavage in response to different anticancer drugs, which leads to increased apoptosis rates and increased cleaved PARP in different cancer cell lines (Zhang et al, 2007). Therefore, Id1/3-PA7-treated ES-2 and PA-1 cells were analysed for PARP cleavage compared with untreated or TrxA-treated cells. We detected PARP cleavage by western blotting in cell extracts of Id1/3-PA7-treated cells using anti-PARP antibody, which recognises uncleaved PARP (113 kDa) and cleaved PARP (85 kDa) (Figure 5A and B). PARP cleavage was counteracted in Id1/3-PA7-treated cells by ectopically overexpressed Id1 and Id3 (Figure 5C and D).
Apoptotic poly-ADP ribose polymerase (PARP) cleavage induced by Id1/3-PA7. (A, B) Untreated and TrxA- or Id1/3-PA7-treated (5 μg per 106 cells) ES-2 and PA-1 cells were analysed for PARP cleavage by western blotting. PARP cleavage was detected in cell extracts of Id1/3-PA7-treated cells. (C, D) In Id1/3-PA7-treated cells, PARP cleavage was counteracted by ectopically overexpressed Id1 and Id3.
Discussion
Id1 and Id3 are considered as potentially versatile therapeutic targets. In various forms of tumours, Id1 and Id3 are overexpressed extensively and in an overlapping pattern (Fong et al, 2000; Coppe et al, 2003). The level of Id protein expression correlates with poor differentiation, enhanced malignant potential, aggressive clinical behaviour of different tumours, including epithelial ovarian tumours, and is a strong predicator of shorter survival (Schindl et al, 2003). Depending on the tumour art and stage, partial loss of Id functions is sufficient to have a therapeutic effect, as shown in Id1+/−Id3−/− knockout mice (Lyden et al, 1999; de Candia et al, 2003). It has also been shown that Id1 and Id3 overexpression correlates with loss of CDKN2A expression in different tumours (Polsky et al, 2001; Lee et al, 2003), suggesting that in some settings Id1/3-induced cell proliferation is mediated through the inactivation of the CDKN2A-pRb pathway. Therefore, we investigated the biological effect of the peptide aptamer Id1/3-PA7 in Id1/Id3-overexpressing and CDKN2A-positive ovarian cancer cells, ES-2 and PA-1. In this paper, we demonstrated that inhibition of Id1 and Id3 by the peptide aptamer Id1/3-PA7 induced cell-cycle arrest and apoptosis. It significantly increased the endogenous expression level of CDKN2A in a dose-dependent manner, which is paralleled by the cleavage of PARP. In contrast to this, the expression levels of CDKN1A and CDKN1B were moderately increased after treatment with only high doses of Id1/3-PA7 (5 and 7.5 μg per 106 cells) (Supplementary data 2). However, these moderately increased expression levels of mRNA could not be confirmed at protein level. The treatment of PA-1 and ES-2 cells with lower doses of Id1/3-PA7 (1–4 μg per 106 cells) has not changed the expression levels of CDKN1A and CDKN1B (Supplementary data 2). This could indicate that in PA-1 and ES-2 ovarian cancer cells, Id1/3-PA7 might preferentially inhibit the heterodimerisation of Id1 and Id3 with ETS proteins, which are the positive regulators of CDKN2A. Previously, we have shown that Id1/3-PA7 regulates the expression of CDKN1A and CDKN1B in a dose-dependent manner in breast cancer cells MCF-7 and MDA-MB-231, which contain the homozygous deletions of the gene CDKN2A (Kamb et al, 1994). These results together suggest that Id1 and Id3, depending on the tumour type, might differentially regulate the expression of tumour suppressor genes CDKN2A, CDKN1A and CDKN1B.
Besides the CDKN/pRb pathways, Id1/3-PA7 could interestingly be used to analyse EGFR pathway-dependent functions, as Id proteins, especially Id1, induce upregulation of EGFR in different tumour cells, very frequently in androgen-independent prostate cancer cells. Thus, Id1/3-PA7 could have effects on androgen-independent proliferation of prostate cancer cells. Overexpression of Id1 is associated with progression of prostate cancer and Id1-induced androgen-independent prostate cancer cell growth is correlated with upregulation of EGFR (Ouyang et al, 2002a, 2002b; Ling et al, 2004).
In our previous work (Mern et al, 2010), we demonstrated using a mammalian two-hybrid system that Id1/3-PA7 interacts with Id1 and Id3 but does not bind to Id2, Id3 and the interacting partners of Id proteins, such as E47, MyoD, ETS1, ETS2 and S5A. Furthermore, we have shown that the biological effects of Id1/3-PA7, in analysed breast and ovarian cancer cells, were counteracted by forced expression of Id1 and Id3. These results indicate that the biological effects of Id1/3-PA7 are based on the functional blocking of Id1 and Id3. We hope that future in vivo application of the peptide aptamer Id1/3-PA7 in tumour-bearing Id1/3 transgenic mice and in Id1+/−Id3−/− knockout mice will give rise to more information regarding its in vivo stability, functional specificity and potential toxicity.
Overexpression of Id proteins, especially of Id1, has been found to be correlated with the progression of different types of solid tumours (Perk et al, 2005; Ling et al, 2006). Their low postnatal expression and their roles in tumourigenesis and tumour neoangiogenesis mark them as attractive targets for anticancer therapy (Perk et al, 2005). Therefore, we suggest that Id1/3-PA7, as inhibitor of Id1 and Id3, could have the potential to be used as a new tool for targeted tumour therapy.
Change history
29 March 2012
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Alani RM, Young AZ, Shifflett CB (2001) Id1 regulation of cellular senescence through transcriptional repression of p16Ink4a. PNAS 98: 7812–7816
Bartel P, Chien CT, Sternglanz R, Fields S (1993) Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14: 920–924
Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61: 49–59
Chen C, Okayama H (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7: 2745–2752
Chien CT, Bartel PL, Sternglanz R, Fields S (1991) The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA 88: 9578–9582
Coppe JP, Smith AP, Desprez PY (2003) Id proteins in epithelial cells. Exp Cell Res 285: 131–145
de Candia P, Solit DB, Giri D, Brogi E, Siegel PM, Olshen AB, Muller WJ, Rosen N, Benezra R (2003) Angiogenesis impairment in Id-deficient mice cooperates with an Hsp90 inhibitor to completely suppress HER2/neu-dependent breast tumours. Proc Natl Acad.Sci USA 100: 12337–12342
Elliott G, O'Hare P (1997) Intercellular trafficking and protein delivery by a herpes virus structural protein. Cell 88: 223–233
Fields S, Song O (1989) A novel genetic system to detect protein-protein interactions. Nature 340: 245–246
Fong S, Debs RJ, Desprez PY (2000) Id genes and proteins as promising targets in cancer therapy. Trends Mol Med 10: 387–392
Han S, Guo C, Hong L, Liu J, Han Z, Liu C, Wang J, Wu K, Ding J, Fan D (2004) Expression and significances of Id1 helix-loop-helix protein overexpression in gastric cancer. Cancer Lett 216: 63–71
Henke E, Perk J, Vider J, de Candia P, Chin Y, Solit DB, Ponomarev V, Cartegni L, Manova K, Rosen N, Benezra R (2008) Peptide-conjugated antisense oligonucleotides for targeted inhibition of a transcriptional regulator in vivo. Nat Biotechnol 26: 91–100
Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA (1994) The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev 8: 1270–1284
Israel MA, Hernandez MC, Florio M, Andres-Barguin PJ, Mantani A, Carter JH, Julin CM (1999) Id gene expression as a key mediator of tumour cell biology. Cancer Res 59: 1726–1730
Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH (1994) A cell cycle regulator potentially involved in genesis of many tumor types. Science 264: 436–440
Koh K, Masafumi K, Futoshi U, Takayoshi M, Tadashi K, Jack AR, Noriaki T, Toshiyoshi F (2002) Activation of caspase-3 and cleavage of Rb are associated with p16-mediated apoptosis in human non-small cell lung cancer cells. Oncogene 21: 2108–2113
Kusume T, Tsuda H, Kawabata M, Inoue T, Umesaki N, Suzuki T, Yamamoto K (1999) The p16-cyclin D1/CDK4-pRb pathway and clinical outcome in epithelial ovarian cancer. Clin Cancer Res 5: 4152–4157
Lee TK, Man K, Ling M, Wang X, Wong Y, Lo C, Poon RT, Ng IO, Fan S (2003) Over-expression of Id-1 induces cell proliferation in hepatocellular carcinoma through inactivation of p16INK4a/RB pathway. Carcinogenesis 24: 1729–1736
Lin CQ, Singh J, Murata K, Itahana Y, Parrinello S, Liang SH, Gillett CE, Campisi J, Desprez, PY (2000) Role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res 60: 1332–1340
Ling MT, Wang X, Lee DT, Tam PC, Tsao SW, Wong YC (2004) Id-1 expression induces androgen-independent prostate cancer cell growth through activation of epidermal growth factor receptor (EGF-R). Carcinogenesis 25: 517–525
Ling MT, Wang X, Ouyang XS, Xu K, Tsao SW, Wong YC (2003) Id-1 expression promotes cell survival through activation of NF-kappaB signalling pathway in prostate cancer cells. Oncogene 22: 4498–4508
Ling MT, Wang X, Zhang X, Wong YC (2006) The multiple roles of Id-1 in cancer progression. Differentiation 74: 481–487
Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R (1999) Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumor xenografts. Nature 401: 670–677
Mern DS, Hoppe-Seyler K, Hoppe-Seyler F, Hasskarl J, Burwinkel B (2010) Targeting Id1 and Id3 by a specific peptide aptamer induces E-box promoter activity, cell cycle arrest, and apoptosis in breast cancer cells. Breast Cancer Res Treat, doi: 10.1007/s10549-010-0810-6
Narita K, Choudhury A, Dobrenis K (2001) Protein transduction of Rab9 in Niemann- Pick C cells reduces cholesterol storage. FASEB 10: 1558–1560
Norton JD (2000) ID helix-loop-helix proteins in cell growth, differentiation and tumourigenesis. J Cell Sci 113: 3897–3905
Norton JD, Atherton GT (1998) Coupling of cell growth control and apoptosis functions of Id proteins. Mol Cell Biol 18: 2371–2381
Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E (2001) Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409: 1067–1070
Ouyang XS, Wang X, Lee DT, Tsao SW, Wong YC (2002a) Over expression of ID-1 in prostate cancer. J Urol 167: 2598–2602
Ouyang XS, Wang X, Ling MT, Wong HL, Tsao SW, Wong YC (2002b) Id-1 stimulates serum independent prostate cancer cell proliferation through inactivation of p16 (INK4a)/pRB) pathway. Carcinogenesis 23: 721–725
Perk J, Iavarone A, Benezra R (2005) Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer 5: 603–614
Phelan A, Elliott G, O'Hare, P (1998) Intercellular delivery of functional p53 by the herpes virus protein VP22. Nat Biotechnol 16: 440–443
Polsky D, Young AZ, Busan KJ, Alani RM (2001) The transcriptional repressor of p16/Ink4a, Id1, is up-regulated in early melanomas. Cancer Res 61: 6008–6011
Prabhu S, Ignatova A, Park ST, Sun XH (1997) Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol 17: 5888–5896
Ruas M, Peters G (1998) The p16INK4a/CDKN2A tumour suppressor and its relatives. Biochim Biophys Acta 1378: 115–117
Ruzinova MB, Benezra R (2003) Id proteins in development, cell cycle and cancer. Trends Cell Biol 13: 410–418
Schindl M, Oberhuber G, Obermair A, Schoppmann SF, Karner B, Birner P (2001) Overexpression of Id-1 protein is a marker for unfavourable prognosis in early-stage cervical cancer. Cancer Res 61: 5703–5706
Schindl M, Schoppmann SF, Ströbel T, Heinzl H, Leisser C, Horvat R, Birner P (2003) Level of Id-1 protein expression correlates with poor differentiation, enhanced malignant potential, and more aggressive clinical behavior of epithelial ovarian tumours. Clin Cancer Res 9: 779–785
Schoppmann SF, Schindl M, Bayer G, Aumayr K, Dienes J, Horvat R, Rudas M, Gnant M, Jakesz R, Birner P (2003) Overexpression of Id-1 is associated with poor clinical outcome in node negative breast cancer. Int J Cancer 104: 677–682
Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1- phase progression. Genes Dev 13: 1501–1512
Wang Q, Tsao SW, Fu S, Xue W, Meng X, Feng H, Wong YC, Wang X (2004) Overexpression of Id-1 in gastric adenocarcinoma: implication for a novel diagnostic marker. Anticancer Res 24: 881–886
Weintraub H, Davis R, Lockshon D, Lassar A (1990) Muscle-specific transcriptional activation by MyoD. Proc Natl Acad Sci USA 87: 5623–5627
Zhang X, Ling MT, Wong YC, Wang X (2007) Evidence of a novel antiapoptotic factor: role of inhibitor of differentiation or DNA binding (Id-1) in anticancer drug-induced apoptosis. Cancer Sci 98: 308–314
Zhang X, Ling MT, Feng H, Wong YC, Tsao SW, Wang X (2004) Id-1 stimulates cell proliferation through activation of EGFR in ovarian cancer cells. BJC 91: 2042–2047
Acknowledgements
We thank R Benezra (Sloan-Kettering Institute for Cancer Research, New York, USA) for providing plasmids pGL4.1-4Rtk-luc and pcDNA3-E47, and S Schmitt (Core Facility Flow Cytometry, DKFZ) and S Poppelreuter (Carl Zeiss MicroImaging GmbH) for supporting cell-cycle profiling and fluorescence microscopy. This work was supported by the Deutsche Forschungsgemeinschaft (Grant HA3185/2-1and 2-3); the Helmholtz Society (Grant VH-NG-213); the German Cancer Research Center; and the Dietmar-Hopp Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Mern, D., Hasskarl, J. & Burwinkel, B. Inhibition of Id proteins by a peptide aptamer induces cell-cycle arrest and apoptosis in ovarian cancer cells. Br J Cancer 103, 1237–1244 (2010). https://doi.org/10.1038/sj.bjc.6605897
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6605897
Keywords
This article is cited by
-
Id proteins: emerging roles in CNS disease and targets for modifying neural stemcell behavior
Cell and Tissue Research (2022)
-
Evolution of KIPPIS as a versatile platform for evaluating intracellularly functional peptide aptamers
Scientific Reports (2021)
-
CDKN2A inhibits cell proliferation and invasion in cervical cancer through LDHA-mediated AKT/mTOR pathway
Clinical and Translational Oncology (2021)
-
ID1 confers cancer cell chemoresistance through STAT3/ATF6-mediated induction of autophagy
Cell Death & Disease (2020)
-
Epigenetic role of the nuclear factor NF-Y on ID gene family in endometrial tissues of women with endometriosis: a case control study
Reproductive Biology and Endocrinology (2019)