Abstract
In higher plants, jasmonate ZIM-domain (JAZ) proteins negatively regulate the biosynthesis of anthocyanins by interacting with bHLH transcription factors. However, it is largely unknown if and how other regulators are involved in this process. In this study, the apple MdJAZ2 protein was characterized in regards to its function in the negative regulation of anthocyanin accumulation and peel coloration. MdJAZ2 was used as a bait to screen a cDNA library using the yeast two-hybrid method. The hypersensitive induced reaction (HIR) proteins, MdHIR2 and MdHIR4, were obtained from this yeast two-hybrid. The ZIM domain of MdJAZ2 and the PHB domain of the MdHIR proteins are necessary for their interactions. The interactions were further verified using an in vitro pull-down assay. Subsequently, immunoblotting assays demonstrated that MdHIR4 enhanced the stability of the MdJAZ2-GUS protein. Finally, a viral vector-based transformation method showed that MdHIR4 inhibited anthocyanin accumulation and fruit coloration in apple by modulating the expression of genes associated with anthocyanin biosynthesis.
Similar content being viewed by others
Introduction
Fruit color is often a main indicator used to evaluate the fruit’s economic value. In many fruits, such as apple, grapevine and peach, anthocyanins play a crucial role in the coloration of the peel and the flesh. Anthocyanins are derivatives of glucosides. They belong to the flavonoid compound family and ubiquitously exist in the vacuole of cells in the flower, fruit, root, stem, and leaf. There are generally six anthocyanin pigments, cyanidin, delphinidin, pelargonidin, peonidin, petunidin and malvidin, found in fruits. Cyanidin is the most common pigment and is produced in over 82% of researched fruits and berries1. Delphinidin and its methylated derivatives, such as petunidins and malvidins, appear as dark bluish and purple colors, but cyanidins and pelargonidins are the main pigments in bright red-colored fruits1. There are other factors, such as co-pigmentation and pH, which affect the perceived hue of a tissue2.
In higher plants, including apple and other fruit trees, the main biosynthesis pathway of anthocyanins is the flavonoid pathway3. The synthetic enzymes and their encoding genes, also called structural genes, have already been extensively identified and investigated4. As is well known, the expression of the structural genes are regulated by various transcription factors (TFs) such as HY5 and MYB-bHLH-WD40 (MBW)5,6,7.
MYB TFs are the most intensively studied components of the MBW complex associated with anthocyanin biosynthesis in fruit trees8,9,10. In apple, MdMYB10, MdMYB1, MdMYB9, MdMYB11, MdMYB110a, MdMYB3 and MdMYBA activate the expression of the anthocyanin structural genes, while MdMYB15, MdMYB16, MdMYB17, MdMYB27, MdMYB28, MdMYB49, MdMYB50 and MdMYB111 repress them10,11,12,13,14,15. Furthermore, MdMYB1 directly regulates the expression of the MdDFR and MdUFGT genes16. In addition to the MYB TFs, another type of MBW TFs, known as bHLH or MYC, are involved in the regulation of the anthocyanin structural genes. MdbHLH3 and MdbHLH33 not only directly regulate the expression of the structural genes but also bind to the promoters of MYB genes, such as MdMYB1, MdMYB9, MdMYB10, MdMYB11, and modulate their transcription11,12,15. A WD repeat protein, MdTTG1, which is homologous to Arabidopsis TRANSPARENT TESTA GLABRA1, is also involved in the regulation of anthocyanin accumulation in apple17,18.
Various environmental factors, such as light and temperature, influence the biosynthesis of anthocyanins19. The MBW complex also works as regulatory machinery for anthocyanin accumulation in response to various developmental and environmental cues. In apple, MdMYB1 and MdbHLH3, MBW components, are regulated at transcriptional and posttranslational levels to modulate anthocyanin accumulation in response to light and temperature12,20. In addition, plant hormones such as auxin, ethylene, gibberellin, jasmonic acid (JA), cytokinins and abscisic acid modulate anthocyanin synthesis21,22,23. Jasmonate ZIM-domain (JAZ) proteins are the key regulators in the JA signaling pathway24. In the classic Arabidopsis model of the JA signaling pathway, when the JA level is relatively low JAZ proteins accumulate and interact with the bHLH TF MYC2, consequently attenuating the expression of the downstream JA-responsive genes25. When the JA level is relatively higher, however, the receptor SCF-COI1 complex binds with the JAZ protein to form the new SCF-COI1-JAZ complex26. The JAZ protein is then ubiquitinated by E3 ubiquitin ligase and degraded by 26S proteasome27. Finally, JA responsive genes are transcribed28,29.
In plants, JAs are crucial hormones involved in plant development and growth, secondary metabolism and defense against biotic and abiotic stresses30. The JAZ proteins play a principal role in various functions of JA. For example, they interact with the R2R3-MYB TFs, such as MYB21 and MYB24, to affect JA-regulated stamen development in Arabidopsis31. They also interact with the bHLH TFs, MYC2, MYC3 and MYC4, to repress JA-induced defense against bacterial pathogens and insect herbivory32.
In Arabidopsis, the JAs also induce anthocyanin biosynthesis by up-regulating TF genes, such as PAP1, PAP2 and GL3, which modulate the expression of the anthocyanin biosynthetic genes DFR, LDOX and UF3GT33. Furthermore, the JAZ proteins interact with the bHLHs, such as GL3, EGL3, and TT8, and with the MYB TFs, such as MYB75 and GL1, which regulate the expression of the MBW complexes associated with anthocyanin biosynthesis, attenuating their transcriptional function and, thereby, inhibiting anthocyanin biosynthesis34. In apple, MdbHLH3 promotes anthocyanin biosynthesis by directly upregulating the expression of not only structural genes, such as MdDFR and MdUFGT but also regulatory MYB genes, including MdMYB1, MdMYB9 and MdMYB11. Additionally, MdJAZ2 interacts with MdbHLH3 and inhibits its transcriptional function, thereby, working as a negative regulator for anthocyanin biosynthesis12,15.
In Arabidopsis, a few of proteins are characterized by their interaction with the JAZ proteins, which influences their functions. DELLA proteins compete with MYC2 for binding to the JAZs to promote the expression of downstream genes35. JAZ proteins directly interact with EIN3/EIL1 and recruit HDA6 to suppress the transcriptional activity of EIN3/EIL1 to regulate plant development and defense responses36. In this study, a yeast two hybridization approach was conducted using MdJAZ2 as bait to screen through an apple fruit peel cDNA library. Hypersensitive induced reaction (HIR) protein, MdHIR2 and MdHIR4, were found to interact with MdJAZ2. The interaction of the MdHIRs with the MdJAZ proteins and their function in the regulation of anthocyanin biosynthesis was verified and characterized. Our findings provide evidence that the MdHIRs acts upstream to the MdJAZ2-MdbHLH3 pathway to negatively regulate anthocyanin accumulation in apple.
Results
MdJAZ2 inhibits anthocyanin accumulation in apple peel
To examine if MdJAZ2 influences anthocyanin accumulation and peel coloration in apple fruit, a viral vector-based transient transformation method was conducted to enhance the expression level of MdJAZ2 in the apple peel20. The viral overexpression vector pIR-MdJAZ2, plus a helper plasmid, IL-60-BS, were injected into the fruit peel of cultivar ‘Red delicious,’ while the empty vector pIR, plus IL-60-BS, served as the control. The expression analysis demonstrated that pIR-MdJAZ2 injection noticeably enhanced the expression of MdJAZ2 gene in apple fruit peel than the pIR control (Fig. 1A). Subsequently, anthocyanin content was measured in the fruit peel around the injection sites. Anthocyanin content in the apple peel injected with pIR-MdJAZ2 was much lower than in the apple peel injected with the empty control (Fig. 1B). As a result, the injection of pIR-MdJAZ2 resulted in a loss of red colouration in the apple skin, compared with the empty control (Fig. 1C).
(A) Quantitative real-time PCR analysis of the expression of MdJAZ2 in the fruit peel surrounding the injection sites. (B) The anthocyanin content of the fruit peel around the injection sites. Anthocyanins were extracted from 8 pieces fruit skin, 1 cm2 in size for each from one individual fruit. (C) The phenotype of the peel surrounding the injection sites. The apple fruit were injected with a viral based overexpression vector, pIR-MdJAZ2, while the empty vector, pIR, was used as the control. The injected apples were kept in an illumination incubator under white light at 17 °C for 4 days. (D) Quantitative real-time PCR analysis of the regulatory and structural genes associated with anthocyanin biosynthesis in the fruit peel around the injection sites. The data are shown as the mean ± SE, which were calculated based on 3 replicates. Mean differences in the bars are significant at P0.05 level with different letters, not significant at P0.05 level with the same letters. Repeat in following figure.
We previously reported that MdJAZ2 interacts with MdbHLH3 and inhibits the expression of MdMYB1, MdMYB9, MdMYB11 and their downstream anthocyanin structural target genes15. Therefore, the transcript levels of these genes were examined with real-time quantitative RT-PCR in the apple peel around the injection sites. The results indicated that MdJAZ2 transient overexpression slightly influenced the expression of MdbHLH3, but significantly repressed the transcript levels of MdMYB1, MdMYB9 and MdMYB11 (Fig. 1D). Consequently, the reduced expression of these genes repressed the expression levels of the anthocyanin structural genes to different degrees. Affected genes included MdCHS, MdCHI, MdANR, MdDFR, MdUFGT, MdF3H, MdANS and MdFLS (Fig. 1D).
Therefore, MdJAZ2 functions as a negative regulator of anthocyanin accumulation and fruit coloration by repressing the expression of the anthocyanin regulatory and structural genes in apple.
Yeast two-hybrid (Y2H) screening of an apple peel cDNA library reveals a putative MdJAZ2-interacting protein, MdHIR2
Y2H screening was conducted to screen MdJAZ2-interacting proteins. As a result, from a library with a primary library titer of 8.16 × 106 cfu/mL of which 95% of the clones had inserts, five positive colonies were obtained. Among the positive colonies a target cDNA was found that corresponds to the gene MDP0000653461.This gene encodes a putative protein that is similar to the hypersensitive induced reaction (HIR) proteins in Arabidopsis. There are a total of four HIR genes in Arabidopsis, AtHIR1, AtHIR2, AtHIR3 and AtHIR4. In addition to MDP0000653461, four more MdHIR genes were found in the apple genome, MDP0000295316, MDP0000630084, MDP0000138908 and MDP0000122340. Reverse Transcription PCR (RT-PCR) and sequence analysis demonstrated that all five apple MdHIR genes transcribed cDNAs that encode putative MdHIR proteins (Fig. S1). The predicted MdHIR proteins were used for sequence alignment analysis with AtHIRs. The sequence alignment analysis showed that the five predicted MdHIR proteins are highly similar in amino acid sequence to the AtHIRs (Fig. 2A).
(A) Comparison of the predicted amino acid sequences of the HIR proteins in apple and Arabidopsis. The amino acid consensus sequences are highlighted in black. Md, Malus domestica; At, Arabidopsis thaliana (AtHIR1, AT5G62740; AtHIR2, AT1G69840; AtHIR3, AT3G01290; AtHIR4, AT5G51570). The HIR proteins contained highly conserved PHB (prohibitin homologues) domains, which are indicated as a red line. (B) A phylogenetic tree of the HIR proteins in Malus domestica and Arabidopsis.
In addition, protein domain searches were performed using the programs PFAM and SMART. These searches found that the five predicted MdHIR proteins contain an SPFH/Band7/PHB domain, which is highly conserved in the four AtHIRs (Fig. 2A). Subsequently, a phylogenetic tree was constructed using the four AtHIRs and the five MdHIRs. Based on the phylogenetic tree, the five MdHIRs were named MdHIR1–1 (MDP0000295316), MdHIR1-2 (MDP0000630084), MdHIR2 (MDP0000653461), MdHIR3 (MDP0000138908) and MdHIR4 (MDP0000122340) (Fig. 2B).
MdHIR2 and MdHIR4 interact with MdJAZ proteins
To verify the interaction between the MdHIRs and the MdJAZs, yeast two-hybrid assays were carried out. The full-length cDNA of each of the MdHIRs was inserted into the vector pGBT9 (BD-MdHIRs) as bait. The full-length CDSs of MdJAZ1, MdJAZ2, MdJAZ3, MdJAZ4, MdJAZ5, MdJAZ6 and MdJAZ8 were cloned with RT-PCR, and then inserted into the vector pGAD424 (AD-MdJAZs) as prey. Subsequently, each combination of the BD-MdHIRs and the AD-MdJAZs was co-expressed in yeast cells. The transformants were cultured on -Trp/-Leu/-His/-Ade screening medium and stained with X-α-gal. Yeast strains containing BD-MdHIR2 plus AD-MdJAZ1, AD-MdJAZ2 or AD-MdJAZ4, and BD-MdHIR4 plus all seven of the AD-MdJAZs were positive for X-α-gal activity when grown on -Trp/-Leu/-His/-Ade screening medium. Yeast strains containing BD-MdHIRs or BD-MdHIRs plus the empty pGAD424 vector were negative for X-α-gal activity. Therefore, MdHIR2 interacted with MdJAZ1, MdJAZ2 and MdJAZ4, while MdHIR4 interacted with all seven of the MdJAZs tested (Fig. 3A). Interestingly, it was also found that MdHIR4 interacted with itself and four other MdHIR proteins (Fig. 3B).
(A) Yeast two-hybrid assays for the interactions between the MdJAZs and the MdHIRs. Seven apple MdJAZs were fused with the GAL4 activation domain (AD) in the pGAD424 vector, while 5 MdHIRs were fused with the GAL4 DNA binding domain (BD) in the pGBT9 vector. Interactions of the MdHIR proteins with the AD domain in the pGAD424 empty vector were used as the negative controls. Interactions are indicated by the blue color on SD/-Ade/-His/-Trp/-Leu/X-Gal medium. (B) Homo- and heterodimerization between the MdHIR4 protein, MdHIR4-N and the MdHIR1-1, MdHIR1-2, MdHIR2, MdHIR3 or MdHIR4 proteins. (C) Schematic diagram of the protein structure of MdJAZ2. The diagram shows the conserved ZIM and Jas domains. The domains found in MdJAZ2 were fused with the AD domain. (D) Schematic diagram of the protein structures for the MdHIR proteins. The diagram shows the conserved PHB domain. Different parts of MdHIR2 and MdHIR4 were fused with the BD domain. (E,F) Yeast two-hybrid assays of the interactions between the different domains of the MdHIR and MdJAZ2 proteins. (G,H) In vitro pull-down assays of the interaction of the MdJAZ2-His and MdHIR-GST proteins. Purified recombinant MdJAZ2-His protein was used to pull down MdHIR2-GST or MdHIR4-GST protein. The purified His tag was used to pull down the MdHIR-GST proteins as the negative control.
To determine which region of the MdHIR proteins is necessary for interaction with the MdJAZ proteins, MdHIR2 and MdHIR4 were divided into the N-terminal PHB domain (MdHIR2N, MdHIR4N) and the C-terminus (MdHIR2C, MdHIR4C), while MdJAZ2 was divided into the N-terminal ZIM domain (MdJAZ2ZIM) and the C-terminal Jas domain (MdJAZ2Jas). The corresponding cDNA fragments of MdHIR2N and MdHIR2C were inserted into pGBT9 (BD-MdHIR2N, BD-MdHIR4N and BD-MdHIR2C, BD-MdHIR4C) as baits, while MdJAZ2ZIM and MdJAZ2Jas were inserted into pGAD424 (AD-MdJAZ2ZIM and AD-MdJAZ2Jas) as preys. Yeast two-hybrid assays demonstrated that the N-terminal ZIM domain of the MdJAZ2 protein is necessary for its interaction with MdHIR2 and MdHIR4, while the N-terminal PHB domain of the MdHIR proteins is required for interaction with MdJAZ2 (Fig. 3C–F).
To further verify the interactions between MdHIR2, MdHIR4 and MdJAZ2 an in vitro pull-down assay was performed with MdHIR2-GST, MdHIR4-GST and MdJAZ2-HIS proteins that were expressed in and purified from the Escherichia coli strain BL21. The results showed that MdHIR2-GST and MdHIR4-GST, but not GST alone, interacted with the MdJAZ2-HIS protein (Fig. 3G,H).
Additionally, yeast two-hybrid and pull-down assays were also performed to examine the interactions between the four AtHIRs and the twelve AtJAZs in Arabidopsis. The yeast two-hybrid assays indicated that either AtHIR1 or AtHIR4 interacted with AtJAZ3, AtJAZ4 or AtJAZ9, while either AtHIR2 or AtHIR3 interacted with only AtJAZ3 (Fig. S2A). Subsequently, pull down assays confirmed the interaction between AtHIR1 and AtJAZ3 or AtJAZ9 (Fig. S2B,C).
MdHIR4 enhances the stability of the MdJAZ2 protein in apple callus
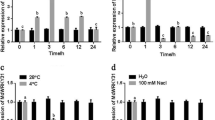
To explore how MdHIR4 influences MdJAZ2, apple callus was transformed using an Agrobacterium-mediated method. Two constructs, pRI101-35S::MdJAZ2-GUS and pRI101-35S::MdHIR4, were made and used for transformation, while the empty vector, pRI101-35S::GUS, was used as a control. As a result, three types of transgenic apple calli, 35S::GUS, 35S::MdJAZ2-GUS and 35S::MdJAZ2-GUS+35S::MdHIR4 were obtained. Quantitative real-time PCR was conducted to verify that the genes were expressed in the corresponding transgenic calli. The results showed that the 35S::MdJAZ2-GUS transgenic callus generated more MdJAZ2 transcripts, and 35S::MdJAZ2-GUS+35S::MdHIR4 transgenic callus exhibited higher levels of MdJAZ2 and MdHIR4 transcripts, than the 35S::GUS control (Fig. 4A,B). In addition, the expression level of MdJAZ2 was similar in 35S::MdJAZ2-GUS and 35S::MdJAZ2-GUS+35S::MdHIR4 (Fig. 4A), and the expression level of MdHIR4 gene was similar in 35S::GUS and 35S::MdJAZ2 calli, indicating that MdHIR4 and MdJAZ2 did not influence each another at the transcriptional level (Fig. S3A).
(A,B) Quantitative real-time PCR analysis of the relative expression of the MdJAZ2 and MdHIR4 genes in three transgenic apple calli, 35S::GUS, 35S::MdJAZ2-GUS and 35S::MdHIR4+35S::MdJAZ2-GUS. (C) The immunoblotting assay using an anti-GUS antibody to determine the degradation of the MdJAZ2-GUS protein in the 35S::MdJAZ2-GUS transgenic apple callus after being treated with or without 50 μM Me-JA. (D) The immunoblotting assay using an anti-GUS antibody to determine the abundance of the MdJAZ2-GUS and GUS proteins in the 35S::MdJAZ2-GUS, 35S::MdJAZ2-GUS+35S::MdHIR4 and 35S::GUS transgenic ‘Orin’ calli.
Additionally, immunoblotting assays were conducted with an anti-GUS antibody to examine the abundance of MdJAZ2-GUS in the tested transgenic calli. The response of MdJAZ2-GUS to jasmonate signal was measured and indicated that MdJAZ2-GUS was degraded upon Me-JA treatment (Fig. 4C). The influence of MdHIR4 on the stability of MdJAZ2-GUS was examined. The results indicated that compared with the GUS protein in the 35S::GUS control calli, both the 35S::MdJAZ2-GUS and 35S::MdJAZ2-GUS+35S::MdHIR4 transgenic calli produced MdJAZ2-GUS, a fusion protein that has a higher molecular weight than GUS alone (Fig. 4D). In addition, there is a smear of proteins in the 35S::MdJAZ2-GUS calli, which suggests a degradation of the larger molecular weight MdJAZ2-GUS fusion protein (Fig. 4D). However, there was almost no degradation product obviously found in the 35S::MdJAZ2-GUS+35S::MdHIR4 transgenic calli (Fig. 4C,D). This indicates that the 35S::MdJAZ2-GUS+35S::MdHIR4 transgenic calli accumulated more MdJAZ2-GUS protein than the 35S::JAZ2-GUS transgenic calli, suggesting that MdHIR4 enhanced the stability of the MdJAZ2-GUS protein.
MdHIR4 represses the accumulation of anthocyanins in the apple peel
To examine if and how MdHIR4 influences the accumulation of anthocyanins in the apple fruit peel, a TRV(Tobacco Rattle Virus)-based VIGS technique was used to transiently suppress the expression of MdHIR4 and MdJAZ2 in the peel of bagged fruit immediately following their detachment from the apple tree. The plasmids TRV-MdJAZ2, TRV-MdHIR4 or TRV-MdJAZ2+TRV-MdHIR4 were injected into the fruit peel and treated the apple leaf. Quantitative real-time PCR analysis showed that the injection or treatment correspondingly suppressed the expression levels of the MdJAZ2 and MdHIR4 genes in the peel and leaf (Fig. 5A,B; Fig. S3A,B). However, the TRV-induced MdJAZ2 suppression did not influence the expression of MdHIR4, while MdHIR4 did not influence MdJAZ2 (Fig. 5A and B; Fig. S3C).
(A,B) The expression levels of the MdJAZ2 and MdHIR4 genes in the fruit peel around the injection sites. (C) The anthocyanin content in the fruit peels around the injection sites. Anthocyanins were extracted from 8 pieces, 1 cm2 in size for each from one individual fruit. (D) Apple fruit coloration around the injection sites. Antisense MdHIR4 and MdJAZ2 cDNAs were used to construct Tobacco rattle virus (TRV)-based VIGS vectors. The resultant vectors TRV-MdHIR4, TRV-MdJAZ2 and their combination, TRV-MdHIR4+TRV-MdJAZ2, were used in the injection of the fruit, while the empty TRV vector was used as the control. (E) Quantitative real-time PCR analysis of the expression levels of the MdCHS, MdCHI, MdMYB9, MdMYB11, MdMYB1, MdANR, MdUFGT, MdbHLH3, MdANS and MdFLS genes in the fruit peel around the injection sites. Md18S was used as a loading control.
As a result, the suppression of MdJAZ2 or MdHIR4 alone noticeably promoted anthocyanin accumulation and fruit/leaf coloration in the peel and leaf. A co-injection with TRV-MdJAZ2+TRV-MdHIR4 resulted in the production of more anthocyanins and the redder color in the fruit peel and leaf than a single injection of either TRV-MdJAZ2 or TRV-MdHIR4 (Fig. 5C,D; Fig. S3C,D). Furthermore, the relative expression of the regulatory genes, MdMYB1, MdMYB9 and MdMYB11, as well as the structural genes, MdCHS, MdCHI, MdF3H, MdDFR, MdANS, MdUFGT, MdANR and MdFLS, were determined through quantitative real-time PCR of the fruit peel around the injection sites. The results indicated that the suppression of MdHIR4 and MdJAZ2 remarkably enhanced the expression levels of these ten regulatory and structural genes (Fig. 5E).
Additionally, a viral vector-based transformation method was carried out to enhance the expression of the MdHIR4 and MdJAZ2 genes. The plasmids pIR, pIR-MdHIR4 and pIR-MdJAZ2+pIR-MdHIR4 were injected into the fruit peel. The results showed that MdHIR4 negatively regulated anthocyanin accumulation and peel coloration in the apple fruit (Fig. 6).
(A) The viral based fusion expression vectors pIR-MdHIR4 and pIR-MdHIR4+pIR-MdJAZ2 plus the helper plasmid IL-60-BS were injected into the apple fruit. The empty pIR vector plus the helper plasmid IL-60-BS was used as the control. The fruit peel coloration was observed 4 days after exposure to white light at 17 °C. (B,C) Quantitative real-time PCR analysis of the expression levels of MdJAZ2 and MdHIR4 in the fruit peels around the injection sites. (D) Anthocyanin content in the fruit peels around the injection sites. (E) qRT-PCR analysis of MdMYB1, MdbHLH3, MdMYB9, MdMYB11, MdCHS, MdCHI, MdF3H, MdDFR, MdANR, MdUFGT, MdANS and MdFLS in the apple fruit peels around the injection sites. Md18S was used as a loading control.
Discussion
Plants produce anthocyanins in almost all organs, such as fruit, flowers and leaves, in response to the ripening and maturation processes, and in response to biotic and abiotic stresses due to its antioxidant activity37,38. As is well known, jasmonic acid (JA) is an important phytohormone that induces anthocyanin biosynthesis via the JA signaling pathway. On the contrary, JAZ proteins interact with the WD40/bHLH/MYB complex and act as negative regulators for anthocyanin accumulation in various plant species15,34. In this study, we identified the MdJAZ-interacting proteins, the MdHIRs, through a yeast two-hybrid screen and then functionally characterized and identified them as negative regulators of anthocyanin biosynthesis in apple.
The HIR proteins ubiquitously exist in plant species. In Arabidopsis, there are four HIR genes, HIR1, HIR2, HIR3 and HIR4, in the genome. Similarly, in the apple genome there are five MdHIR genes, MdHIR1-1, MdHIR1-2, MdHIR2, MdHIR3 and MdHIR4 (Fig. 2). Additionally, the HIR genes have been cloned and identified in various plants, such as tobacco, maize, barley, rice, wheat, pepper and legumes. In these plant species, the HIR proteins contain highly conserved amino acid sequences, and possess the SPFH domain (also named the PHB domain)39. They belong to a protein superfamily that controls cell proliferation, ion channel regulation and cell death40,41.
In this study, it was found that the PHB domain of MdHIR2 and MdHIR4 interacted with the N-terminal ZIM domain of the MdJAZ proteins (Fig. 3E and F). The ZIM (also named the TIFY) domain primarily mediates the homo- and heteromeric interactions between most of the JAZs in Arabidopsis and is involved in JA signaling output42. In addition, it also binds to Novel Interactor of JAZ (NINJA), which contains an EAR motif, and mediates the interaction between the JAZ proteins and TOPLESS to negatively regulate JA signaling43. Jasmonate does not affect the stability of NINJA, and NINJA overexpression does not affect JAZ3 stability43. The MdHIR proteins interacted with the ZIM domain of the MdJAZ proteins and inhibited their degradation in apple (Figs 3C–F and 4D).
In addition to the ZIM domain, the JAZ proteins also contain a C-terminal Jas motif that is required for the interactions between the JAZ proteins and COI1 and MYC2. The degradation of the JAZ proteins is induced by JA, and depends on the interaction between JAZ and the SCFCOI1 protein complex32. Jasmonate induces anthocyanin accumulation by promoting the degradation of JAZ proteins in plants15,34. The MdHIR proteins inhibited the degradation of the MdJAZ proteins (Fig. 4C,D), thereby negatively regulating the biosynthesis of anthocyanins in apple. A model summarizing our findings regarding the regulatory pathway through which the MdHIR proteins inhibit the biosynthesis of anthocyanins is presented in Fig. 7. Additionally, the AtHIRs interacted with the AtJAZ proteins in Arabidopsis (Fig. S2), suggesting that this mechanism could work in other plant species.
When Jasmonate levels are low in the apple fruit the MdJAZ2 protein interacts with the MdbHLH3 protein and inhibits the promotion of anthocyanin structural genes. When Jasmonate levels are high in the apple fruit the MdJAZ2 protein is degraded by the SCFCOI1 complex, which releases the transcription factor MdbHLH3. MdbHLH3 binds to the promoter of the anthocyanin structural genes and induces anthocyanin biosynthesis. However, the MdHIR proteins interact with the ZIM domain of the MdJAZ2 protein, but do not influence the interaction between MdbHLH3 and the Jas domain of the MdJAZ2 protein. Consequently, the MdHIR proteins inhibit the SCFCOI1-induced degradation of the MdJAZ2 proteins, thereby strengthening the association of MdJAZ2 and MdbHLH3, and inhibiting the release of the MdbHLH3 protein. As a result, MdHIR4 negatively regulates anthocyanin biosynthesis and fruit coloration in the apple fruit.
In plants, the HIR proteins are involved in the hypersensitive response (HR). The HR is one of the various defense mechanisms mounted by plants in response to pathogen attack. This response is now almost universally accepted as a form of programmed cell death (PCD) characterized by the rapid death of plant cells at the site of pathogen infection. It generally causes localized cell death and results in necrotic lesions around infection sites in different plant organs. In barley and wheat, the Hv-HIR1, Hv-HIR2, Hv-HIR3 and Hv-HIR4 genes are induced by pathogens and are involved in the induction of the HR44,45. In Arabidopsis, the HIR proteins physically associate with the immune receptor RPS2 in plant immune responses46. In rice, a novel simple extracellular leucine-rich repeat (eLRR) domain protein, OsLRR1, enters the endosomal pathway and interacts with OsHIR1 to participate in PCD47. In wheat, the hypersensitive-induced reaction genes, TaHIR1 and TaHIR3, play positive roles in resistance to the stripe rust fungus48. In pepper, CaLRR1 specifically binds to the plasma membrane (PM)-localized CaHIR1 to regulate PCD in leaves in response to an infection by Xanthomonas campestris pv. Vesicatoria49.
The HR is one of the most characteristic plant defenses against biotrophic pathogens. Salicylic acid (SA) plays a primary role in the activation of disease resistance mechanisms frequently associated with the HR50. The resistance (R) protein, which is a pathogen-encoded avirulence protein, can trigger the HR51. The R protein-mediated HR and SA-mediated basal resistance are generally considered effective against biotrophic pathogens, but ineffective against necrotrophic pathogens. Instead, plant resistance to necrotrophic pathogens is often mediated by jasmonic acid (JA) signaling, a process also involved in plant responses to wounding52.
During the HR, ROS production activates PCD53. Interestingly, anthocyanin works as an active scavenger of ROS in plant cells54. During PCD in lace plant (Aponogeton madagascariensis) leaves, the first visible change observed is the reduction of visible anthocyanin55, suggesting that anthocyanin may be involved in PCD. Therefore, the MdHIR-mediated inhibition of anthocyanin accumulation may be conducive to the occurrence of HIR and PCD.
Both isoflavonoid phytoalexin concomitants and anthocyanins are biosynthesized through the phenylpropanoid metabolic pathway. They belong to two different branches of this pathway, thereby being theoretically competitive with each other. For example, there in soybean is a strong bias towards decreasing the synthesis of anthocyanins and proanthocyanins, but increasing the synthesis of isoflavonoid phytoalexin concomitants during the resistance response56. It seems reasonable to suppose that the MdHIR-mediated inhibition of anthocyanin and proanthocyanin accumulation promotes the production of isoflavonoid phytoalexin concomitants, which may be conducive to PCD and HR. In addition, MdHIR proteins stabilize MdJAZ2 proteins (Fig. 4C,D), while JA promotes the degradation of the JAZ proteins. Therefore, JA should inhibit the function of the MdHIR proteins. As a result, JA seems to restrict cell death processes associated with hypersensitive induced reaction (HIR) in response to SA, ethylene, and ROS57, and thus is part of the machinery that prevents excessive damage to host tissues.
Material and Methods
Plant materials
Apple fruits (Malus domestica Borkh.) were bagged at 30 days after full bloom (DAFB). The bagged fruits of ‘Red Delicious’ cultivar were harvested from a adult tree at 140 DAFB, while those of ‘Fuji’ at 170 DAFB. After injection, fruit was placed in an illumination incubator for three days at 17 °C. The callus of the ‘Orin’ cultivar was grown on MS medium containing 1.5 mg L−1 6-benzylaminopurine (6-BA) and 0.5 mg L−1 indole-3-acetic acid (IAA) at 25 °C in the dark. The tissue cultures of the ‘Gala’ cultivar in vitro were cultured on MS medium supplemented with 0.2 mg L−1 IAA and 1.5 mg L−1 6-BA at 25 °C under a 16-h light/8-h dark photoperiod.
Determination of total anthocyanins
Anthocyanins were extracted from 8 pieces of apple skin, each of 1 cm2 in size from one individual fruit, in 1 mL 1% (v/v) HCl-methanol for 24 h at room temperature in the dark. The upper aqueous phase was separated by centrifugation for 5 min at 13000 g and was subjected to spectrophotometric quantification at 650, 620 and 530 nm using a UV–Vis spectrophotometer (Shimadzu UV-2450, Kyoto, Japan). The content of the anthocyanins was determined using the following formula: OD = (A530–A620)–0.1 (A650–A620)58.
Gene cloning and expression analysis
Total RNA was extracted from plant material for gene cloning and expression analyses using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For quantitative real-time PCR analyses, we used the clone enzyme Kit (Transgene, Beijing, China) and SYBR Green MasterMix (SYBR Premix EX Taq TM, Dalian, China), according to the manufacturer’s instructions. The primer sequences used for quantitative real-time PCR analyses are listed in Supplemental Table S1.
DNA constructs and genetic transformation
The full length coding regions of MdJAZ2 and MdHIR4 were amplified from the cDNA of Malus domestica using standard molecular biology protocols and enzymatic digestion technology (Invitrogen, Carlsbad, CA, USA). The MdHIR4 PCR product was recombined with vector pCXSN-HA59 to create the pCXCN-MdHIR4 plasmid. The MdJAZ2 PCR product was cloned into pMD18-T (Takara Bio, Atsu, Japan) to create a fusion protein of the GUS gene and the coding region of MdJAZ2, for convenient detection of MdJAZ2. The MdJAZ2-GUS fusion was recombined into the vector pRI101-AN (Takara Bio, Atsu, Japan), to form the pRI-MdJAZ2-GUS plasmid. In both expression vectors the promoter was the cauliflower mosaic virus (CaMV) 35S promoter. All the specific primers and restriction enzymes used are listed in Supplemental Table 1. The two plasmids were introduced into the Agrobacterium tumefaciens strain LBA4404, and the resultant Agrobacterium tumefaciens transformants were used to transform the apple callus as described by An et al.18.
Yeast two-hybrid (Y2H) screening and assays
To screen for proteins that interact with MdJAZ2, the full-length CDS of the MdJAZ2 gene was inserted into pGBT9 vector which contain the DNA binding domain of GAL4. The resulting construct, MdJAZ2-BD, was used as bait vector to screen an apple cDNA library. The cDNA library was made with total RNAs isolated from apple fruit peel and constructed by Oebiotech Company (Shanghai, China).
Y2H assays were carried out as described by Xie et al. (2012). All of the coding regions of the JAZ genes used in this study were amplified from the cDNA of Arabidopsis and Malus domestica. The JAZ genes were recombined with the vector pGAD424 to generate constructs containing the JAZ genes fused to the GAL4 activation domain for Y2H analysis. The coding regions of the HIR genes used in this study were amplified from the cDNA of Arabidopsis and Malus domestica. They were then recombined with the vector pGBT9 to generate constructs of the HIR gene fused to the GAL4 DNA binding domain for Y2H analysis. The specific primers and restriction enzymes that were used are listed in Supplemental Table 1.
Yeast transformants were exhaustively screened on synthetic defined (SD) media (-Leu/-Trp/-His/-Ade) according to the manufacturer’s instructions (Clontech, Palo Alto, CA, USA). The JAZ-AD and HIR-BD plasmids were co-transformed into the Y2HGOLD yeast cell strain using the lithium acetate method and were cultured at 30 °C. The resulting yeast transformants were filtered on medium lacking Trp and Leu (-Trp/-Leu), and putative transformants were subsequently transferred to medium lacking Trp, Leu, His and Adenine (-Leu/-Trp/-His/-Ade) with and without X-alpha-gal.
Pull-down assays
The full-length coding sequences of MdJAZ2, AtJAZ3, AtJAZ9 and MdHIR2, MdHIR4, AtHIR1 were cloned into the pGEX-4T-1 and pET32a vectors, respectively, to generate the JAZ-HIS and HIR-GST constructs used for the pull-down assays. All the specific primers and restriction enzymes that were used are listed in Supplemental Table 1.
For the in vitro pull-down experiments, expression of the proteins MdJAZ2/AtJAZ3/AtJAZ9-HIS and MdHIR2/MdHIR4/AtHIR1-GST was induced and the proteins were purified from E. coli BL21 cells. The purified protein mixtures of the JAZ and HIR genes were incubated for 1 h at 4 °C, 80 rpm, and purified using a His purification kit (Cwbio, Beijing, China), according to the manufacturer’s instructions. Finally, the proteins were detected through Western blot analysis using anti-His and anti-GST antibodies.
Construction of the viral vectors and the Agrobacterium infiltration of the apple fruit
Viral vectors were used as described by Li et al.20. to observe the effects of MdJAZ2 and the MdHIR genes overexpression or suppression in apple fruit. The full-length sequences of MdJAZ2 and MdHIR4 were cloned into the pIR vector under the control of the 35S promoter. The overexpression constructs were named pIR-MdJAZ2 and pIR-MdHIR4. To generate antisense expression vectors for MdJAZ2 and MdHIR4, the fragment of MdJAZ2 and MdHIR4 used in the above-mentioned genetic transformation was cloned into the tobacco rattle virus (TRV) vector in an antisense orientation under the control of the 35S promoter. All the specific primers and restriction enzymes that were used are listed in Supplemental Table 1. The resultant vectors were named TRV-MdJAZ2, and TRV-MdHIR4. The resultant viral vectors were used for injection into the apple fruit as described by Xie et al.12.
The degradation of the protein
The callus of the ‘Orin’ cultivar, which was transformed with the pRI-MdJAZ2-GUS fusion gene, was grown on the culture medium for 15 days and treated with 50 mM MG132 (proteinase inhibitor, Sigma-Aldrich) overnight in the dark. Then, the calli were washed clean and treated with 50 μM Me-JA. Untreated calli was used as a control. The JAZ protein was detected through Western blotting using an anti-GUS antibody (BGI, Beijing, China). The ‘Orin’ callus co-transformed with 35S::MdJAZ2-GUS and 35S::MdHIR4 was obtained and used to testing for the function of the HIR protein in regards to the stability of JAZ. The amount of JAZ protein was detected in the 35S::MdJAZ2-GUS callus and compared with the amount of JAZ protein in the 35S::MdJAZ2-GUS+35S::MdHIR4 callus by means of Western blotting using the anti-GUS tag antibody (BGI, Beijing, China).
Additional Information
How to cite this article: Chen, K.-Q. et al. MdHIR proteins repress anthocyanin accumulation by interacting with the MdJAZ2 protein to inhibit its degradation in apples. Sci. Rep. 7, 44484; doi: 10.1038/srep44484 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Macheix, J. J. & Fleuriet, A. Fruit phenolics. CRC press (1990).
Gauche, C., Malagoli, E. D. S. & Bordignon Luiz, M. T. Effect of pH on the copigmentation of anthocyanins from Cabernet Sauvignon grape extracts with organic acids. Scientia Agricola 67, 41–46 (2010).
Holton, T. A. & Cornish, E. C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7, 1071 (1995).
Jaakola, L. et al. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 130, 729–739 (2002).
Lee, J. et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749 (2007).
Feller, A., Machemer, K., Braun, E. L. & Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116 (2011).
Xu, W., Dubos, C. & Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends in Plant Science 20, 176–185 (2015).
Takos, A. M. et al. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232 (2006).
Chagné, D. et al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 161, 225–239 (2013).
Ban, Y. et al. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant and Cell Physiol 48, 958–970 (2007).
Espley, R. V. et al. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427 (2007).
XIE, X. B. et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell & Environment 35, 1884–1897 (2012).
Umemura, H., Otagaki, S., Wada, M., Kondo, S. & Matsumoto, S. Expression and functional analysis of a novel MYB gene, MdMYB110a_JP, responsible for red flesh, not skin color in apple fruit. Planta 238, 65–76 (2013).
Vimolmangkang, S., Han, Y., Wei, G. & Korban, S. S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 13, 176 (2013).
An, X. H. et al. MdMYB9 and MdMYB11 are Involved in the Regulation of the JA-Induced Biosynthesis of Anthocyanin and Proanthocyanidin in Apples. Plant Cell Physiol. 56, 650–662 (2015).
Zhang, H. L. et al. In silico analysis and expression confirmation of the regulation of fruit coloration by transcriptional factor MdMBY1 in Delicious apple. Acta Horticulturae Sinica 36, 1581–1588 (2009).
Brueggemann, J., Weisshaar, B. & Sagasser, M. A WD40-repeat gene from Malus × domestica is a functional homologue of Arabidopsis thaliana TRANSPARENT TESTA GLABRA1. Plant Cell Reports 29, 285–294 (2010).
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F. & Hao, Y. J. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. Journal of Plant Physiology 169, 710–717 (2012).
Carbone, F. et al. Developmental, genetic and environmental factors affect the expression of flavonoid genes, enzymes and metabolites in strawberry fruits. Plant Cell & Environment 32, 1117–1131 (2009).
Li, Y. Y. et al. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022 (2012).
Deikman, J. & Hammer, P. E. Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol. 108, 47–57 (1995).
Jeong, S. T., Goto-Yamamoto, N., Kobayashi, S. & Esaka, M. Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Science 167, 247–252 (2004).
Loreti, E. et al. Gibberellins, jasmonate and abscisic acid modulate the sucrose‐induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytologist 179, 1004–1016 (2008).
Thines, B. et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665 (2007).
Chico, J. M. et al. Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. The Plant Cell 26, 1967–1980 (2014).
Yan, J. et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. The Plant Cell 21, 2220–2236 (2009).
Devoto, A. et al. COI1 links jasmonate signalling and fertility to the SCF ubiquitin–ligase complex in Arabidopsis. The Plant Journal 32, 457–466 (2002).
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007).
Farmer, E. E. Plant biology: jasmonate perception machines. Nature 448, 659–660 (2007).
Browse, J. Jasmonate: an oxylipin signal with many roles in plants. Vitamins & Hormones 72, 431–456 (2005).
Song, S. et al. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000–1013 (2011).
Fernández-Calvo, P. et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715 (2011).
Shan, X., Zhang, Y., Peng, W., Wang, Z. & Xie, D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. Journal of Experimental Botany 60, 3849–3860 (2009).
Qi, T. et al. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814 (2011).
Hou, X., Lee, L. Y. C., Xia, K., Yan, Y. & Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Development Cell 19, 884–894 (2010).
Zhu, Z. et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. 108, 12539–12544 (2011).
Kondo, S., Tsuda, K., Muto, N. & Ueda, J. E. Antioxidative activity of apple skin or flesh extracts associated with fruit development on selected apple cultivars. Scientia Horticulturae 96, 177–185 (2002).
Zhou, X., Hua, D., Chen, Z., Zhou, Z. & Gong, Z. Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 60, 79–90 (2009).
Nijtmans, L. G. J., Sanz, M. A., Grivell, L. A. & Coates, P. J. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cellular & Molecular Life Sciences Cmls 59, 143–155 (2002).
Dangl, J. L. & Jones, J. D. Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 (2001).
Nadimpalli, R., Yalpani, N., Johal, G. S. & Simmons, C. R. Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. Journal of Biological Chemistry 275, 29579–29586 (2000).
Chung, H. S. & Howe, G. A. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21, 131–145 (2009).
Pauwels, L. & Goossens, A. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100 (2011).
Rostoks, N., Schmierer, D., Kudrna, D. & Kleinhofs, A. Barley putative hypersensitive induced reaction genes: genetic mapping, sequence analyses and differential expression in disease lesion mimic mutants. Theoretical & Applied Genetics 107, 1094–1101 (2003).
Zhang, G. et al. Cloning and Characterization of a Novel Hypersensitive-induced Reaction Gene from Wheat Infected by Stripe Rust Pathogen. Journal of Phytopathology 157, 722–728 (2009).
Qi, Y. et al. Physical association of Arabidopsis hypersensitive induced reaction proteins (HIRs) with the immune receptor RPS2. Journal of Biological Chemistry 286, 31297–31307 (2011).
Zhou, L. et al. A novel simple extracellular leucine-rich repeat (eLRR) domain protein from rice (OsLRR1) enters the endosomal pathway and interacts with the hypersensitive-induced reaction protein 1 (OsHIR1). Plant Cell & Environment 32, 1804–1820 (2009).
Duan, Y. et al. Wheat hypersensitive-induced reaction genes TaHIR1 and TaHIR3 are involved in response to stripe rust fungus infection and abiotic stresses. Plant Cell Reports 32, 273–283 (2013).
Jung, H. W. & Hwang, B. K. The leucine-rich repeat (LRR) protein, CaLRR1, interacts with the hypersensitive induced reaction (HIR) protein, CaHIR1, and suppresses cell death induced by the CaHIR1 protein. Molecular Plant Pathology 8, 503–514 (2007).
Dangl, J. L., Dietrich, R. A. & Richberg, M. H. Death don’t have no mercy: cell death programs in plant-microbe interactions. Plant Cell 8, 1793 (1996).
Martin, G. B., Bogdanove, A. J. & Sessa, G. Understanding the functions of plant disease resistance proteins. Annual Review of Plant Biology 54, 23–61 (2003).
Spoel, S. H., Johnson, J. S. & Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proceedings of the National Academy of Sciences, 104(47), 18842-18847. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 104, 18842–18847 (2007).
Heath, M. C. Hypersensitive response-related death. In Programmed Cell Death in Higher Plants. 77–90. Springer: Netherlands, (2000).
Bi, X. et al. Anthocyanin contributes more to hydrogen peroxide scavenging than other phenolics in apple peel. Food Chemistry 152, 205–209 (2014).
Wertman, J., Lord, C. E., Dauphinee, A. N. & Gunawardena, A. H. The pathway of cell dismantling during programmed cell death in lace plant (Aponogeton madagascariensis) leaves. BMC Plant Biol. 12, 115 (2012).
Zabala, G. et al. Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biol. 6, 26 (2006).
Overmyer, K., Brosché, M. & Kangasjärvi, J. Reactive oxygen species and hormonal control of cell death. Trends Plant Science 8, 335–342 (2003).
Lee, H. S. & Wicker, L. Quantitative changes in anthocyanin pigments of lychee fruit during refrigerated storage. Food Chemistry 40, 263–270 (1991).
Chen, S., Songkumarn, P., Liu, J. & Wang, G. L. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121 (2009).
Acknowledgements
The authors would like to thank Dr. Ilan Sela for IL-60-BS and pIR binary vectors, Dr. Takaya Moriguchi of National Institute of Fruit Tree Science, Japan, for ‘Orin’ apple callus. This work was supported by grants from NSFC (31471854, 31272142, 31325024) and Ministry of Education of China (IRT15R42).
Author information
Authors and Affiliations
Contributions
Y.J.H. and K.Q.C. designed research; K.Q.C. and X.Y.Z. performed research; X.Y.Z., X.H.A., and C.X.Y. contributed new reagents tools; Y.T., X.Y.Z. and D.D.L. analyzed data; Y.J.H., X.Y.Z. and K.Q.C. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chen, KQ., Zhao, XY., An, XH. et al. MdHIR proteins repress anthocyanin accumulation by interacting with the MdJAZ2 protein to inhibit its degradation in apples. Sci Rep 7, 44484 (2017). https://doi.org/10.1038/srep44484
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44484
This article is cited by
-
MdHIR4 transcription and translation levels associated with disease in apple are regulated by MdWRKY31
Plant Molecular Biology (2019)
-
Genome-wide survey and analysis of the TIFY gene family and its potential role in anthocyanin synthesis in Chinese sand pear (Pyrus pyrifolia)
Tree Genetics & Genomes (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.