Abstract
Locusta has strong fly wings to ensure its long distance migration, but the molecular mechanism that regulates the Locusta wing development is poorly understood. To address the developmental mechanism of the Locusta flying wing, we cloned the Dpp target gene spalt (sal) and analyzed its function in wing growth in the Locusta. The Locusta wing size is apparently reduced with vein defects when sal is interfered by injection of dsRNA, indicating that sal is required for locust wing growth and vein formation. This function is conserved during the Drosophila wing development. To better understand sal’s function in wing growth, we then used Drosophila wing disc as a model for further study. We found that sal promotes cell proliferation in the whole wing disc via positive regulation of a microRNA bantam. Our results firstly unravel sal’s function in the Locusta wing growth and confirm a highly conserved function of sal in Locusta and Drosophila.
Similar content being viewed by others
Introduction
Locusta migratoria is one of the global destructive migratory agricultural pests. Swarm formation and long-distance flight behavior are the two major reasons why Locusta plagues are so destructive even today1,2. During the migratory stage, these insects can keep flying for hours over hundreds of kilometers2. Therefore, besides gene expansion associated sufficient energy supply3, well developed and functional wings are required to adapt their migratory behavior4,5.
Locusta is an incomplete metamorphosis insect whose young nymph resembles the adult with visible developing forewings and hindwings6. The wing shape and size are varied among geographical populations. Based on forewing shape, grasshopper Trilophidia annulata can be divided into geographical groups7 and mircoRNAs are associated with the polymorphism of adult wings8. Biomechanical study shows that the fan-like distribution of veins improve fracture toughness in Locusta9,10. However, little is known about the molecular mechanism of how the Locusta wing develops into such delicate structure.
Current understanding of insect wing development mechanism is mainly from the fruitfly Drosophila melanogaster. In comparison, the fruitfly, a Diptera insect, undergoes complete metamorphosis. The adult wings are developed from larval wing imaginal discs which are formed from late embryogenesis and undergo intricate cell proliferation, differentiation and morphogenesis during larval and pupal stage inside the body. The pattern formation is delicately regulated by organizers located in the anterior/posterior (A/P) and dorsal/ventral (D/V) boundaries which secrete signal molecules including the long-range morphogens Decapentaplegic (Dpp) and Wingless (Wg)11,12, and short-range morphogen Hedgehog (Hh)13. These morphogens form gradients to regulate the expression of their target genes and control almost all aspects of wing development12.
The Dpp morphogen gradients controls cell growth through negatively regulation of the transcriptional suppressor brinker (brk), which downregulates bantam (ban), a developmentally regulated mircoRNA14,15,16. ban promotes cell proliferation autonomously and suppresses proliferation-induced apoptosis17. Various Dpp targets identified, such as optomotor-blind (omb), spalt (sal), and vestigial (vg), are repressed by brk15,17,18,19. Among them, omb represses ban expression in the medial regions of the wing disc to promote cell proliferation while plays an opposite role in the lateral regions20. The two Drosophila sal homologues spalt major (salm) and spalt related (salr) are also highly expressed in the central region of the wing pouch and mediate Dpp signaling pathway by promoting cell proliferation21,22. salm and salr have similar function but are not fully redundant23. For example, salm, but not salr, is evolutionally conserved in insects as a main regulator of fibrillar flight muscle fate24. Downstream the Salm/Salr complex, the vein-specific expression of the knirps and iroquois gene complexes and a serious of candidate genes are regulated for wing vein patterning and growth25,26. sal also participates in defining fly notum and wing hinge27 and the melanin pigmentation of wing eyespots in butterflies28,29,30. Therefore, sal may play a conserved role in insect wing development to generate the fast oscillating wings and whether sal functions through ban remains to be illustrated.
To better understand the developmental mechanism of Locusta flying wings, we cloned Locusta sal cDNAs and investigated the role of sal during wing blade and vein formation. By RNA interference (RNAi), we found that sal is required for wing growth and vein formation, which is conserved between the Locusta and Drosophila. Then, using the Drosophila wing disc model we dissected sal function in wing growth and found that sal promotes cell proliferation rate during larval wing development. Furthermore, mircoRNA bantam mediates the role of sal in cell proliferation regulation.
Materials and Methods
Drosophila strains and Locusta
All fruit flies used were Drosophila melanogaster. Flies were raised at 25 °C. Three transgenes UAS-salm27, UAS-salmRNAi (Tsinghua Fly Center THU3581) and UAS-salrRNAi (THU3147) were used to manipulate the expression level of sal. The efficiency of these RNAi lines has been recently verified by Tang et al.31. The Gal4 lines used were ci-Gal432, sal-Gal433, and ap-Gal434 to drive the expression of the UAS transgene. The bantam enhancer reporter line was br-C12-lacZ35.
Locusta migratoria was raised at 28 °C with 60% relative humidity.
Identification and sequence analysis of spalt from L. migratoria transcriptome
The available salm and salr gene sequences from Drosophila were used as references to screen L. migratoria transcriptome database we previously established6. The potential candidates of L. migratoria sal genes were confirmed by searching the BLASTX algorithm against the non-redundant NCBI nucleotide database using a cut-off E-value of 10−5.
Based on the Drosophila Spalt protein sequences, the deduced Locusta Spalt protein domains were determined by using DNAMAN software (Lynnon Biosoft). Other insect Spalt and Spalt-like proteins are obtained from NCBI (https://www.ncbi.nlm.nih.gov/protein). Global protein alignment and neighbor-joining phylogenic tree were performed by Geneious R9.
RNA interference
The primers to make the double strand RNA probes were as follows:
Lmsal411-F 5′-ATGTTGCAGCGGCGTGCACAAGAGG-3′
Lmsal411-R 5′-TTGTCCCAGTTGTGCCAGCAGTGGA-3′
Lmsal468-F 5′-GCCATAGACCCTGCTAAGGACCCAG-3′
Lmsal468-R 5′-GTCCTTCACCTCTGCAGCTTGTATC-3′
GFP-F 5′-CACAAGTTCAGCGTGTCCG-3′
GFP-R 5′-GTTCACCTTGATGCCGTTC-3′
Lmsal411-dsRNA, Lmsal468-dsRNA, and GFP-dsRNA were injected into the 4th instar nymphs using 20 μg dsRNAs per insect. Nymphs were raised at 28 °C for 10 days until they grew up to the 5th instar. Imaging the whole body and wing discs using SONY DSC-HX1camera before the wing discs were lysed.
Quantitative Real-Time RT-PCR Analysis
Total RNAs were extracted with Trizol reagent from the 5th nymph wing discs, digested with DNase I, and reverse transcribed using the FastQuant cDNA Synthesis kit (Tiangen Biotech), then quantitative real-time RT-PCR was performed using Go Taq qPCR Master Mix (Promega). The primers for quantitative RT-PCR were as follows:
Lmsal411q-F 5′-GAGAATGCCAGCCAGGGTC-3′
Lmsal411q-R 5′-CGGTGCTTGAAGAAGGGTT-3′
Lmsal468q-F 5′-ACCAAAACATCGGAGACA-3′
Lmsal468q-R 5′-ATAGGACACGGTGGCAGA-3′
Relative transcript levels were assessed using the Comparative CT method. β-actin was used as an internal control.
BrdU staining
BrdU staining was performed as previously described20,36. Dissected third instar wing discs were co-cultured with BrdU (1:100) in Schneider’s medium for 40–50 min at 25 °C. Then, samples were fixed in 4% formaldehyde and washed in PBT before immunostaining.
Immunohistochemistry
The primary antibodies used were mouse anti-BrdU, 1:100 (MBL) and mouse anti-β-galactosidase1:2000 (Promega Z3783), and the secondary antibody was anti-mouse DyLight 549 (1:200, Agrisera). Images were obtained using an Olympus FV10-ASW laser scanning confocal microscope and processed with Adobe Photoshop 8.0.
Wing size measurement
Image-J program was used to measure the wing areas and vein distances. High resolution images were opened in Image-J program, a straight line was drawn at the wanted sites between L2 and L4 veins to calculate the distance; and a wanted region as outlined along the L2 and L4 veins (for Drosophila wing) or the wing margin (for Locusta wing) to calculate the areas.
Results and Discussion
Identification of Locusta sal genes
Based on the transcriptome of the Locusta wing disc in our previous study6, we blasted the cDNA sequences with salm and salr in Drosophila and identified two orthologous unigenes a12971 and a6922, which are 2087 bp and 1073 bp, respectively, in the Locusta (Sup. Fig. 1). We named these two putative genes as Lmsal411 and Lmsal468, respectively. Although these two unigenes were incomplete spalt gene sequences, both putative translations were predicted (Sup. Fig. 1). Protein sequence alignment showed that these two putative translations were 23.8% identity. Lmsal411, which was aligned with the anterior part of Drosophila Sal, was 33.1% identical to both Salm and Salr, while Lmsal468, aligned with the posterior part, showed a little higher identity (42.1% and 35.5%, respectively) (Fig. 1). The ZnF-C2H2 motifs showed a highly conservation in Lmsal411 (87.0% identity) and Lmsal468 (72.4% identity), indicating a conservative DNA binding motif of transcription factors. In addition, the phylogenetic analysis from different insect species revealed that Lmsal411 clustered with Drosophila sal, while Lmsal468 with bees (Fig. 2). Therefore, due to the less identity between these two translations and higher similarity to Drosophila salm, we speculate that these two unigenes may be different loci of the same Locusta spalt.
Above analysis implies that Locusta sal may share partial functions with that of Drosophila. In the Drosophila wing, the transcription factors Salm and Salr participate in wing growth21 and vein formation18,25. However, the regional growth effects of Sal on wing growth and cell proliferation rate have not yet intensively investigated.
Locusta sal regulates wing growth
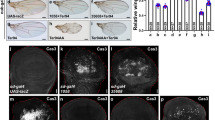
To test the functional conservation of sal, we performed RNA interference experiments in Locusta using Lmsal genes we identified. After 10 days of injection, the mRNA levels of each gene were measured using quantitative real-time RT-PCR. The mRNA expression levels of Lmsal411 and Lmsal468 were significantly reduced to 62.7 ± 9.1% (p < 0.05) and 36.1 ± 8.5% (p < 0.01), respectively, when the corresponding gene was knocked-down (Fig. 3A and B). We noted that Lmsal411 mRNA was reduced to 30.3 ± 21.8% when interfering with Lmsal468, and vise versa, Lmsal468 mRNA was reduced to 33.5 ± 11.3% when interfering with another (Fig. 3A and B). Thus, the expression of these two unigenes are dependent on each other, implying that they belong to either the same gene or one gene complex. As when knocking-down either gene, the efficiency of interference of both genes were similar and had no statistic difference (Fig. 3A and B), and taken together with the above analysis of sequence identity, it is more likely that they are belonging to the same gene. When Lmsal411 was interfered in 4th instar, the wings of early 5th instar nymphs were remarkably reduced to 45% of the control (15.4 vs 27.5 mm2) (Fig. 3D and F). Consistently, interfering with Lmsal468 resulted in 69% loss of wing area (8.6 vs 27.5 mm2) (Fig. 3E and F). The phenotypes of Lmsal468-RNAi were much more obvious than that of Lmsal411-RNAi (Fig. 3D and E) probably due to the higher RNAi efficiency of the former. Moreover, the hindwing concomitantly reduced with the forewings, while the body size was not apparently affected (Fig. 3D and E). The veins in the posterior part was slightly affected in Lmsal468-RNAi nymphs (arrow in Fig. 3E’). This result indicates that Locusta sal regulates wing growth and vein pattern formation.
(A and B) Relative expression of the two genes in the wing discs of 5th instar nymphs after Lmsal411 dsRNAi (A) or Lmsal468 dsRNAi (B) was injected into 4th instar nymphs for 10 days. (C) The body and wing size are normal after GFP-dsRNA control injection. (D) The wing is smaller in Lmsal411-dsRNA injected locusts. (E) The wing is much smaller in Lmsal468-dsRNA injected locusts. (C’,D’and E’) show the higher magnification in (C,D and E), respectively. (F) Quantification of wing area. In each genotype, five nymphs were injected which give similar results as shown. *, ** and *** represent significant difference with the GFP-dsRNA control (pairwise comparison of t-tests, p < 0.05, 0.01, and 0.001, respectively). ns means no significant difference. Scale bar represents 4 mm.
Drosophila sal promotes adult wing growth
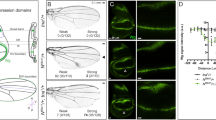
To further confirm sal is functionally conserved in other insects to promote wing growth, we regionally up or down-regulated salm and salr in Drosophila wings and examined the adult wing size. Drosophila wing has five longitudinal veins, which are L1-L5 from the anterior to posterior part. Between L1 and L4 is the anterior part and other areas are the posterior wing. The distance from L2 to L4, which is indicated by a dashed line in Fig. 4A, was measured to monitor the size of the anterior part of the wing. The flies containing ci-Gal4, which is expressed in the anterior part of the wing, was used as a control and showed normal size and morphology. When salm was overexpressed, the relative distance was significantly increased (1.07 ± 0.01), while when knocked-down salm or salr, the distance was strikingly reduced (0.85 ± 0.01 and 0.94 ± 0.01, respectively) (Fig. 4B–D and I). To confirm this result, we used another driver sal-Gal4, which is expressed in the medial pouch to repeat the manipulation. Again, the relative area enclosed by L2 and L4 (sal-Gal4 expression region) was increased in salm overexpression wings and reduced in salm and salr knockdown wings (Fig. 4F–H and J). In addition, sal regulated the vein formation in all tested lines except ci-Gal4>salm which is consistent with previous studies21,25,26. These data confirm that Drosophila sal regulates wing growth and vein pattern formation. Thus, sal function in promoting wing growth is conserved in the fruitfly and locust.
(A) Adult wing of ci-Gal4 control shows the normal wing. L1‒L5 indicates the five longitudinal veins. The dashed line from the distal end of L2 crossing L3 and extending to L4 indicates the distance between L2 and L4. (B) The wing is larger in salm overexpressed flies. (C and D) Wings are smaller when salm or salr is knocked-down and the L2 and L3 veins are fused. (E) Adult wing of sal-Gal4 control shows the normal wing. The dashed line circles the whole area between L2 and L4. (F) The anterior wing is larger in salm overexpressed flies. (G and H) Wings are smaller when salm or salr is knocked-down and the L2 and L3 veins are fused, similar with ci-Gal4 driving wings. (I) Relative distance between L2 and L4 represented by dashed line in (A). (J) Relative areas circled by L2 and L4 represented by dashed line in (E). The number in (I) and (J) shows the quantification number. ***Represents significant difference (pairwise comparison of t-tests, p < 0.001).
sal promotes cell proliferation in the wing disc
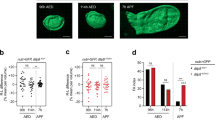
To unravel the mechanism of wing growth regulated by sal, we use the Drosophila wing disc as the research model because it is easy of molecular labeling. We examined the cell proliferation rate in Drosophila wing discs by Bromodeoxyuridine (BrdU) incorporation which is widely used in cell proliferation studies. BrdU is a thymidine analog that can be incorporated into DNA during DNA synthesis in culture, and then be detected by specific anti-BrdU antibodies following with quantitative analysis of the anti-BrdU fluorescent intensities. In wild-type control, BrdU level is evenly distributed in the wing discs20. Elevating Salm level by expressing UAS-salm in the ci-Gal4 or ap-Gal4 domain, the level of BrdU level was increased (Fig. 5A and B). Within the pouch, the fluorescence intensity of BrdU was increased in the salm expressing regions, indicating an enhanced proliferation rate (Fig. 5A and B). Vice versa, compromising salm and salr by expressing UAS-salm-RNAi or UAS-salr-RNAi in the sal-Gal4 or ci-Gal4 domain, the level of BrdU staining was reduced which means that the cell proliferation rate was decreased. These data demonstrate that Drosophila sal promotes cell proliferation in the early wing developmental stages.
(A) The cell proliferation rate is increased when salm is overexpressed in ci-Gal4 region. (B) The cell proliferation rate is increased when salm is overexpressed in ap-Gal4 region. (C and D) The cell proliferation rate is repressed when salr is knocked-down. (E) The cell proliferation rate is repressed when salm is knocked-down. GFP shows the Gal4 expressing domain. White boxes define the area of fluorescence quantification of BrdU staining.
sal promotes microRNA bantam expression
We previously found that one of the Dpp target genes omb represses ban expression in medial regions of the Drosophila wing discs while plays an opposite role in lateral regions20. Subsequently we took the advantages of transgenic lines available for Drosophila to do further investigation, because we are lacking of genetic tools to do it on the Locusta wing to test whether and how sal regulates the expression of ban. ban transcription was monitored by br-C12-lacZ35 which was mainly expressed in the hinge/blade folds surrounding the wing pouch (Fig. 6A). Expression of UAS-salm in the medial region of the wing disc driven by dpp-Gal4 apparently upregulated ban transcription level (arrows in Fig. 6B and B’). To confirm this result, clones were generated in the wing disc. Consistently, ban was upregulated in both medial regions and lateral region (Fig. 6C–C”). Therefore, sal promotes ban expression in the wing disc in a non-regional specific manner, unlike the manner of omb.
(A) The expression pattern of br-C12-lacZ reporter in the wild-type wing disc. (B) ban is elevated in salm overexpression cells (GFP positive) in both pouch and notum regions. (C) salm overexpression clones show upregulated ban level in the whole wing disc. (C”) is the magnification of the arrow-indicated clone in (C).
Previous studies have illustrated several models to explain how Dpp gradient controls a uniform cell proliferation rate in the wing disc37. Dpp restricts the transcription factor Brinker (Brk) domain to the lateral wing disc where suppresses the growth promoter ban15. While Brk activity is not essential for the medial wing disc growth38,39,40. Omb is a main mediator of Dpp signaling in the control of cell proliferation rate. Omb represses ban in medial regions, however, promotes it in lateral regions20. Distinct to omb, sal promotes ban expression in the whole wing disc (Fig. 6). Therefore, sal may mediate partial functions of Dpp in growth control. Other possibility is that sal may mediate the roles of other upstream factors such as Lines41, Wingless42, and Ubx in Drosophila43 as well as Tribolium44.
Additional Information
How to cite this article: Wang, D. et al. spalt is functionally conserved in Locusta and Drosophila to promote wing growth. Sci. Rep. 7, 44393; doi: 10.1038/srep44393 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Enserink, M. Entomology. Can the war on locusts be won? Science 306, 1880–1882, doi: 10.1126/science.306.5703.1880 (2004).
Lovejoy, N. R., Mullen, S. P., Sword, G. A., Chapman, R. F. & Harrison, R. G. Ancient trans-Atlantic flight explains locust biogeography: molecular phylogenetics of Schistocerca. Proc Biol Sci 273, 767–774, doi: 10.1098/rspb.2005.3381 (2006).
Wang, X. et al. The locust genome provides insight into swarm formation and long-distance flight. Nat Commun 5, 2957, doi: 10.1038/ncomms3957 (2014).
Dirks, J. H., Parle, E. & Taylor, D. Fatigue of insect cuticle. J Exp Biol 216, 1924–1927, doi: 10.1242/jeb.083824 (2013).
Wootton, R. J., Herbert, R. C., Young, P. G. & Evans, K. E. Approaches to the structural modelling of insect wings. Philos Trans R Soc Lond B Biol Sci 358, 1577–1587, doi: 10.1098/rstb.2003.1351 (2003).
Liu, S. et al. De novo transcriptome analysis of wing development-related signaling pathways in Locusta migratoria manilensis and Ostrinia furnacalis (Guenee). PLoS One 9, e106770, doi: 10.1371/journal.pone.0106770 (2014).
Bai, Y., Dong, J. J., Guan, D. L., Xie, J. Y. & Xu, S. Q. Geographic variation in wing size and shape of the grasshopper Trilophidia annulata (Orthoptera: Oedipodidae): morphological trait variations follow an ecogeographical rule. Sci Rep 6, 32680, doi: 10.1038/srep32680 (2016).
Li, R. et al. MicroRNAs of the mesothorax in Qinlingacris elaeodes, an alpine grasshopper showing a wing polymorphism with unilateral wing form. Bull Entomol Res 106, 225–232, doi: 10.1017/S0007485315000991 (2016).
Dirks, J. H. & Taylor, D. Veins improve fracture toughness of insect wings. PLoS One 7, e43411, doi: 10.1371/journal.pone.0043411 (2012).
Rajabi, H., Darvizeh, A., Shafiei, A., Taylor, D. & Dirks, J. H. Numerical investigation of insect wing fracture behaviour. J Biomech 48, 89–94, doi: 10.1016/j.jbiomech.2014.10.037 (2015).
Podos, S. D. & Ferguson, E. L. Morphogen gradients: new insights from DPP. Trends Genet 15, 396–402 (1999).
Tabata, T. & Takei, Y. Morphogens, their identification and regulation. Development 131, 703–712, doi: 10.1242/dev.01043 (2004).
Strigini, M. & Cohen, S. M. A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development 124, 4697–4705 (1997).
Hipfner, D. R., Weigmann, K. & Cohen, S. M. The bantam gene regulates Drosophila growth. Genetics 161, 1527–1537 (2002).
Martin, F. A., Perez-Garijo, A., Moreno, E. & Morata, G. The brinker gradient controls wing growth in Drosophila. Development 131, 4921–4930, doi: 10.1242/dev.01385 (2004).
Muller, B., Hartmann, B., Pyrowolakis, G., Affolter, M. & Basler, K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell 113, 221–233 (2003).
Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B. & Cohen, S. M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 (2003).
de Celis, J. F., Barrio, R. & Kafatos, F. C. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature 381, 421–424, doi: 10.1038/381421a0 (1996).
Grimm, S. & Pflugfelder, G. O. Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science 271, 1601–1604 (1996).
Zhang, X., Luo, D., Pflugfelder, G. O. & Shen, J. Dpp signaling inhibits proliferation in the Drosophila wing by Omb-dependent regional control of bantam. Development 140, 2917–2922, doi: 10.1242/dev.094300 (2013).
Organista, M. F. & De Celis, J. F. The Spalt transcription factors regulate cell proliferation, survival and epithelial integrity downstream of the Decapentaplegic signalling pathway. Biol Open 2, 37–48, doi: 10.1242/bio.20123038 (2013).
Barrio, R. & de Celis, J. F. Regulation of spalt expression in the Drosophila wing blade in response to the Decapentaplegic signaling pathway. Proc Natl Acad Sci USA 101, 6021–6026, doi: 10.1073/pnas.0401590101 (2004).
Barrio, R., de Celis, J. F., Bolshakov, S. & Kafatos, F. C. Identification of regulatory regions driving the expression of the Drosophila spalt complex at different developmental stages. Dev Biol 215, 33–47, doi: 10.1006/dbio.1999.9434 (1999).
Schonbauer, C. et al. Spalt mediates an evolutionarily conserved switch to fibrillar muscle fate in insects. Nature 479, 406–409, doi: 10.1038/nature10559 (2011).
de Celis, J. F. & Barrio, R. Function of the spalt/spalt-related gene complex in positioning the veins in the Drosophila wing. Mech Dev 91, 31–41 (2000).
Organista, M. F. et al. The Spalt Transcription Factors Generate the Transcriptional Landscape of the Drosophila melanogaster Wing Pouch Central Region. PLoS Genet 11, e1005370, doi: 10.1371/journal.pgen.1005370 (2015).
Grieder, N. C., Morata, G., Affolter, M. & Gehring, W. J. Spalt major controls the development of the notum and of wing hinge primordia of the Drosophila melanogaster wing imaginal disc. Dev Biol 329, 315–326, doi: 10.1016/j.ydbio.2009.03.006 (2009).
Futahashi, R., Shirataki, H., Narita, T., Mita, K. & Fujiwara, H. Comprehensive microarray-based analysis for stage-specific larval camouflage pattern-associated genes in the swallowtail butterfly, Papilio xuthus. BMC Biol 10, 46, doi: 10.1186/1741-7007-10-46 (2012).
Monteiro, A. et al. Distal-less regulates eyespot patterns and melanization in Bicyclus butterflies. J Exp Zool B Mol Dev Evol 320, 321–331, doi: 10.1002/jez.b.22503 (2013).
Shirai, L. T. et al. Evolutionary history of the recruitment of conserved developmental genes in association to the formation and diversification of a novel trait. BMC Evol Biol 12, 21, doi: 10.1186/1471-2148-12-21 (2012).
Tang, W., Wang, D. & Shen, J. Asymmetric distribution of Spalt in Drosophila wing squamous and columnar epithelia ensures correct cell morphogenesis. Sci Rep 6, 30236, doi: 10.1038/srep30236 (2016).
Croker, J. A., Ziegenhorn, S. L. & Holmgren, R. A. Regulation of the Drosophila transcription factor, Cubitus interruptus, by two conserved domains. Dev Biol 291, 368–381, doi: 10.1016/j.ydbio.2005.12.020 (2006).
Cruz, C., Glavic, A., Casado, M. & de Celis, J. F. A gain-of-function screen identifying genes required for growth and pattern formation of the Drosophila melanogaster wing. Genetics 183, 1005–1026, doi: 10.1534/genetics.109.107748 (2009).
Calleja, M., Moreno, E., Pelaz, S. & Morata, G. Visualization of gene expression in living adult Drosophila. Science 274, 252–255 (1996).
Oh, H. & Irvine, K. D. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev Cell 20, 109–122, doi: 10.1016/j.devcel.2010.12.002 (2011).
Sui, L., Pflugfelder, G. O. & Shen, J. The Dorsocross T-box transcription factors promote tissue morphogenesis in the Drosophila wing imaginal disc. Development 139, 2773–2782, doi: 10.1242/dev.079384 (2012).
Affolter, M. & Basler, K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet 8, 663–674, doi: 10.1038/nrg2166 (2007).
Gibson, M. C. & Perrimon, N. Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science 307, 1785–1789, doi: 10.1126/science.1104751 (2005).
Shen, J. & Dahmann, C. Extrusion of cells with inappropriate Dpp signaling from Drosophila wing disc epithelia. Science 307, 1789–1790, doi: 10.1126/science.1104784 (2005).
Schwank, G., Yang, S. F., Restrepo, S. & Basler, K. Comment on “Dynamics of dpp signaling and proliferation control”. Science 335, 401; author reply 401, doi: 10.1126/science.1210997 (2012).
Nusinow, D., Greenberg, L. & Hatini, V. Reciprocal roles for bowl and lines in specifying the peripodial epithelium and the disc proper of the Drosophila wing primordium. Development 135, 3031–3041, doi: 10.1242/dev.020800 (2008).
Hatini, V., Green, R. B., Lengyel, J. A., Bray, S. J. & Dinardo, S. The Drumstick/Lines/Bowl regulatory pathway links antagonistic Hedgehog and Wingless signaling inputs to epidermal cell differentiation. Genes Dev 19, 709–718, doi: 10.1101/gad.1268005 (2005).
Weatherbee, S. D., Halder, G., Kim, J., Hudson, A. & Carroll, S. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev 12, 1474–1482 (1998).
Tomoyasu, Y., Wheeler, S. R. & Denell, R. E. Ultrabithorax is required for membranous wing identity in the beetle Tribolium castaneum. Nature 433, 643–647, doi: 10.1038/nature03272 (2005).
Acknowledgements
We thank Tsinghua Fly Center, the Bloomington Stock Center and Vienna Drosophila RNAi Center for fly stocks. This research was supported by the National Natural Science Foundation of China [NSFC31372255] and the 973 Program [2013CB127603].
Author information
Authors and Affiliations
Contributions
J.S. developed the concept and designed the experiments. D.W., J.L., S.L. and H.Z. performed the experiments. D.W., L.Z., W.S., and J.S. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, D., Li, J., Liu, S. et al. spalt is functionally conserved in Locusta and Drosophila to promote wing growth. Sci Rep 7, 44393 (2017). https://doi.org/10.1038/srep44393
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44393
This article is cited by
-
Knockout of crustacean leg patterning genes suggests that insect wings and body walls evolved from ancient leg segments
Nature Ecology & Evolution (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.