Featured
-
-
Article
| Open Access Inducing multiple nicks promotes interhomolog homologous recombination to correct heterozygous mutations in somatic cells
Inducing multiple nicks promotes interhomolog homologous recombination to correct heterozygous mutations in somatic cells
Gene editing is still hampered by unintended genomic alterations. Here the authors propose a method for correcting heterozygous mutations that employs multiple nicks induced by Cas9 nickase and the homologous chromosome as an endogenous repair template (NICER): this rarely induces unintended genomic alterations.
- Akiko Tomita
- , Hiroyuki Sasanuma
- & Shinichiro Nakada
-
Article
| Open Access tgCRISPRi: efficient gene knock-down using truncated gRNAs and catalytically active Cas9
tgCRISPRi: efficient gene knock-down using truncated gRNAs and catalytically active Cas9
CRISPRi is used for gene silencing in mammalian cells. Here the authors report a gene-suppression/activation strategy using active Cas9 complexed with truncated gRNAs (tgCRISPRi/a) without causing DNA cleavage: they use this to repress or activate expression of several target genes throughout somatic tissues in Drosophila melanogaster.
- Ankush Auradkar
- , Annabel Guichard
- & Ethan Bier
-
Article
| Open Access A split and inducible adenine base editor for precise in vivo base editing
A split and inducible adenine base editor for precise in vivo base editing
TadA deaminases widely used in many base editors lack post-translational control in cells. Here the authors report a split adenine base editor (sABE) using chemically induced dimerisation (CID) to control the catalytic activity of TadA8e and show this can be used for PCSK9 gene editing in the mouse liver.
- Hongzhi Zeng
- , Qichen Yuan
- & Xue Gao
-
Article
| Open Access A strategy for Cas13 miniaturization based on the structure and AlphaFold
A strategy for Cas13 miniaturization based on the structure and AlphaFold
Small Cas enzymes are required for therapeutic use. Here the authors report an Interaction, Dynamics and Conservation (IDC) strategy for protein miniaturisation and use this to generate five compact variants of Cas13 based on a combination of IDC strategy and AlphaFold2.
- Feiyu Zhao
- , Tao Zhang
- & Zhanjun Li
-
Article
| Open Access A generalizable Cas9/sgRNA prediction model using machine transfer learning with small high-quality datasets
A generalizable Cas9/sgRNA prediction model using machine transfer learning with small high-quality datasets
Current bacterial sgRNA activity models struggle with accurate predictions and generalizations. Here the authors report crisprHAL, a machine learning architecture that can be trained on existing datasets, and shows good sgRNA activity prediction accuracy can generalize predictions to different bacteria.
- Dalton T. Ham
- , Tyler S. Browne
- & David R. Edgell
-
Article
| Open Access A Type II-B Cas9 nuclease with minimized off-targets and reduced chromosomal translocations in vivo
A Type II-B Cas9 nuclease with minimized off-targets and reduced chromosomal translocations in vivo
SpCas9 unintended editing is a major concern. Here the authors report an off-target method using Duplex Sequencing with increased sensitivity for Cas9 mutation detection; they also identify a Cas9 variant of the II-B subfamily with intrinsic high fidelity (PsCas9) and see improved specificity.
- Burcu Bestas
- , Sandra Wimberger
- & Marcello Maresca
-
Article
| Open Access Programmable RNA detection with CRISPR-Cas12a
Programmable RNA detection with CRISPR-Cas12a
Cas12a is widely used in diagnostic platforms. Here the authors show that Cas12a can be programmed to directly detect RNA substrates, this is due to the 3’-end of the crRNA tolerating both RNA and DNA substrates: they use this to report a method, SAHARA, to detect RNA sequences.
- Santosh R. Rananaware
- , Emma K. Vesco
- & Piyush K. Jain
-
Article
| Open Access Prediction of base editor off-targets by deep learning
Prediction of base editor off-targets by deep learning
Base editors can induce unwanted off-target effects. Here the authors design libraries of gRNA-off-target pairs and perform a screen to obtain editing efficiencies for ABE and CBE: they use the datasets to train DL models (ABEdeepoff and CBEdeepoff) which can predict mutation tolerance at potential off-targets.
- Chengdong Zhang
- , Yuan Yang
- & Yongming Wang
-
Article
| Open Access One-step generation of tumor models by base editor multiplexing in adult stem cell-derived organoids
One-step generation of tumor models by base editor multiplexing in adult stem cell-derived organoids
CRISPR base editing technologies can be used for disease modelling. Here the authors use various base editing tools to generate tumour models in human adult stem cell-derived hepatocyte, endometrial and intestinal organoids.
- Maarten H. Geurts
- , Shashank Gandhi
- & Hans Clevers
-
Article
| Open Access Treatment of monogenic and digenic dominant genetic hearing loss by CRISPR-Cas9 ribonucleoprotein delivery in vivo
Treatment of monogenic and digenic dominant genetic hearing loss by CRISPR-Cas9 ribonucleoprotein delivery in vivo
Liposome-mediated gene editing was used to abolish a mutation in gene Atp2b2 and recover hearing in a mouse model of dominant deafness. Editing was also used to target two mutations to recover hearing. The study detected large deletions due to editing.
- Yong Tao
- , Veronica Lamas
- & Zheng-Yi Chen
-
Article
| Open Access Simultaneous inhibition of DNA-PK and Polϴ improves integration efficiency and precision of genome editing
Simultaneous inhibition of DNA-PK and Polϴ improves integration efficiency and precision of genome editing
Low efficiency of target DNA integration remains a challenge in genome engineering. Here the authors perform large-scale compound library and genetic screens to identify targets that enhance gene editing: they see that combined DNA-PK and Polϴ inhibition with potent compounds increases editing efficiency and precision.
- Sandra Wimberger
- , Nina Akrap
- & Marcello Maresca
-
Article
| Open Access Cas9-mediated knockout of Ndrg2 enhances the regenerative potential of dendritic cells for wound healing
Cas9-mediated knockout of Ndrg2 enhances the regenerative potential of dendritic cells for wound healing
Chronic wounds impose a significant burden to a broad patient population. Here, the authors use CRISPR/Cas9 to enhance the regenerative capacity of dendritic cells by knocking out the gene Ndrg2, and show that seeding these engineered dendritic cells on hydrogels constitutes an effective therapy for chronic wounds in diabetic and non-diabetic conditions.
- Dominic Henn
- , Dehua Zhao
- & Geoffrey C. Gurtner
-
Article
| Open Access Sequential roles for red blood cell binding proteins enable phased commitment to invasion for malaria parasites
Sequential roles for red blood cell binding proteins enable phased commitment to invasion for malaria parasites
Malaria parasites invade erythrocytes to proliferate, but visualizing this rapid process is challenging. Here the authors use live imaging and genome-editing of P. knowlesi to dissect invasion and establish the roles of two vital parasite proteins.
- Melissa N. Hart
- , Franziska Mohring
- & Robert W. Moon
-
Article
| Open Access Insertion sequence transposition inactivates CRISPR-Cas immunity
Insertion sequence transposition inactivates CRISPR-Cas immunity
CRISPR-Cas immunity systems safeguard prokaryotic genomes by inhibiting the invasion of mobile genetic elements. Here, the authors show that insertion sequences can efficiently insert into cas genes, thus inactivating CRISPR defenses and increasing bacterial susceptibility to foreign DNA invasion.
- Yong Sheng
- , Hengyu Wang
- & Qianjin Kang
-
Article
| Open Access Cell cycle arrest and p53 prevent ON-target megabase-scale rearrangements induced by CRISPR-Cas9
Cell cycle arrest and p53 prevent ON-target megabase-scale rearrangements induced by CRISPR-Cas9
ON-target genotoxicity in gene editing is generally underestimated. Here the authors report Fluorescence-Assisted Megabase-scale Rearrangements Detection (FAMReD) systems to detect and characterize rare large loss of heterozygosity: they show that ON-target genotoxicity can be prevented by p53 and cell cycle arrest.
- G. Cullot
- , J. Boutin
- & A. Bedel
-
Article
| Open Access Striated muscle-specific base editing enables correction of mutations causing dilated cardiomyopathy
Striated muscle-specific base editing enables correction of mutations causing dilated cardiomyopathy
Dilated cardiomyopathy is the second most common cause for heart failure. Here the authors combine CRISPR base editors with the muscle-targeting viral vector AAVMYO to repair patient mutations in the cardiac splice factor Rbm20 in two mouse models.
- Markus Grosch
- , Laura Schraft
- & Lars M. Steinmetz
-
Article
| Open Access Dynamic interplay between target search and recognition for a Type I CRISPR-Cas system
Dynamic interplay between target search and recognition for a Type I CRISPR-Cas system
The details of CRISPR-Cas target search are unresolved. Here the authors analyse the target search process of the Type I CRISPR-Cas complex Cascade: they show that target search and target recognition are tightly linked, and DNA supercoiling and limited 1D diffusion play a role.
- Pierre Aldag
- , Marius Rutkauskas
- & Ralf Seidel
-
Article
| Open Access Tumour mutations in long noncoding RNAs enhance cell fitness
Tumour mutations in long noncoding RNAs enhance cell fitness
The role of mutations within long noncoding RNAs (lncRNAs) exons on tumour cell fitness remains to be explored. Here, the authors investigate the landscape of driver lncRNAs in primary and metastatic samples and validate the functional significance of single nucleotide variants in the NEAT1 oncogene in vitro and in vivo.
- Roberta Esposito
- , Andrés Lanzós
- & Rory Johnson
-
Article
| Open Access Template-jumping prime editing enables large insertion and exon rewriting in vivo
Template-jumping prime editing enables large insertion and exon rewriting in vivo
Retrotransposons replicate their genetic information through target-primed reverse transcription (TPRT). Here the authors report a template-jumping prime editor (TJ-PE) to act similarly to TPRT and achieve insertions of large DNA fragments at endogenous sites: they rewrite a mutated exon in the mouse liver.
- Chunwei Zheng
- , Bin Liu
- & Wen Xue
-
Article
| Open Access CasTuner is a degron and CRISPR/Cas-based toolkit for analog tuning of endogenous gene expression
CasTuner is a degron and CRISPR/Cas-based toolkit for analog tuning of endogenous gene expression
Understanding dosage-sensitive processes requires quantitative modulation of protein abundance. Here the authors report a CRISPR-based methodology for analog tuning of endogenous gene expression, CasTuner, and show homogeneous gene expression tuning across mouse and human cells.
- Gemma Noviello
- , Rutger A. F. Gjaltema
- & Edda G. Schulz
-
Article
| Open Access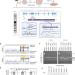 Increased renal elimination of endogenous and synthetic pyrimidine nucleosides in concentrative nucleoside transporter 1 deficient mice
Increased renal elimination of endogenous and synthetic pyrimidine nucleosides in concentrative nucleoside transporter 1 deficient mice
Concentrative nucleoside transporters (CNTs) are cellular nucleoside influx systems, but their in vivo roles are poorly defined. By generating CNT1 knockout (KO) mice, here the authors show a role of CNT1 in the renal reabsorption of endogenous and synthetic nucleosides.
- Avinash K. Persaud
- , Matthew C. Bernier
- & Rajgopal Govindarajan
-
Article
| Open Access HMGN1 enhances CRISPR-directed dual-function A-to-G and C-to-G base editing
HMGN1 enhances CRISPR-directed dual-function A-to-G and C-to-G base editing
Limited work has been done on concurrent C-to-G and A-to-G base editing. Here the authors test how a number of chromatin-associated factors affect base editing and show that HMGN1 enhanced the efficiency; by fusing HMGN1 to GBE and ABE they develop a CRISPR-based dual-function A-to-G and C-to-G base editor (GGBE).
- Chao Yang
- , Zhenzhen Ma
- & Xueli Zhang
-
Article
| Open Access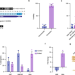 Direct correction of haemoglobin E β-thalassaemia using base editors
Direct correction of haemoglobin E β-thalassaemia using base editors
The authors demonstrate efficient and direct correction of the DNA mutation causing Haemoglobin E β-thalassaemia with CRISPR Cas9 base editors. The work includes profiling of off-target effects using deep neural networks.
- Mohsin Badat
- , Ayesha Ejaz
- & James O. J. Davies
-
Article
| Open Access ARRDC5 expression is conserved in mammalian testes and required for normal sperm morphogenesis
ARRDC5 expression is conserved in mammalian testes and required for normal sperm morphogenesis
Male organisms produce huge numbers of sperm each day, with defects in the process resulting infertility. Here they show that arrestin domain-containing 5 (ARRDC5) is a testis-specific molecule required for mammalian spermatogenesis.
- Mariana I. Giassetti
- , Deqiang Miao
- & Jon M. Oatley
-
Article
| Open Access Associate toxin-antitoxin with CRISPR-Cas to kill multidrug-resistant pathogens
Associate toxin-antitoxin with CRISPR-Cas to kill multidrug-resistant pathogens
CRISPR-regulated toxin-antitoxin (CreTA), safeguards CRISPR-Cas immune systems. Here the authors characterize a bacterial CreTA and use this to generate a proof-of-concept antimicrobial strategy, ATTACK, which associates TA and CRISPR-Cas to kill multidrug resistant pathogens.
- Rui Wang
- , Xian Shu
- & Ming Li
-
Article
| Open Access Engineered CRISPR-OsCas12f1 and RhCas12f1 with robust activities and expanded target range for genome editing
Engineered CRISPR-OsCas12f1 and RhCas12f1 with robust activities and expanded target range for genome editing
Cas12f proteins are small and sought after for therapeutic applications. Here the authors report six bacterial Cas12f1 proteins with nuclease activity in mammalian cells, perform sgRNA and protein engineering to generate variants with enhanced editing and broader PAMs, apply an inducible version in vivo.
- Xiangfeng Kong
- , Hainan Zhang
- & Hui Yang
-
Article
| Open Access Prime editing with genuine Cas9 nickases minimizes unwanted indels
Prime editing with genuine Cas9 nickases minimizes unwanted indels
Cas9 nickases (nCas9s) produce nicks or single-strand breaks in the DNA. Here the authors analyse the on- and off-target nicks generated by these nickases, and show that nCas9 (H840A) but not nCas9 (D10A) can cleave both strands and produce unwanted DNA double-strand breaks.
- Jaesuk Lee
- , Kayeong Lim
- & Jin-Soo Kim
-
Article
| Open Access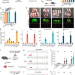 Cytosine base editors induce off-target mutations and adverse phenotypic effects in transgenic mice
Cytosine base editors induce off-target mutations and adverse phenotypic effects in transgenic mice
The potential off-target effects of long-term expression of base editors in vivo are unclear. Here the authors report SAFETI, Systematic evaluation Approach For gene Editing tools by Transgenic mIce, to examine off-target effects of base editors over time in mice, and see abnormal side effects.
- Nana Yan
- , Hu Feng
- & Erwei Zuo
-
Comment
| Open AccessA gene drive is a gene drive: the debate over lumping or splitting definitions
We address a controversy over use of the term “gene drive” to include both natural and synthetic genetic elements that promote their own transmission within a population, arguing that this broad definition is both practical and has advantages for risk analysis.
- Stephanie L. James
- , David A. O’Brochta
- & Omar S. Akbari
-
Article
| Open Access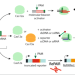 CRISPR-induced DNA reorganization for multiplexed nucleic acid detection
CRISPR-induced DNA reorganization for multiplexed nucleic acid detection
Nucleic acid sensing involving CRISPR technologies is powerful but has certain limitations, such as PAM sequence requirements and limited multiplexing. Here, authors report a CRISPR-based barcoding technology which enables multiple outputs from any target sequence, based on cis- and trans-cleavage.
- Margot Karlikow
- , Evan Amalfitano
- & Keith Pardee
-
Article
| Open Access Limitations of gene editing assessments in human preimplantation embryos
Limitations of gene editing assessments in human preimplantation embryos
DNA repair in response to DSBs in the preimplantation embryo is hard to analyze. Here the authors show that over 25% of pre-existing heterozygous loci in control single blastomere samples appeared as homozygous after whole genome amplification, therefore, they validated gene editing seen in human embryos in ESCs.
- Dan Liang
- , Aleksei Mikhalchenko
- & Shoukhrat Mitalipov
-
Article
| Open Access Improving adenine and dual base editors through introduction of TadA-8e and Rad51DBD
Improving adenine and dual base editors through introduction of TadA-8e and Rad51DBD
There is a low efficiency of A-to-G base conversion in at specific positions using base editors. Here the authors fuse ABE8e with the Rad51 DNA-binding domain to generate a hyperactive ABE allowing improved A-to-G editing efficiency at the region proximal to the PAM and improved simultaneous A/C conversion efficiency.
- Niannian Xue
- , Xu Liu
- & Xiaohui Zhang
-
Article
| Open Access CvkR is a MerR-type transcriptional repressor of class 2 type V-K CRISPR-associated transposase systems
CvkR is a MerR-type transcriptional repressor of class 2 type V-K CRISPR-associated transposase systems
RNA-guided, CRISPR-associated transposons hold great promise for precision genome editing. Here, the authors provide genetic, biochemical and structural data how their activity is regulated in situ by CvkR, an unusual MerR family regulator.
- Marcus Ziemann
- , Viktoria Reimann
- & Wolfgang R. Hess
-
Article
| Open Access DddA homolog search and engineering expand sequence compatibility of mitochondrial base editing
DddA homolog search and engineering expand sequence compatibility of mitochondrial base editing
There is a need to improve and expand mitochondrial base editing tools. Here the authors identify a DddA homolog from Simiaoa sunii (Ddd_Ss) which can efficiently deaminate cytosine in dsDNA; they develop cytosine base editors and introduce mutations at multiple mitochondrial DNA loci.
- Li Mi
- , Ming Shi
- & Yangming Wang
-
Article
| Open Access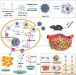 A cooperative nano-CRISPR scaffold potentiates immunotherapy via activation of tumour-intrinsic pyroptosis
A cooperative nano-CRISPR scaffold potentiates immunotherapy via activation of tumour-intrinsic pyroptosis
Delivery of immune therapy drugs to tumours might be hampered by their limited bioavailability and the difficulty of targeting complex exogenous compounds. Here authors trigger immunologic cell death, via activating tumour-cell-intrinsic pathways via CRISPR-based nanotechnology to enable efficient anti-tumour immune response in mouse models of melanoma.
- Ning Wang
- , Chao Liu
- & Changyang Gong
-
Article
| Open Access Systematically attenuating DNA targeting enables CRISPR-driven editing in bacteria
Systematically attenuating DNA targeting enables CRISPR-driven editing in bacteria
Genome editing in bacteria normally requires efficient recombination and high transformation efficiencies, which often isn’t. Here the authors report that systematically attenuating DNA targeting activity enables RecA-mediated repair in different bacteria, allowing chromosomal cleavage to drive editing.
- Daphne Collias
- , Elena Vialetto
- & Chase L. Beisel
-
Article
| Open Access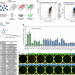 B1 SINE-binding ZFP266 impedes mouse iPSC generation through suppression of chromatin opening mediated by reprogramming factors
B1 SINE-binding ZFP266 impedes mouse iPSC generation through suppression of chromatin opening mediated by reprogramming factors
Induced pluripotent stem cell (iPSC) reprogramming is inherently inefficient. Here the authors identify 24 reprogramming roadblock genes through a CRISPR/Cas9-mediated genome-wide knockout screen including a KRAB-ZFP Zfp266, knockout of which consistently enhances murine iPSC generation.
- Daniel F. Kaemena
- , Masahito Yoshihara
- & Keisuke Kaji
-
Article
| Open Access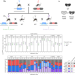 Closing the gap to effective gene drive in Aedes aegypti by exploiting germline regulatory elements
Closing the gap to effective gene drive in Aedes aegypti by exploiting germline regulatory elements
CRISPR/Cas9-based homing gene drives have emerged as a potential new approach to mosquito control. Here the authors use transgenic lines with germline-specific regulatory elements to express Cas9 and achieve up to 94% inheritance bias, closing the gap between A. aegyptidrives and the highly efficient drives observed in Anopheles species.
- Michelle A. E. Anderson
- , Estela Gonzalez
- & Luke Alphey
-
Article
| Open Access Development of a versatile nuclease prime editor with upgraded precision
Development of a versatile nuclease prime editor with upgraded precision
Strategies to improve the specificity of nuclease-based prime editor (PEn) are needed. Here the authors report a 53BP1-inhibitory ubiquitin variant-assisted PEn platform (uPEn) to inhibit NHEJ and enable precise prime editing for generation of insertions, deletions and replacements.
- Xiangyang Li
- , Guiquan Zhang
- & Xingxu Huang
-
Article
| Open Access CRISPR screens reveal genetic determinants of PARP inhibitor sensitivity and resistance in prostate cancer
CRISPR screens reveal genetic determinants of PARP inhibitor sensitivity and resistance in prostate cancer
Identifying prostate cancer patients who may respond well to PARP inhibitors is important for their success in the clinic. Here, using a genome-wide CRISPR-Cas9 knockout screen, the authors identify MMS22L as a biomarker for sensitivity to PARP inhibition in BRCA1/2-proficient prostate cancer.
- Takuya Tsujino
- , Tomoaki Takai
- & Li Jia
-
Article
| Open Access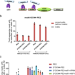 Enhancement of a prime editing system via optimal recruitment of the pioneer transcription factor P65
Enhancement of a prime editing system via optimal recruitment of the pioneer transcription factor P65
Prime editing represents a great advance to the genome editing field but is currently limited by the editing efficiency. Here the authors look to improve the efficiency by recruiting target proteins and show that the transcription factor P65 could enhance the desired editing outcomes at different gene loci.
- Ronghao Chen
- , Yu Cao
- & Xueli Zhang
-
Review Article
| Open Access Assessing and advancing the safety of CRISPR-Cas tools: from DNA to RNA editing
Assessing and advancing the safety of CRISPR-Cas tools: from DNA to RNA editing
CRISPR-Cas tools have shown exceptional promise in genome engineering over the past decade. Here the authors review the development of CRISPR-Cas9/Cas12/Cas13 nucleases, DNA base editors, prime editors, and RNA base editors, as well as their editing precision, off-target effects, and clinical considerations.
- Jianli Tao
- , Daniel E. Bauer
- & Roberto Chiarle
-
Article
| Open Access Therapeutic adenine base editing of human hematopoietic stem cells
Therapeutic adenine base editing of human hematopoietic stem cells
Here, Liao and colleagues apply adenine base editor ABE8e and its PAM-less variant ABE8e-SpRY to β-thalassemia patient hematopoietic stem cells in the form of ribonucleoprotein complexes, resulting in efficient long-term editing and β-thalassemia alleviation.
- Jiaoyang Liao
- , Shuanghong Chen
- & Yuxuan Wu
-
Article
| Open Access Genetic conversion of a split-drive into a full-drive element
Genetic conversion of a split-drive into a full-drive element
CRISPR-based gene-drives can carry the Cas9 and guide RNA (gRNA) components in a single-linked cassette or in separate elements inserted into different genomic loci. Here the authors genetically transform and compare full versus split drives, with the former performing less efficiently than predicted.
- Gerard Terradas
- , Jared B. Bennett
- & Ethan Bier
-
Article
| Open Access The effect of DNA polymorphisms and natural variation on crossover hotspot activity in Arabidopsis hybrids
The effect of DNA polymorphisms and natural variation on crossover hotspot activity in Arabidopsis hybrids
The effect of sequence polymorphisms on recombination is not fully understood. Here, the authors develop the seed-typing method to map crossovers (COs) by sequencing and show that COs occur preferentially within polymorphic hotspots and that this effect depends on the mismatch repair gene MSH2 in Arabidopsis.
- Maja Szymanska-Lejman
- , Wojciech Dziegielewski
- & Piotr A. Ziolkowski
-
Article
| Open Access A genome-wide CRISPR screen identifies WDFY3 as a regulator of macrophage efferocytosis
A genome-wide CRISPR screen identifies WDFY3 as a regulator of macrophage efferocytosis
Efferocytosis describes the engulfment and clearance of apoptotic cells by phagocytes. Here the authors identify in primary mouse macrophage WDFY3 as a regulator for efferocytosis, in which c-terminal WDFY3 is sufficient to modulate degradation while full-length WDFY3 is required to modulate the uptake of apoptotic cells.
- Jianting Shi
- , Xun Wu
- & Hanrui Zhang
-
Article
| Open Access Sequence-specific capture and concentration of viral RNA by type III CRISPR system enhances diagnostic
Sequence-specific capture and concentration of viral RNA by type III CRISPR system enhances diagnostic
Many viral diagnostic approaches require nucleic acid extraction and amplification prior to detection. Here, the authors develop a method based on type III CRISPR systems which allows sequence specific capture, concentration, and detection of viral RNA directly from patient samples.
- Anna Nemudraia
- , Artem Nemudryi
- & Blake Wiedenheft
-
Article
| Open Access PEAC-seq adopts Prime Editor to detect CRISPR off-target and DNA translocation
PEAC-seq adopts Prime Editor to detect CRISPR off-target and DNA translocation
It is still a challenge to accurately identify off-target endonuclease edits. Here the authors report PEAC-seq using a Prime Editor to insert a tag to the editing sites and enrich the tagged regions with site-specific primers for sequencing: they show that PEAC-seq could identify DNA translocations.
- Zhenxing Yu
- , Zhike Lu
- & Lijia Ma
-
Article
| Open Access The coordination of anti-phage immunity mechanisms in bacterial cells
The coordination of anti-phage immunity mechanisms in bacterial cells
Bacteria are equipped with diverse immune strategies to fight bacteriophage infections, including restriction nucleases, abortive infection and CRISPR-Cas systems. Here, Arias et al. use mathematical models of immune responses in individual bacterial cells to highlight the importance of the timing and coordination of different antiviral systems, and present hypotheses that may inspire future research.
- Clemente F. Arias
- , Francisco J. Acosta
- & Cristina Fernández-Arias

 A cleavage rule for selection of increased-fidelity SpCas9 variants with high efficiency and no detectable off-targets
A cleavage rule for selection of increased-fidelity SpCas9 variants with high efficiency and no detectable off-targets