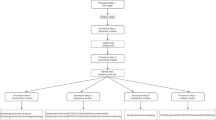
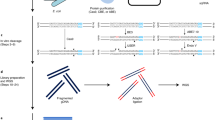
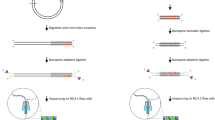
Abstract
Ribonucleotides are the most common noncanonical nucleotides incorporated into the genome of replicating cells. They are efficiently removed by ribonucleotide excision repair initiated by RNase H2 cleavage. In the absence of RNase H2, such embedded ribonucleotides can be used to track DNA polymerase activity in vivo. To determine their precise location in Saccharomyces cerevisiae, we developed embedded ribonucleotide sequencing (emRiboSeq), which uses recombinant RNase H2 to selectively create ligatable 3′-hydroxyl groups, in contrast to alternative methods that use alkaline hydrolysis. EmRiboSeq allows reproducible, strand-specific and potentially quantitative detection of embedded ribonucleotides at single-nucleotide resolution. For the genome-wide mapping of other noncanonical bases, RNase H2 can be replaced with specific nicking endonucleases in this protocol; we term this method endonuclease sequencing (EndoSeq). With the protocol taking <5 d to complete, these methods allow the in vivo study of DNA replication and repair, including the identification of replication origins and termination regions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Clausen, A.R., Zhang, S., Burgers, P.M., Lee, M.Y. & Kunkel, T.A. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase delta. DNA Repair (Amst.) 12, 121–127 (2013).
Goksenin, A.Y. et al. Human DNA polymerase epsilon is able to efficiently extend from multiple consecutive ribonucleotides. J. Biol. Chem. 287, 42675–42684 (2012).
Gosavi, R.A., Moon, A.F., Kunkel, T.A., Pedersen, L.C. & Bebenek, K. The catalytic cycle for ribonucleotide incorporation by human DNA Pol lambda. Nucleic Acids Res. 40, 7518–7527 (2012).
Kasiviswanathan, R. & Copeland, W.C. Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J. Biol. Chem. 286, 31490–31500 (2011).
Nick McElhinny, S.A. et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA 107, 4949–4954 (2010).
Nick McElhinny, S.A. et al. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 6, 774–781 (2010).
Nick McElhinny, S.A. & Ramsden, D.A. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 23, 2309–2315 (2003).
Reijns, M.A. et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149, 1008–1022 (2012).
Yao, N.Y., Schroeder, J.W., Yurieva, O., Simmons, L.A. & O'Donnell, M.E. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc. Natl. Acad. Sci. USA 110, 12942–12947 (2013).
Caldecott, K.W. Molecular biology. Ribose—an internal threat to DNA. Science 343, 260–261 (2014).
Dalgaard, J.Z. Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet. 28, 592–597 (2012).
Potenski, C.J. & Klein, H.L. How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 42, 10226–10234 (2014).
Vaisman, A. & Woodgate, R. Redundancy in ribonucleotide excision repair: competition, compensation, and cooperation. DNA Repair (Amst.) 29, 74–82 (2015).
Wallace, B.D. & Williams, R.S. Ribonucleotide triggered DNA damage and RNA-DNA damage responses. RNA Biol. 11, 1340–1346 (2014).
Williams, J.S. & Kunkel, T.A. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst.) 19, 27–37 (2014).
Martin, M.J., Garcia-Ortiz, M.V., Esteban, V. & Blanco, L. Ribonucleotides and manganese ions improve non-homologous end joining by human Polmu. Nucleic Acids Res. 41, 2428–2436 (2013).
Ghodgaonkar, M.M. et al. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol. Cell 50, 323–332 (2013).
Lujan, S.A., Williams, J.S., Clausen, A.R., Clark, A.B. & Kunkel, T.A. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol. Cell 50, 437–443 (2013).
Allen-Soltero, S., Martinez, S.L., Putnam, C.D. & Kolodner, R.D. A Saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol. Cell. Biol. 34, 1521–1534 (2014).
Cho, J.E., Kim, N., Li, Y.C. & Jinks-Robertson, S. Two distinct mechanisms of topoisomerase 1-dependent mutagenesis in yeast. DNA Repair (Amst.) 12, 205–211 (2013).
Gunther, C. et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Invest. 125, 413–424 (2015).
Hiller, B. et al. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J. Exp. Med. 209, 1419–1426 (2012).
Kalhorzadeh, P. et al. Arabidopsis thaliana RNase H2 deficiency counteracts the needs for the WEE1 checkpoint kinase but triggers genome instability. Plant Cell 26, 3680–3692 (2014).
Kim, N. et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332, 1561–1564 (2011).
Pizzi, S. et al. Reduction of hRNase H2 activity in Aicardi-Goutières syndrome cells leads to replication stress and genome instability. Hum. Mol. Genet. 24, 649–658 (2015).
Crow, Y. et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 38, 910–916 (2006).
Reijns, M.A. & Jackson, A.P. Ribonuclease H2 in health and disease. Biochem. Soc. Trans. 42, 717–725 (2014).
Schellenberg, M.J., Tumbale, P.P. & Williams, R.S. Molecular underpinnings of aprataxin RNA/DNA deadenylase function and dysfunction in neurological disease. Prog. Biophys. Mol. Biol. 117, 157–165 (2015).
Tumbale, P., Williams, J.S., Schellenberg, M.J., Kunkel, T.A. & Williams, R.S. Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity. Nature 506, 111–115 (2014).
Reijns, M.A. et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature 518, 502–506 (2015).
Sparks, J.L. et al. RNase H2-initiated ribonucleotide excision repair. Mol. Cell 47, 980–986 (2012).
Williams, J.S. et al. Evidence that processing of ribonucleotides in DNA by topoisomerase 1 is leading-strand specific. Nat. Struct. Mol. Biol. 22, 291–297 (2015).
Clausen, A.R. et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat. Struct. Mol. Biol. 22, 185–191 (2015).
Daigaku, Y. et al. A global profile of replicative polymerase usage. Nat. Struct. Mol. Biol. 22, 192–198 (2015).
Koh, K.D., Balachander, S., Hesselberth, J.R. & Storici, F. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nat. Methods 12, 251–257 (2015).
Koh, K.D., Hesselberth, J. & Storici, F. Ribose-seq: ribonucleotides in DNA to Illumina library. Protoc. Exchange 10.1038/protex.2015.044 (19 May 2015).
Jinks-Robertson, S. & Klein, H.L. Ribonucleotides in DNA: hidden in plain sight. Nat. Struct. Mol. Biol. 22, 176–178 (2015).
Bailly, V., Derydt, M. & Verly, W.G. Delta-elimination in the repair of AP (apurinic/apyrimidinic) sites in DNA. Biochem. J. 261, 707–713 (1989).
Bryan, D.S., Ransom, M., Adane, B., York, K. & Hesselberth, J.R. High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. 24, 1534–1542 (2014).
Potenski, C.J., Niu, H., Sung, P. & Klein, H.L. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature 511, 251–254 (2014).
Vaisman, A. et al. Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair. PLoS Genet. 9, e1003878 (2013).
Williams, J.S. et al. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell 49, 1010–1015 (2013).
Clark, A.B., Lujan, S.A., Kissling, G.E. & Kunkel, T.A. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair (Amst.) 10, 476–482 (2011).
Huang, M.E., Rio, A.G., Nicolas, A. & Kolodner, R.D. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100, 11529–11534 (2003).
Lazzaro, F. et al. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell 45, 99–110 (2012).
Reijns, M.A. et al. The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. J. Biol. Chem. 286, 10530–10539 (2011).
Acknowledgements
We thank A. Gallacher for technical assistance throughout the development of this protocol and T. Kunkel (National Institute of Environmental Health Sciences) for sharing yeast strains. This work was supported by funding from the Medical Research Council (MRC; Centenary Award to M.A.M.R.), the MRC and Medical Research Foundation (to M.S.T.) and the MRC and Lister Institute of Preventive Medicine (to A.P.J.).
Author information
Authors and Affiliations
Contributions
M.A.M.R., M.S.T. and A.P.J. conceived and designed the original protocol. M.A.M.R. and J.D. modified and updated the protocol to its current state. M.S.T. performed all computational analyses. J.D. and M.A.M.R. wrote the manuscript with assistance from M.S.T. and A.P.J.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ding, J., Taylor, M., Jackson, A. et al. Genome-wide mapping of embedded ribonucleotides and other noncanonical nucleotides using emRiboSeq and EndoSeq. Nat Protoc 10, 1433–1444 (2015). https://doi.org/10.1038/nprot.2015.099
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.099
This article is cited by
-
G-quadruplex and 8-oxo-7,8-dihydroguanine across the genome: methodologies and crosstalk
Genome Instability & Disease (2022)
-
Signatures of TOP1 transcription-associated mutagenesis in cancer and germline
Nature (2022)
-
Genome-wide mapping of genomic DNA damage: methods and implications
Cellular and Molecular Life Sciences (2021)
-
Manual and automated preparation of single-stranded DNA libraries for the sequencing of DNA from ancient biological remains and other sources of highly degraded DNA
Nature Protocols (2020)
-
Next-generation sequencing approaches for the study of genome and epigenome toxicity induced by sulfur mustard
Archives of Toxicology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.