Abstract
Proton transport is indispensable for cell life. It is believed that molecular mechanisms of proton movement through different types of proton-conducting molecules have general universal features. However, elucidation of such mechanisms is a challenge. It requires true-atomic-resolution structures of all key proton-conducting states. Here we present a comprehensive function-structure study of a light-driven bacterial inward proton pump, xenorhodopsin, from Bacillus coahuilensis in all major proton-conducting states. The structures reveal that proton translocation is based on proton wires regulated by internal gates. The wires serve as both selectivity filters and translocation pathways for protons. The cumulative results suggest a general concept of proton translocation. We demonstrate the use of serial time-resolved crystallography at a synchrotron source with sub-millisecond resolution for rhodopsin studies, opening the door for principally new applications. The results might also be of interest for optogenetics since xenorhodopsins are the only alternative tools to fire neurons.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Atomic models built using X-ray crystallography data have been deposited in the RCSB Protein Data Bank with PDB codes 7ZMY (for the ground state at pH 8.2 at 100 K), 7ZN3 (for the L state at pH 8.2 at 100 K), 7ZN0 (for the M state at pH 8.2 at 100 K), 7ZN8 (for the ground state at pH 7.0 at 100 K), 7ZN9 (for the M state at pH 7.0 at 100 K), 7ZNA (for the ground state at pH 5.2 at 100 K), 7ZNB (for the M state at pH 5.2 at 100 K), 7ZNC (for the ground state at pH 7.6 at 100 K in the absence of sodium), 7ZND (for the M state at pH 7.6 at 100 K in the absence of sodium), 7ZNE (for the ground state at pH 8.2 at 293 K with 7.5-ms-long exposure), 7ZNI (for the 7.5–15-ms snapshot at pH 8.2 at 293 K), 7ZNG (for the ground state at pH 8.2 at 293 K with 500-μs-long exposure) and 7ZNH (for the 250–750-μs snapshot at pH 8.2 at 293 K). Extrapolated structure factors and phases for the intermediate states of BcXeR are available from Zenodo (https://doi.org/10.5281/zenodo.7612803). Publicly available structures of NsXeR (PDB ID: 6EYU) and HsBR (PDB IDs: 5ZIL, 7Z0D, 7Z0E, 6RPH) were used for analysis. Source data are provided with this paper.
References
Gushchin, I. & Gordeliy, V. in Membrane Protein Complexes: Structure and Function (eds. Harris, J. R. & Boekema, E. J.) 19–56 (Springer, 2018).
Oesterhelt, D. & Stoeckenius, W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. N. Biol. 233, 149–152 (1971).
Sahel, J.-A. et al. Partial recovery of visual function in a blind patient after optogenetic therapy. Nat. Med. 27, 1223–1229 (2021).
Inoue, K. et al. A natural light-driven inward proton pump. Nat. Commun. 7, 13415 (2016).
Inoue, S. et al. Spectroscopic characteristics of Rubricoccus marinus xenorhodopsin (RmXeR) and a putative model for its inward H+ transport mechanism. Phys. Chem. Chem. Phys. 20, 3172–3183 (2018).
Shevchenko, V. et al. Inward H+ pump xenorhodopsin: mechanism and alternative optogenetic approach. Sci. Adv. 3, e1603187 (2017).
Weissbecker, J. et al. The voltage dependent sidedness of the reprotonation of the retinal schiff base determines the unique inward pumping of xenorhodopsin. Angew. Chem. Int. Ed. 60, 23010–23017 (2021).
Alcaraz, L. D. et al. The genome of Bacillus coahuilensis reveals adaptations essential for survival in the relic of an ancient marine environment. Proc. Natl Acad. Sci. USA 105, 5803–5808 (2008).
Chizhov, I. et al. Spectrally silent transitions in the bacteriorhodopsin photocycle. Biophys. J. 71, 2329–2345 (1996).
Landau, E. M. & Rosenbusch, J. P. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl Acad. Sci. USA 93, 14532–14535 (1996).
Luecke, H., Schobert, B., Richter, H.-T., Cartailler, J.-P. & Lanyi, J. K. Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 291, 899–911 (1999).
Hasegawa, N., Jonotsuka, H., Miki, K. & Takeda, K. X-ray structure analysis of bacteriorhodopsin at 1.3 Å resolution. Sci. Rep. 8, 13123 (2018).
Gerwert, K., Freier, E. & Wolf, S. The role of protein-bound water molecules in microbial rhodopsins. Biochim. Biophys. Acta 1837, 606–613 (2014).
Weinert, T. et al. Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography. Science 365, 61–65 (2019).
Mous, S. et al. Dynamics and mechanism of a light-driven chloride pump. Science 375, 845–851 (2022).
Wickstrand, C., Dods, R., Royant, A. & Neutze, R. Bacteriorhodopsin: would the real structural intermediates please stand up? Biochim. Biophys. Acta 1850, 536–553 (2015).
Kouyama, T., Kawaguchi, H., Nakanishi, T., Kubo, H. & Murakami, M. Crystal structures of the L1, L2, N, and O States of pharaonis halorhodopsin. Biophys. J. 108, 2680–2690 (2015).
Nango, E. et al. A three-dimensional movie of structural changes in bacteriorhodopsin. Science 354, 1552–1557 (2016).
Borshchevskiy, V. et al. True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins. Nat. Struct. Mol. Biol. 29, 440–450 (2022).
Freier, E., Wolf, S. & Gerwert, K. Proton transfer via a transient linear water-molecule chain in a membrane protein. Proc. Natl Acad. Sci. 108, 11435–11439 (2011).
Kuppuraj, G., Dudev, M. & Lim, C. Factors governing metal−ligand distances and coordination geometries of metal complexes. J. Phys. Chem. B 113, 2952–2960 (2009).
Szalewicz, K. In Encyclopedia of Physical Science and Technology 3rd edn (ed. Meyers, R. A.) 505–538 (Academic Press, 2003).
Jung, K.-H., Trivedi, V. D. & Spudich, J. L. Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 47, 1513–1522 (2003).
Volkov, O. et al. Structural insights into ion conduction by channelrhodopsin 2. Science 358, eaan8862 (2017).
Ho, B. K. & Gruswitz, F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Studier, F. W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Drachev, L. A. et al. Direct measurement of electric current generation by cytochrome oxidase, H+-ATPase and bacteriorhodopsin. Nature 249, 321–324 (1974).
Drachev, L. A., Kaulen, A. D., Khitrina, L. V. & Skulachev, V. P. Fast stages of photoelectric processes in biological membranes. Eur. J. Biochem. 117, 461–470 (1981).
Rokitskaya, T. I. et al. Rhodopsin channel activity can be evaluated by measuring the photocurrent voltage dependence in planar bilayer lipid membranes. Biochemistry (Mosc.) 86, 409–419 (2021).
Kovalev, K. et al. Molecular mechanism of light-driven sodium pumping. Nat. Commun. 11, 2137 (2020).
Bratanov, D. et al. Unique structure and function of viral rhodopsins. Nat. Commun. 10, 4939 (2019).
von Stetten, D. et al. In crystallo optical spectroscopy (icOS) as a complementary tool on the macromolecular crystallography beamlines of the ESRF. Acta Crystallogr. D Biol. Crystallogr. 71, 15–26 (2015).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Tickle, I. J. et al. STARANISO http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi (Global Phasing Ltd., 2016).
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Vogeley, L. et al. Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 Å. Science 306, 1390–1393 (2004).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Weierstall, U. et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 5, 3309 (2014).
White, T. A. et al. CrystFEL: a software suite for snapshot serial crystallography. J. Appl. Crystallogr. 45, 335–341 (2012).
Gevorkov, Y. et al. XGANDALF—extended gradient descent algorithm for lattice finding. Acta Crystallogr. Sect. Found. Adv. 75, 694–704 (2019).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Borshchevskiy, V. I., Round, E. S., Popov, A. N., Büldt, G. & Gordeliy, V. I. X-ray-radiation-induced changes in bacteriorhodopsin structure. J. Mol. Biol. 409, 813–825 (2011).
Paithankar, K. S. & Garman, E. F. Know your dose: RADDOSE. Acta Crystallogr. D Biol. Crystallogr. 66, 381–388 (2010).
Acknowledgements
We acknowledge the Structural Biology Group of the European Synchrotron Radiation Facility (ESRF) and the European Molecular Biology Laboratory (EMBL) unit in Hamburg at Deutsche Elektronen-Synchrotron (DESY) for granting us access to the synchrotron beamlines. Structural and spectroscopic data obtained at the ESRF were recorded and analyzed between 2020 and 2021. This work was supported by the common program of the Agence Nationale de la Recherche (ANR), France and Deutsche Forschungsgemeinschaft, Germany (ANR-15-CE11-0029-02), and by funding from Frankfurt: Cluster of Excellence Frankfurt Macromolecular Complexes (to E.B.), by the Max Planck Society (to E.B.), and by the Commissariat à l’Energie Atomique et aux Energies Alternatives (Institut de Biologie Structurale)–Helmholtz-Gemeinschaft Deutscher Forschungszentren (Forschungszentrum Jülich) Special Terms and Conditions 5.1 specific agreement. V.G. greatly acknowledges his HGF professorship. This work used the icOS and HTX platforms at the Grenoble Instruct-ERIC center (ISBG; UAR 3518 CNRS-CEA-UJF-EMBL) within the Grenoble Partnership for Structural Biology (PSB). Platform access was supported by FRISBI (ANR-10-INBS-0005-02) and GRAL, a project at the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003). Protein expression and spectroscopic analysis were supported by the Russian Science Foundation (RSF) (project 21-64-00018). This work was supported by the project ANR-19-CE11-0026. K.K. has been supported by EMBL Interdisciplinary Postdoctoral Fellowship (EIPOD4) under Marie Sklodowska-Curie Actions Cofund (grant agreement number 847543). The work of A.A. was supported by funding from the German Research Foundation to T. Moser via the Multiscale Bioimaging - Cluster of Excellence (EXC 2067/1-390729940). Time-resolved crystallography data treatment was supported by RFBR (project 19-29-12022). A.K. acknowledges support from the Ministry of Science and Higher Education of the Russian Federation (agreement 075-03-2023-106, project FSMG-2021-0002). A. Rogachev acknowledges the Ministry of Science and Higher Education of the Russian Federation (project no. 075-01645-22-06/720000F.99.1.BЗ85AV67000). X-ray data collection was supported by the Russian Ministry of Science and Higher Education (grant no. 075-15-2021-1354). The research of electrogenic behavior of BcXeR in proteoliposomes was carried out at the expense of the grant of the Russian Science Foundation No. 22-14-00104 (to S.S.).
Author information
Authors and Affiliations
Contributions
K.K., A.A. and F.T. contributed equally and each has the right to list himself first in bibliographic documents. A.A. used bioinformatics to find the BcXeR gene, proposed it as a target and performed initial functional tests in E. coli; F.T. did cloning, expression and purification of the protein and its mutants; F.T. did functional studies of the proteins in an E. coli cell suspension; A.A. supervised the expression and purification; S.S. performed studies of electrogenic behavior of BcXeR in proteoliposomes; D.S. and F.T. performed flash-photolysis studies on protein in solution and in crystals; I.C. supervised flash-photolysis experiments; K.K. crystallized the protein; R.A. helped with the crystallization; S.B., R.A. and A. Royant performed cryotrapping of the intermediates at the icOS Lab and recorded the spectra of the states in crystals; K.K. performed cryotrapping of the intermediates in crystals at the P14 beamline; M.A. installed the laser setup at the P14 beamline for the cryotrapping and TR studies; K.K. collected and processed the diffraction data, and solved and refined the structures; G.B. helped with data collection, structure refinement, and analysis; M.N. provided online monitoring of X-ray diffraction images during TR crystallography data collection at 1 kHz; D.v.S. provided scripts for serial data processing and help with serial data analysis; V.S., N.I., E.B., T.R.S., A. Rogachev, F.R.-V., V.B. and G.B. helped with data analysis; A.K. animated the transitions between the intermediates, under the supervision of I.G.; A.A. performed bioinformatic analysis with the help of R.R.; F.T., A.A., K.K. and V.G. analyzed the results and prepared the manuscript with the important contribution of G.B. and with input from all the other authors. V.G. supervised and analyzed the results of experiments and proposed the conception of molecular mechanism of inward proton transport.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Keiichi Inoue and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Carolina Perdigoto and Katarzyna Ciazynska, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
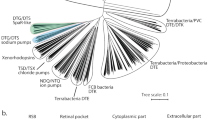
Extended Data Fig. 1 Xenorhodopsin homologs and their proton-pumping activity.
a. Phylogenetic tree of close homologs of XeRs obtained using a Hidden Markov Model. In total, 76 genes of MRs closely-related to XeRs were identified. The tree is divided into three major clades of proteins: bacteria’s xenorhodopsins, archaea’s xenorhodopsins, and a group of close homologs of Anabaena Sensory Rhodopsin (ASR). Representative proteins (including BcXeR) are highlighted with red dots in each clade. AaSR – Sensory rhodopsin from Aliterella atlantica (GenBank: KPQ35581.1), PpaSR – Sensory rhodopsin from Phormidesmis priestleyi Ana (NCBI Reference Sequence: WP_045056952.1). b. Light-induced pH changes in E.coli cell suspensions expressing representative proteins highlighted in panel A. Data for PoXeR, RmXeR, and ASR were adopted from the respective publications4,5,21. Data for NsXeR, AlkXeR, and HrvXeR were have been reported previously6. Data for BcXeR, PpaSR, and AaSR were obtained in the present study (see Methods ‘Measurements of pump activity in the E.coli cells suspension’ for details).
Extended Data Fig. 2 Spectroscopy of BcXeR.
a. Absorption spectra of BcXeR at various pH. b. Arrhenius plot of BcXeR rate constants. c. Electrometric time-resolved measurements of the membrane potential and comparison with spectroscopic data. d. The photocycle scheme of BcXeR in crystals. e. The photocycle scheme of BcXeR in solution. f. The photocycle scheme of NsXeR6. g. The photocycle scheme of HsBR9. h. The pH dependence of the time-resolved traces of BcXeR in solution at 390 nm characteristic for the M state.
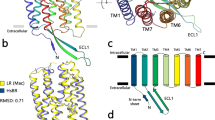
Extended Data Fig. 3 Trimers of proton-pumping microbial rhodopsins.
View from the extracellular side is shown on the top panels. View from the cytoplasmic side is shown on the bottom panel. The trimers of BcXeR (present work), NsXeR (PDB ID: 6EYU6), and HsBR (PDB ID: 5ZIL12) are shown. The N-terminal α-helix of NsXeR is colored blue. Retinal cofactor is shown with sticks and is colored teal.
Extended Data Fig. 4 Examples of electron density maps of BcXeR obtained at 100 K.
a. The cofactor retinal region in the ground state of BcXeR at pH 8.2 at 100 K. 2Fo-Fc electron density maps are contoured at the level of 1.5σ and are shown with a black mesh. b. The cofactor retinal region in the M state of BcXeR at pH 8.2 at 100 K. 2Fo-Fc electron density maps are contoured at the level of 1.2σ and are shown with a black mesh. c. The PTG region in the ground state of BcXeR at pH 8.2 in the presence of sodium at 100 K. 2Fo-Fc electron density maps are contoured at the level of 1.5σ and are shown with a black mesh. d. The PTG region in the M state of BcXeR at pH 8.2 in the presence of sodium at 100 K. 2Fo-Fc electron density maps are contoured at the level of 1.2σ and are shown with a black mesh. e. The region of L81 in the L state of BcXeR at 100 K. Folight-Fodark difference electron density maps are contoured at the level of 3.5σ and are shown with a green (positive) and a red (negative) mesh. The maps indicate the flip of L81 and the appearance of three water molecules mediating the linear proton wire between the RSB and S211 in the L state (shown with black dashed lines).
Extended Data Fig. 5 Amino acid conservativity analysis of XeRs and mutational analysis of BcXeR.
a. Amino acids mutated in the present study. Colors correspond to groups presented on panel D. b. Alignment of structurally important regions in representative proteins (shown with the red dots in panel A), which significantly impact xenorhodopsin function. Structurally important regions were selected according to the high-resolution structure of BcXeR presented in this work. The selected regions contain amino acids surrounding the RSB, proton acceptor/proton release group, intracellular and extracellular sides of the protein, DT-pair (which is commonly known as a DC-pair in channelrhodopsins, D156/C128 of channelrhodopsin-2), and trimerization interface. Amino acids are enumerated according to the BcXeR. c. Analysis of structurally important region conservation among each of the clades. Numbers N on the left of the panel indicate a number of proteins in each clade. Regions of the proteins correspond to those in panel B. d. Relative proton pumping activity of BcXeR mutants in E.coli suspension during illumination. pH change in wild type is considered as unity.
Extended Data Fig. 6 Common structural features of BcXeR and HsBR.
a. Structural alignment of retinal binding pockets of BcXeR (green) and HsBR (violet). View from the side of the RSB and K207 (K216). b. View from the side of the β-ionone ring. c. Overall side view of BcXeR. d. Overall side view of HsBR. Internal cavities are shown with a blue surface. The similar hydrophobic gates in BcXeR and HsBR are shown with red arrows and residues forming the gates are shown in red. The proton transit group (PTG) of BcXeR and the proton release group (PRG) of HsBR are highlighted in yellow. Hydrophobic/hydrophilic membrane core boundaries are shown with gray horizontal lines. e. Structural alignment of the central region of helices G of BcXeR (violet) and HsBR (green). π-bulge regions are indicated with a black arrow. Hydrogen bonds stabilizing the π-bulges are shown with dashed lines. Names of the corresponding residues in HsBR are shown in parentheses.
Extended Data Fig. 7 Double conformations in the ground and M states of BcXeR.
a. 2Fo-Fc electron density maps contoured at the level of 1.0σ demonstrate the presence of the second conformation of L136, W173, and S206 in the M state at 100 K. b. Fo-Fc difference electron density maps built omitting the second conformation in the structure of the M state at 100 K contoured at the level of 3.0σ. The maps indicate the presence of the second conformation of the residues L136, W173, and S206. c. 2Fo-Fc electron density maps near Wat402 in the ground state contoured at the level of 1.0σ. The distance between two conformations is shown with a dashed line and is given in Å. d. 2Fo-Fc electron density maps near Wat402 in the L state contoured at the level of 1.0σ. The strong negative peak in the Fo-Fc difference electron density maps near Wat402 in the L state contoured at the level of 3.0σ (red mesh) built with the model fitted with 100% occupancy of each of the water molecules support the two alternative conformations of Wat402 in the L state. The difference maps were built with 1 cycle of refinement with fixed atom positions and B factors set equal to the mean values of the nearby residues (H6 and R70). The distance between two conformations is shown with a dashed line and is given in Å. e. 2Fo-Fc electron density maps near Wat402 and Wat403 in the M state contoured at the level of 1.0σ. The strong positive peak in the Fo-Fc difference electron density maps near Wat402 and Wat403 in the M state contoured at the level of 3.0σ (green mesh) built with the model fitted with 50% occupancy of each of the water molecules support the two separate water molecules in the M state. The difference maps were built with 1 cycle of refinement with fixed atom positions and B factors set equal to the mean values of the nearby residues (H6 and R70). The distance between the two conformations is shown with a dashed line and is given in Å.
Extended Data Fig. 8 Examples of electron density maps of BcXeR obtained at 293 K.
a. The cofactor retinal region of the ground state of BcXeR at pH 8.2 at 293 K (500 μs X-ray exposures). 2Fo-Fc electron density maps are contoured at the level of 1.2σ. b. The cofactor retinal region of the 250–750-μs snapshot (L state) of BcXeR at pH 8.2 at 293 K. Foextrapolated electron density maps are contoured at the level of 1.5σ. c. \({{\rm{F}}}_{{\rm{o}}}^{{\rm{extrapolated}}}\) electron density maps contoured at the level of 1.5σ around the L81, L136, and W173 corresponding to the 250–750-μs snapshot (L state) of BcXeR at pH 8.2 at 293 K. Original (ground state) positions of the residues are shown in green. d. The cofactor retinal region of the ground state of BcXeR at pH 8.2 at 293 K (7.5 ms X-ray exposures). 2Fo-Fc electron density maps are contoured at the level of 1.0σ. e. The cofactor retinal region of the 7.5-15-ms snapshot (M state) of BcXeR at pH 8.2 at 293 K. \({{\rm{F}}}_{{\rm{o}}}^{{\rm{extrapolated}}}\) electron density maps are contoured at the level of 1.0σ. f. \({{\rm{F}}}_{{\rm{o}}}^{{\rm{extrapolated}}}\) electron density maps contoured at the level of 1.5σ around the carbonyl oxygen of K207 corresponding to the 250-750-μs snapshot (L state) of BcXeR at pH 8.2 at 293 K. Original (ground state) positions of the residues are shown in green. g. The PTG of the ground state of BcXeR at pH 8.2 at 293 K (500 μs X-ray exposures). 2Fo-Fc electron density maps are contoured at the level of 1.2σ. h. The PTG of the ground state of BcXeR at pH 8.2 at 293 K (7.5 ms X-ray exposures). 2Fo-Fc electron density maps are contoured at the level of 1.0 σ. i. \({{\rm{F}}}_{{\rm{o}}}^{{\rm{extrapolated}}}\) electron density maps contoured at the level of 1.0σ around the carbonyl oxygen of K207 corresponding to the 7.5-15-ms snapshot (M state) of BcXeR at pH 8.2 at 293 K.
Extended Data Fig. 9 Comparison of intermediate states of BcXeR and HsBR.
a. Comparison of the cytoplasmic sides of BcXeR and HsBR (PDB ID: 7Z0D19) in the L state. Water molecules, transiently appearing in the L state are highlighted in yellow. H-bond proton wires connecting the RSB to the PTG and proton donor D96 in case of BcXeR and HsBR, respectively, are shown with black dashed lines. b. Comparison of the RSB in the M state of BcXeR and HsBR (PDB ID: 7Z0E19). c. Absence and presence of large-scale structural rearrangements in BcXeR and HsBR (PDB ID: 6RPH14), respectively at the late steps of their photocycles. Ground (white), L (lightblue), and M (deepblue) state structures of BcXeR and ground (white), M (lightblue), and N (deepblue) states structures of HsBR are aligned to visualize the evolution of the structural changes triggered by retinal isomerization and translated via tryptophan residue in the helix F. Black arrows highlighted in yellow indicate the propagation of structural rearrangements from the retinal to W173 and to helices E and F in case of BcXeR and HsBR, respectively. Gray arrows indicate the proton uptake from the cytoplasmic part in the case of HsBR and, correspondingly, the absence of the uptake in the case of BcXeR.
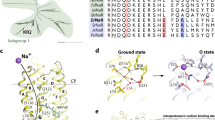
Extended Data Fig. 10 Different conformations of the proton transit group of BcXeR.
Detailed view of the proton transit group (PTG) of BcXeR under different conditions. Conformations of the PTG in the absence of sodium in crystallization conditions are shown in the red section. Ground and L states are shown in green; M states are shown in yellow. Sodium ions are shown with purple spheres. 2Fo-Fc electron density maps are shown for the ground state of BcXeR at pH 8.2 in the presence of sodium and are contoured at the level of 1.5σ. The Na-O distances are shown with bold italic numbers and are given in Å.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Notes 1–6, Fig. 1 and References.
Supplementary Video 1
Evolution of the retinal binding pocket of BcXeR in the course of photocycle. Animation of structural transitions was done using GROMACS. The structure was biased towards each intermediate consecutively using position restraints. The resulting trajectories are schematic.
Supplementary Video 2
The proposed scheme of the light-driven inward proton pumping by BcXeR. Animation of structural transitions was done using GROMACS. The structure was biased towards each intermediate consecutively using position restraints. The resulting trajectories are schematic.
Source data
Source Data Fig. 1
Experimental source data.
Source Data Fig. 2
Experimental source data.
Source Data Extended Data Fig./Table 1
Experimental source data.
Source Data Extended Data Fig./Table 2
Experimental source data.
Source Data Extended Data Fig./Table 5
Experimental source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kovalev, K., Tsybrov, F., Alekseev, A. et al. Mechanisms of inward transmembrane proton translocation. Nat Struct Mol Biol 30, 970–979 (2023). https://doi.org/10.1038/s41594-023-01020-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-023-01020-9