Featured
-
-
Letter |
 Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate
Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate
In the CRISPR type III system, ‘ring’ nucleases possess a metal-independent mechanism that cleaves cyclic oligoadenylate ring molecules to switch off the antiviral state in cells.
- Januka S. Athukoralage
- , Christophe Rouillon
- & Malcolm F. White
-
Letter |
 The NORAD lncRNA assembles a topoisomerase complex critical for genome stability
The NORAD lncRNA assembles a topoisomerase complex critical for genome stability
The long non-coding RNA NORAD interacts with proteins involved in DNA replication and repair, and controls the ability of RBMX to form a ribonucleoprotein complex that helps to maintain genomic stability.
- Mathias Munschauer
- , Celina T. Nguyen
- & Eric S. Lander
-
Letter |
 Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation
Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation
In Schizosaccharomyces pombe, histone H3K9 methylation acts synergistically with short interfering RNA to perpetuate gene silencing during multiple mitotic and meiotic cell divisions.
- Ruby Yu
- , Xiaoyi Wang
- & Danesh Moazed
-
Article |
 Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance
Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance
ZNFX-1 and WAGO-4 localize to germ granules in early Caenorhabditis elegans embryogenesis and later separate to form independent liquid-like droplets, and the temporal and spatial ordering of these droplets may help cells to organize complex RNA processing pathways.
- Gang Wan
- , Brandon D. Fields
- & Scott Kennedy
-
Letter |
 Hierarchical roles of mitochondrial Papi and Zucchini in Bombyx germline piRNA biogenesis
Hierarchical roles of mitochondrial Papi and Zucchini in Bombyx germline piRNA biogenesis
The biogenesis of piRNAs in the silkworm Bombyx is simpler than in Drosophila, with the exonucleases Trim and Nbr having no major role, and the endonuclease Zuc acting at the 3′ rather than the 5′ end.
- Kazumichi M. Nishida
- , Kazuhiro Sakakibara
- & Mikiko C. Siomi
-
Letter |
 Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells
Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells
A sequence that is frequently found in Alu elements drives the localization of some long RNAs to the nucleus in human cells.
- Yoav Lubelsky
- & Igor Ulitsky
-
News & Views |
 Viruses hijack a long non-coding RNA
Viruses hijack a long non-coding RNA
Manipulation of host-cell metabolism is an essential aspect of viral replication cycles. Viral co-option of a cellular long non-protein-coding RNA has now been found to be a key step in this process.
- Nicholas S. Heaton
- & Bryan R. Cullen
-
Article |
 A transfer-RNA-derived small RNA regulates ribosome biogenesis
A transfer-RNA-derived small RNA regulates ribosome biogenesis
A 22-nucleotide fragment of a transfer RNA regulates translation by binding to the mRNA of a ribosomal protein and increasing its expression, and downregulation of the fragment in patient-derived liver tumour cells reduces tumour growth in mice.
- Hak Kyun Kim
- , Gabriele Fuchs
- & Mark A. Kay
-
Letter |
 A viral Sm-class RNA base-pairs with mRNAs and recruits microRNAs to inhibit apoptosis
A viral Sm-class RNA base-pairs with mRNAs and recruits microRNAs to inhibit apoptosis
Herpesvirus saimiri expresses a small nuclear RNA named HSUR2, with which it recruits two cellular microRNAs to repress specific mRNA targets and thereby inhibit apoptosis in infected T cells.
- Carlos Gorbea
- , Tim Mosbruger
- & Demián Cazalla
-
News & Views |
 Rhino gives voice to silent chromatin
Rhino gives voice to silent chromatin
Flies use master lists of DNA sequences from transposons to identify and silence these virus-like, genomic parasites. How the lists themselves escape the fate of their transposon targets has now been solved. See Letter p.54
- Phillip D. Zamore
-
Article |
 A heterochromatin-dependent transcription machinery drives piRNA expression
A heterochromatin-dependent transcription machinery drives piRNA expression
Transcription of Drosophila PIWI-interacting RNA (piRNA) clusters is enforced through RNA polymerase II pre-initiation complex formation within repressive heterochromatin, accomplished through the transcription factor IIA subunit paralogue Moonshiner.
- Peter Refsing Andersen
- , Laszlo Tirian
- & Julius Brennecke
-
News & Views |
 The long and short of a DNA-damage response
The long and short of a DNA-damage response
Ultraviolet light can damage DNA, triggering a general shutdown of gene transcription — yet some genes are activated by UV light. An investigation of this counter-intuitive behaviour reveals a surprising gene-regulation mechanism.
- Antonio Conconi
- & Brendan Bell
-
News & Views |
 More uses for genomic junk
More uses for genomic junk
It emerges that nascent non-coding RNAs transcribed from regulatory DNA sequences called enhancers bind to the enzyme CBP to promote its activity locally. In turn, the activities of CBP stimulate further enhancer transcription.
- Karen Adelman
- & Emily Egan
-
Article |
 An atlas of human long non-coding RNAs with accurate 5′ ends
An atlas of human long non-coding RNAs with accurate 5′ ends
A catalogue of human long non-coding RNA genes and their expression profiles across samples from major human primary cell types, tissues and cell lines.
- Chung-Chau Hon
- , Jordan A. Ramilowski
- & Alistair R. R. Forrest
-
Technology Feature |
 The RNA code comes into focus
The RNA code comes into focus
As researchers open up to the reality of RNA modification, an expanded epitranscriptomics toolbox takes shape.
- Kelly Rae Chi
-
Article |
 Adipose-derived circulating miRNAs regulate gene expression in other tissues
Adipose-derived circulating miRNAs regulate gene expression in other tissues
Adipose tissue is a major source of circulating exosomal miRNAs, which contribute to the regulation of gene expression in distant tissues and organs.
- Thomas Thomou
- , Marcelo A. Mori
- & C. Ronald Kahn
-
Letter |
 mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide
mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide
The polypeptide SPAR is encoded by a long non-coding RNA, localizes to the late endosome and lysosome, and regulates muscle regeneration by inhibiting mTORC1.
- Akinobu Matsumoto
- , Alessandra Pasut
- & Pier Paolo Pandolfi
-
Letter |
 Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development
Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development
Transcription of a long non-coding RNA, known as upperhand (Uph) located upstream of the HAND2 transcription factor is required to maintain transcription of the Hand2 gene by RNA polymerase, and blockade of Uph expression leads to heart defects and embryonic lethality in mice.
- Kelly M. Anderson
- , Douglas M. Anderson
- & Eric N. Olson
-
Letter |
 Local regulation of gene expression by lncRNA promoters, transcription and splicing
Local regulation of gene expression by lncRNA promoters, transcription and splicing
Various cis-regulatory functions of genomic loci that produce long non-coding RNAs are revealed, including instances where their promoters have enhancer-like activity and the lncRNA transcripts themselves are not required for activity.
- Jesse M. Engreitz
- , Jenna E. Haines
- & Eric S. Lander
-
Research Highlights |
RNA spray fights fungus
-
Article |
 m6A RNA methylation promotes XIST-mediated transcriptional repression
m6A RNA methylation promotes XIST-mediated transcriptional repression
The methylation of adenosine residues on the long non-coding RNA XIST is essential for X-chromosome transcriptional repression during female mammalian development.
- Deepak P. Patil
- , Chun-Kan Chen
- & Samie R. Jaffrey
-
News & Views |
 Moulding the ribosome
Moulding the ribosome
Production of the cell's translational apparatus, the ribosome, requires the orchestrated function of hundreds of proteins. A structure of its earliest precursor yields unprecedented insight into ribosome formation.
- Marlene Oeffinger
-
Article |
 Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma
Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma
Disparate modes of suppression of the let-7 microRNA family are selectively and inversely related in neuroblastoma.
- John T. Powers
- , Kaloyan M. Tsanov
- & George Q. Daley
-
Letter |
 Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis
Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis
The activation of lipid X receptors (LXRs) in mouse liver not only promotes cholesterol efflux but also inhibits cholesterol synthesis simultaneously; this is mediated by the lipid-responsive long non-coding RNA LeXis, which is induced by a Western diet and orchestrates crosstalk between LXRs and the cholesterol biosynthetic pathway.
- Tamer Sallam
- , Marius C. Jones
- & Peter Tontonoz
-
News Feature |
 The quiet revolutionary: How the co-discovery of CRISPR explosively changed Emmanuelle Charpentier’s life
The quiet revolutionary: How the co-discovery of CRISPR explosively changed Emmanuelle Charpentier’s life
The microbiologist spent years moving labs and relishing solitude. Then her work on gene-editing thrust her into the scientific spotlight.
- Alison Abbott
-
Letter |
 Melanoma addiction to the long non-coding RNA SAMMSON
Melanoma addiction to the long non-coding RNA SAMMSON
A known oncogene, MITF, resides in a region of chromosome 3 that is amplified in melanomas and associated with poor prognosis; now, a long non-coding RNA gene, SAMMSON, is shown to also lie in this region, to also act as a melanoma-specific survival oncogene, and to be a promising therapeutic target for anti-melanoma therapy.
- Eleonora Leucci
- , Roberto Vendramin
- & Jean-Christophe Marine
-
News Feature |
 CRISPR: gene editing is just the beginning
CRISPR: gene editing is just the beginning
The real power of the biological tool lies in exploring how genomes work.
- Heidi Ledford
-
Letter |
 Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm
Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm
Maternal mRNAs are tethered within the Drosophila germ plasm via base-pairing interactions between mRNAs and piRNPs containing the Aub Piwi protein; the preference for certain mRNAs to be tethered appears to be related to their longer length, which provides more potential piRNP-binding sites, and the results suggest a new role for piRNAs in germ-cell specification independent of their role in transposon silencing.
- Anastassios Vourekas
- , Panagiotis Alexiou
- & Zissimos Mourelatos
-
Article |
 Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions
Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions
Using dual RNA-seq technology to profile RNA expression simultaneously in the bacterial pathogen Salmonella and its host during infection reveals molecular phenotypes of small noncoding RNAs in the infection process.
- Alexander J. Westermann
- , Konrad U. Förstner
- & Jörg Vogel
-
Article |
 DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions
DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions
The ability of the DEAD-box RNA helicase DDX5 to interact with master transcription factor RORγt is dependent on binding of the long noncoding RNA Rmrp; the DDX5–RORγt complex coordinates transcription of selective TH17 genes and is required for the pathogenicity of TH17 cells.
- Wendy Huang
- , Benjamin Thomas
- & Dan R. Littman
-
Article |
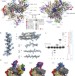 Molecular structures of unbound and transcribing RNA polymerase III
Molecular structures of unbound and transcribing RNA polymerase III
RNA polymerase III (Pol III), the largest eukaryote polymerase yet characterized, transcribes structured small non-coding RNAs; here cryo-electron microscopy structures of budding yeast Pol III allow building of an atomic-level model of the complete 17-subunit complex, both unbound and while elongating RNA.
- Niklas A. Hoffmann
- , Arjen J. Jakobi
- & Christoph W. Müller
-
News & Views |
 Antibiotic tricks a switch
Antibiotic tricks a switch
A screen for compounds that block a bacterial biosynthetic pathway has uncovered an antibiotic lead that shuts off pathogen growth by targeting a molecular switch in a regulatory RNA structure. See Article p.672
- Thomas Hermann
-
Letter |
 Integrator mediates the biogenesis of enhancer RNAs
Integrator mediates the biogenesis of enhancer RNAs
This study demonstrates a role for the Integrator complex in the stimulus-dependent induction of eRNAs and their 3′ processing; together with previously known roles of Integrator in transcription elongation and RNA processing, these results indicate that Integrator has broad functions in the regulation of eukaryotic gene expression.
- Fan Lai
- , Alessandro Gardini
- & Ramin Shiekhattar
-
News Feature |
 THE ELABORATE ARCHITECTURE OF RNA
THE ELABORATE ARCHITECTURE OF RNA
Cells contain an ocean of twisting and turning RNA molecules. Now researchers are working out the structures — and how important they could be.
- Elie Dolgin
-
News & Views |
 Rap and chirp about X inactivation
Rap and chirp about X inactivation
Two new techniques identify proteins that directly interact with a non-protein-coding RNA called Xist to mediate inactivation of one X chromosome in female mammals. See Letter p.232
- Anna Roth
- & Sven Diederichs
-
Letter |
 The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3
The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3
The mechanisms by which Xist, a long non-coding RNA, silences one X chromosome in female mammals are unknown; here a mass spectrometry-based approach is developed to identify several proteins that interact directly with Xist, including the transcriptional repressor SHARP that is required for transcriptional silencing through the histone deacetylase HDAC3.
- Colleen A. McHugh
- , Chun-Kan Chen
- & Mitchell Guttman
-
News & Views |
 Coding in non-coding RNAs
Coding in non-coding RNAs
The discovery of peptides encoded by what were thought to be non-coding – or 'junk' – regions of precursors to microRNA sequences reveals a new layer of gene regulation. These sequences may not be junk, after all. See Letter p.90
- Peter M. Waterhouse
- & Roger P. Hellens
-
Letter |
 Primary transcripts of microRNAs encode regulatory peptides
Primary transcripts of microRNAs encode regulatory peptides
Plant primary microRNA (miRNA) transcripts (pri-miRNAs) are not just a source of miRNAs but can also encode regulatory peptides (miPEPs) that enhance the accumulation, and so the effect, of the corresponding mature miRNAs—an observation that may have agronomical applications.
- Dominique Lauressergues
- , Jean-Malo Couzigou
- & Jean-Philippe Combier
-
Letter |
 N6-methyladenosine marks primary microRNAs for processing
N6-methyladenosine marks primary microRNAs for processing
The addition of the N6-methyladenosine (m6A) mark to primary microRNAs by METTL3 in mammalian cells is found to promote the recognition of these microRNA precursors by DGCR8, a component of the microprocessor complex.
- Claudio R. Alarcón
- , Hyeseung Lee
- & Sohail F. Tavazoie
-
Letter |
 Initiation of translation in bacteria by a structured eukaryotic IRES RNA
Initiation of translation in bacteria by a structured eukaryotic IRES RNA
A eukaryotic viral internal ribosome entry site (IRES) element is described that binds both bacterial and eukaryotic ribosomes and initiates translation in both, demonstrating that RNA structure-based initiation can occur in both these domains of life, although in bacteria the element uses a mechanism that differs from that in eukaryotes.
- Timothy M. Colussi
- , David A. Costantino
- & Jeffrey S. Kieft
-
Letter |
 NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs
NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs
A newly developed method, NAD captureSeq, has been used to show that bacteria cap the 5′-ends of some RNAs to protect against degradation, much as happens with eukaryotic messenger RNAs, although with a different modification: nicotinamide adenine dinucleotide.
- Hana Cahová
- , Marie-Luise Winz
- & Andres Jäschke
-
Letter |
 RNA helicase DDX21 coordinates transcription and ribosomal RNA processing
RNA helicase DDX21 coordinates transcription and ribosomal RNA processing
DEAD-box RNA helicase DDX21 is involved in both the transcription and RNA processing of ribosomal genes in human cells, sensing the transcriptional status of both RNA polymerase I and RNA polymerase II and associating with non-coding RNAs involved in ribonucleoprotein formation, possibly allowing for coordinated regulation of protein synthesis.
- Eliezer Calo
- , Ryan A. Flynn
- & Joanna Wysocka
-
News |
 Cancer cells can ‘infect’ normal neighbours
Cancer cells can ‘infect’ normal neighbours
Tiny RNAs shed by tumours can transform healthy cells into cancerous ones.
- Heidi Ledford
-
Letter |
 Transcriptional interference by antisense RNA is required for circadian clock function
Transcriptional interference by antisense RNA is required for circadian clock function
The transcriptions of frq sense and antisense RNAs are mutually inhibitory and form a double negative feedback loop required for robust and sustained circadian rhythmicity: antisense transcription inhibits sense expression by causing chromatin modifications and premature transcription termination.
- Zhihong Xue
- , Qiaohong Ye
- & Yi Liu
-
Letter |
 Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
The CRISPR/Cas system is an RNA-guided bacterial protection system against foreign nucleic acids of bacterial and archaeal origin; here a high-resolution crystal structure of the CRIPSR RNA–Cas complex shows that the CRIPSR RNA plays an essential role not only in target recognition but also in complex assembly.
- Hongtu Zhao
- , Gang Sheng
- & Yanli Wang
-
Letter |
 A long noncoding RNA protects the heart from pathological hypertrophy
A long noncoding RNA protects the heart from pathological hypertrophy
Here, a long noncoding RNA, termed Mhrt, is identified in the loci of myosin heavy chain (Myh) genes in mice and shown to be capable of suppressing cardiomyopathy in the animals, as well as being repressed in diseased human hearts.
- Pei Han
- , Wei Li
- & Ching-Pin Chang
-
Letter |
 Noncoding RNA transcription targets AID to divergently transcribed loci in B cells
Noncoding RNA transcription targets AID to divergently transcribed loci in B cells
The 11-subunit RNA exosome is thought to regulate the mammalian noncoding transcriptome; here, a mouse model is generated in which the essential Exosc3 subunit of the RNA exosome in B cells is conditionally deleted, revealing a link between sites of genomic RNA exosome function and AID-mediated chromosomal translocations.
- Evangelos Pefanis
- , Jiguang Wang
- & Uttiya Basu
-
Letter |
 Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease
Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease
Crystal structure of the RNA-guided endonuclease Cas9 bound to a guide RNA and a target DNA duplex reveals how base-specific recognition of a short motif known as PAM in the DNA target results in localized strand separation in the DNA immediately upstream of the PAM, allowing the target DNA strand to hybridize to the guide RNA.
- Carolin Anders
- , Ole Niewoehner
- & Martin Jinek
-
Article |
 Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway
Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway
Programmed −1 ribosomal frameshifting (−1 PRF) is a process by which a signal in a messenger RNA causes a translating ribosome to shift by one nucleotide, thus changing the reading frame; here −1 PRF in the mRNA for the co-receptor for HIV-1, CCR5, is stimulated by two microRNAs and leads to degradation of the transcript by nonsense-mediated decay and at least one other decay pathway.
- Ashton Trey Belew
- , Arturas Meskauskas
- & Jonathan D. Dinman

 Excised linear introns regulate growth in yeast
Excised linear introns regulate growth in yeast