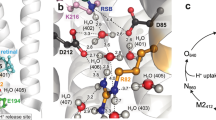
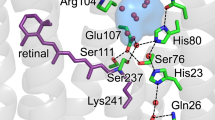
Abstract
A wide variety of mechanisms are used to generate a proton-motive potential across cell membranes, a function lying at the heart of bioenergetics. Bacteriorhodopsin, the simplest known proton pump1, provides a paradigm for understanding this process. Here we report, at 2.1 Å resolution, the structural changes in bacteriorhodopsin immediately preceding the primary proton transfer event in its photocycle. The early structural rearrangements2 propagate from the protein's core towards the extracellular surface, disrupting the network of hydrogen-bonded water molecules that stabilizes helix C in the ground state. Concomitantly, a bend of this helix enables the negatively charged3 primary proton acceptor, Asp 85, to approach closer to the positively charged primary proton donor, the Schiff base. The primary proton transfer event would then neutralize these two groups, cancelling their electrostatic attraction and facilitating a relaxation of helix C to a less strained geometry. Reprotonation of the Schiff base by Asp 85 would thereby be impeded, ensuring vectorial proton transport. Structural rearrangements also occur near the protein's surface, aiding proton release to the extracellular medium.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Oesterhelt, D. & Stoeckenius, W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature New Biol. 233, 149–152 ( 1971).
Edman, K. et al. High-resolution X-ray structure of an early intermediate in the bacteriorhodopsin photocycle. Nature 401, 822–826 (1999).
Braiman, M. S. et al. Vibrational spectroscopy of bacteriorhodopsin mutants: Light-driven proton transport involves protonation changes of aspartic acid residues 85, 96 and 212. Biochemistry 27, 8516– 8520 (1988).
Landau, E. M. & Rosenbusch, J. P. Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proc. Natl Acad. Sci. USA 93, 14532–14535 (1996).
Schlichting, I., Berendzen, J., Phillips, G. N. & Sweet, R. M. Crystal structure of photolysed carbonmonoxy-myoglobin. Nature 371, 808–812 ( 1994).
Genick, U. K., Soltis, S. M., Kuhn, P., Canestrelli, I. L. & Getzoff, E. D. Structure at 0. 85 Å resolution of an early protein photocycle intermediate. Nature 392, 206–209 (1998).
Maeda, A., Sasaki, J., Yamazaki, Y., Needleman, R. & Lanyi, J. K. Interaction of Aspartate-85 with a water molecule and the protonated Schiff base in the L intermediate of bacteriorhodopsin: A Fourier-transform infrared spectroscopy study. Biochemistry, 33, 1713–1717 (1994).
Hendrickson, F. M., Burkard, F. & Glaeser, R. M. Structural characterization of the L-to-M transition of the bacteriorhodopsin photocycle. Biophys. J. 75 , 1446–1454 (1998).
Hadfield, A. & Hajdu, J. A fast and portable microspectrophotometer for protein crystallography. J. Appl. Crystallogr. 26, 839–842 (1993).
Belrhali, H. et al. Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 Å resolution. Structure 7, 909–917 (1999).
Subramaniam, S., Gerstein, M., Oesterhelt, D. & Henderson, R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 12, 1–8 (1993).
Subramaniam, S. et al. Protein conformational changes in the bacteriorhodopsin photocycle. J. Mol. Biol. 287, 145– 161 (1999).
Takei, H., Gat, Y., Rothman, Z., Lewis, A. & Sheves, M. Active site Lysine backbone undergoes conformational changes in the bacteriorhodopsin photocycle. J. Biol. Chem. 269, 7387–7389 (1994).
Luecke, H., Schobert, B., Richter, H. T., Cartailler, J.-P. & Lanyi, J. K. Structural changes in bacteriorhodopsin during ion transport at 2 Angstrom resolution. Science 286, 255–260 (1999).
Luecke, H., Schobert, B., Richter, H. T., Cartailler, J. -P. & Lanyi, J. K. Structure of bacteriorhodopsin at 1. 55 Å resolution. J. Mol. Biol. 291, 899–911 (1999).
Balashov, S. P., Imasheva, E. S., Govindjee, R. & Ebrey, T. G. Titration of aspartate-85 in bacteriorhodopsin: what it says about chromophore isomerization and proton release. Biophys. J. 70, 473–481 (1996).
Althaus, T. & Stockburger, M. Time and pH dependence of the L-to-M transition in the photocycle of bacteriorhodopsin and its correlation with proton release. Biochemistry 37, 2807 –2817 (1998).
Gat, Y. & Sheves, M. A mechanism for controlling the pKa of the retinal protonated Schiff base in retinal proteins. A study with model compounds. J. Am. Chem. Soc. 115, 3772– 3773 (1993).
Balashov, S. P. et al. Glutamate-194 to cysteine mutation inhibits fast light-induced proton release in bacteriorhodopsin. Biochemistry 36 , 8671–8676 (1997).
Hu, J. G., Sun, B. Q., Petkova, A. T., Griffin, R. G. & Herzfeld, J. The predischarge chromophore in bacteriorhodopsin: a 15N solid state NMR study of the L photointermediate. Biochemistry 36, 9316– 9322 (1997).
Khorana, H. G. Two light-transducing membrane proteins: bacteriorhodopsin and the mammalian rhodopsin. Proc. Natl Acad. Sci. USA 90, 1166–1171 (1993).
Heberle, J., Oesterhelt, D. & Dencher, N. A. Decoupling of photo- and proton cycle in the Asp → Glu mutant of bacteriorhodopsin. EMBO J. 12, 3721–3727 (1993).
Nollert, P. & Landau, E. M. Enzymic release of crystals from lipidic cubic phases. Biochem. Soc. Trans. 26, 709–713 (1998).
Otwinowski, Z. in Data Collection and Processing (eds Sawyer, L., Isaacs, N. W. & Bailey S.) 55–62 (DL/SCI/R34, Daresbury Laboratory, Warrington, 1993).
Bourgeois, D. New processing tools for weak and/or spatially overlapped macromolecular diffraction patterns. Acta Crystallogr. D 55, 1733– 1741 (1999).
Collaborative computational project nr4. The CCP4 suite: Programs for protein crystallography. Acta. Crystallogr. D 50, 760–763 ( 1994).
Chandra, N., Ravi Acharya, K. & Moody, P. C. E. Analysis and characterization of data from twinned crystals. Acta Crystallogr. D 55, 1750– 1758 (1999).
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Esnouf, R. M. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. 15, 133–138 (1997).
Merrit, E. A. & Murphy, M. E. P. Raster3D version 2. 0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994).
Acknowledgements
We thank H. Belrhali, D. Bourgeois, J. Hajdu, A. Hardmeyer, J. Navarro, P. Nollert, H. Pettersson and S. Ramaswamy for experimental contributions and discussions, and G. Büldt for providing purple membrane. Support from the EU-BIOTECH, the Swedish Natural Science Research Council (NFR) and the Swiss National Science Foundation's SPP BIOTECH is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Royant, A., Edman, K., Ursby, T. et al. Helix deformation is coupled to vectorial proton transport in the photocycle of bacteriorhodopsin. Nature 406, 645–648 (2000). https://doi.org/10.1038/35020599
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35020599
This article is cited by
-
True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins
Nature Structural & Molecular Biology (2022)
-
Tracking Membrane Protein Dynamics in Real Time
The Journal of Membrane Biology (2021)
-
Collective exchange processes reveal an active site proton cage in bacteriorhodopsin
Communications Biology (2020)
-
Theoretical study on the substituent effect of halogen atom at different position of 7-azaindole-water derivatives: relative stability and excited-state proton-transfer mechanism
Structural Chemistry (2018)
-
Water Pathways in the Bacteriorhodopsin Proton Pump
The Journal of Membrane Biology (2011)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.