Abstract
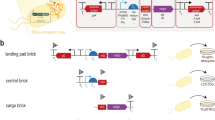
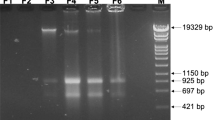
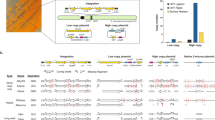
Direct cell-to-cell transfer of genomes from bacteria to yeast facilitates genome engineering for bacteria that are not amenable to genetic manipulation by allowing instead for the utilization of the powerful yeast genetic tools. Here we describe a protocol for transferring whole genomes from bacterial cells to yeast spheroplasts without any DNA purification process. The method is dependent on the treatment of the bacterial and yeast cellular mixture with PEG, which induces cell fusion, engulfment, aggregation or lysis. Over 80% of the bacterial genomes transferred in this way are complete, on the basis of structural and functional tests. Excluding the time required for preparing starting cultures and for incubating cells to form final colonies, the protocol can be completed in 3 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Wang, H.H. et al. Genome-scale promoter engineering by coselection MAGE. Nat. Methods 9, 591–593 (2012).
Noskov, V.N., Segall-Shapiro, T.H. & Chuang, R.Y. Tandem repeat coupled with endonuclease cleavage (TREC): a seamless modification tool for genome engineering in yeast. Nucleic Acids Res. 38, 2570–2576 (2010).
Gibson, D.G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010).
Itaya, M., Tsuge, K., Koizumi, M. & Fujita, K. Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc. Natl. Acad. Sci. USA 102, 15971–15976 (2005).
Benders, G.A. et al. Cloning whole bacterial genomes in yeast. Nucleic Acids Res. 38, 2558–2569 (2010).
Karas, B.J., Tagwerker, C., Yonemoto, I.T., Hutchison, C.A. III & Smith, H.O. Cloning the Acholeplasma laidlawii PG-8A genome in Saccharomyces cerevisiae as a yeast centromeric plasmid. ACS Syn. Biol. 1, 22–28 (2012).
Tagwerker, C. et al. Sequence analysis of a complete 1.66 Mb Prochlorococcus marinus MED4 genome cloned in yeast. Nucleic Acids Res. 40, 10375–10383 (2012).
Karas, B.J. et al. Direct transfer of whole genomes from bacteria to yeast. Nat. Methods 10, 410–412 (2013).
Guerra-Tschuschke, I., Martin, I. & Gonzalez, M.T. Polyethylene glycol-induced internalization of bacteria into fungal protoplasts: electron microscopic study and optimization of experimental conditions. Appl. Environ. Microbiol. 57, 1516–1522 (1991).
van Solingen, P. & van der Plaat, J.B. Fusion of yeast spheroplasts. J. Bacteriol. 130, 946–947 (1977).
Markie, D. A simple assay for optimizing yeast-mammalian cell fusion conditions. Mol. Biotechnol. 6, 99–104 (1996).
Dai, M., Ziesman, S., Ratcliffe, T., Gill, R.T. & Copley, S.D. Visualization of protoplast fusion and quantitation of recombination in fused protoplasts of auxotrophic strains of Escherichia coli. Metab. Eng. 7, 45–52 (2005).
Tarshis, M., Salman, M. & Rottem, S. Fusion of mycoplasmas: the formation of cell hybrids. FEMS Microbiol. Lett. 66, 67–71 (1991).
Akamatsu, T. & Sekiguchi, J. Selection methods in bacilli for recombinants and transformants of intra- and interspecific fused protoplasts. Arch. Microbiol. 134, 303–308 (1983).
Kao, K.N. & Michayluk, M.R. A method for high-frequency intergeneric fusion of plant protoplasts. Planta 115, 13 (1974).
Schaffner, W. Direct transfer of cloned genes from bacteria to mammalian cells. Proc. Natl. Acad. Sci. USA 77, 2163–2167 (1980).
Maehara, T., Itaya, M., Ogura, M. & Tanaka, T. Effect of Bacillus subtilis BsuM restriction-modification on plasmid transfer by polyethylene glycol-induced protoplast fusion. FEMS Microbiol. Lett. 325, 49–55 (2011).
Baigori, M., Sesma, F., de Ruiz Holgado, A.P. & de Mendoza, D. Transfer of plasmids between Bacillus subtilis and Streptococcus lactis. Appl. Environ. Microbiol. 54, 1309–1311 (1988).
Gyuris, J. & Duda, E.G. High-efficiency transformation of Saccharomyces cerevisiae cells by bacterial minicell protoplast fusion. Mol. Cell Biol. 6, 3295–3297 (1986).
Li, L. & Blankenstein, T. Generation of transgenic mice with megabase-sized human yeast artificial chromosomes by yeast spheroplast-embryonic stem cell fusion. Nat. Protoc. 8, 1567–1582 (2013).
Fleischmann, R.D. et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512 (1995).
Noskov, V.N. et al. Assembly of large, high G+C bacterial DNA fragments in yeast. ACS Syn. Biol. 1, 267–273 (2012).
Hutchison, C.A. et al. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286, 2165–2169 (1999).
Gibson, D.G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Smith, H.O., Tomb, J.F., Dougherty, B.A., Fleischmann, R.D. & Venter, J.C. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269, 538–540 (1995).
Kouprina, N. & Larionov, V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 3, 371–377 (2008).
Karas, B.J. et al. Assembly of eukaryotic algal chromosomes in yeast. J. Biol. Eng. 7, 30 (2013).
Lartigue, C. et al. Genome transplantation in bacteria: changing one species to another. Science 317, 632–638 (2007).
Lartigue, C. et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science 325, 1693–1696 (2009).
Larionov, V., Kouprina, N., Solomon, G., Barrett, J.C. & Resnick, M.A. Direct isolation of human BRCA2 gene by transformation-associated recombination in yeast. Proc. Natl. Acad. Sci. USA 94, 7384–7387 (1997).
Replogle, K., Hovland, L. & Rivier, D.H. Designer deletion and prototrophic strains derived from Saccharomyces cerevisiae strain W303-1a. Yeast 15, 1141–1149 (1999).
Tully, J.G., Whitcomb, R.F., Clark, H.F. & Williamson, D.L. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science 195, 892–894 (1977).
Poje, G. & Redfield, R.J. General methods for culturing Haemophilus influenzae. Methods Mol. Med. 71, 51–56 (2003).
Acknowledgements
This work was supported by Synthetic Genomics, Inc. B.J.K. was supported by the Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowships Program and by Synthetic Genomics, Inc. Y.S. was also supported by the US Defense Advanced Research Projects Agency contract no. N66001-12-C-4039 and the US Department of Energy cooperative agreement no. DE-EE0006109.
Author information
Authors and Affiliations
Contributions
B.J.K., J.J., P.D.W., D.G.G., J.I.G., J.C.V., C.A.H., H.O.S. and Y.S. designed the research. B.J.K., J.J., E.I., L.S., L.M., P.D.W., D.G.G., C.A.H. and Y.S. performed experiments. B.J.K., J.J., P.D.W. and Y.S. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
J.C.V. is the Chief Executive Officer and Co-Chief Scientific Officer of Synthetic Genomics, Inc. (SGI). H.O.S. is the Co-Chief Scientific Officer and a member of the Board of Directors of SGI. C.A.H. is the Chairman of the SGI Scientific Advisory Board. D.G.G. is the Vice President for DNA Technology at SGI. All these four authors and the J. Craig Venter Institute hold SGI stock.
Supplementary information
Supplementary Note: Sequence of a yeast vector cassette.
Yeast elements that need to be inserted into bacterial genomes can be amplified from the plasmid pmycYAcTn (accession number GU593054)1. The sequence of CEN6 is underlined, that of the ARS is in italics, the HIS3 promoter is in bold, and the HIS3 coding sequence is in bold and underlined. (PDF 120 kb)
Rights and permissions
About this article
Cite this article
Karas, B., Jablanovic, J., Irvine, E. et al. Transferring whole genomes from bacteria to yeast spheroplasts using entire bacterial cells to reduce DNA shearing. Nat Protoc 9, 743–750 (2014). https://doi.org/10.1038/nprot.2014.045
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.045
This article is cited by
-
The link between yeast cell wall porosity and plasma membrane permeability after PEF treatment
Scientific Reports (2019)
-
Analysis of a novel mutant allele of GSL8 reveals its key roles in cytokinesis and symplastic trafficking in Arabidopsis
BMC Plant Biology (2018)
-
Synthetic biology advances and applications in the biotechnology industry: a perspective
Journal of Industrial Microbiology and Biotechnology (2018)
-
Genome reprogramming for synthetic biology
Frontiers of Chemical Science and Engineering (2017)
-
Ecologically Driven Competence for Exogenous DNA Uptake in Yeast
Current Microbiology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.