Abstract
Sugars play a variety of roles in plants, and their accumulation in seeds and/or surrounding pericarp tissues is distinctly different between grasses and eudicots. However, little is known about the evolutionary pattern of genes involved in sugar accumulation in these two major groups of flowering plants. Here, we compared evolutionary rates, gene duplication, and selective patterns of genes involved in sugar metabolism and transport between grasses and eudicots using six grass species and seven eudicot species as materials. Overall, sugar transporter genes exhibit divergent evolutionary patterns, whereas, sugar metabolism genes showing similar evolutionary pattern between monocots and eudicots. Sugar transporter genes have higher frequencies of recent duplication in eudicots than in grasses and their patterns of evolutionary rate are different. Evidence for divergent selection of these two groups of flowering plants is also observed in sugar transporter genes, wherein, these genes have undergone positive selection in eudicots, but not in grasses. Taken together, these findings suggest that sugar transporter genes rather than sugar metabolism genes play important roles in sugar accumulation in plants, and that divergent evolutionary patterns of sugar transporter genes are associated with the difference of sugar accumulation in storage tissues of grasses and eudicots.
Similar content being viewed by others
Introduction
Monocots and eudicots are the two major classes of angiosperms. Eudicots includes many economic crops such as soybean (Glycine max), castor-oil plant (Ricinus communis), tomato (Solanum lycopersicum), and fruit trees such as apple (Malus × domestica) and orange (Citrus sinensis). Grasses (Poaceae) are the most economically important family of monocots and contain many important cereal crops, including rice (Oryza sativa), foxtail millet (Setaria italica), and corn (Zea mays), among others. Monocots and eudicots show distinct differences in morphology and physiology, such as the structural features and storage compounds of seeds, although little is known about the mechanism underlying the differences1. In general, monocot seeds have endosperms at maturity and serving as storage tissues, whereas, mature seeds of eudicots lack endosperms, and cotyledons serve as major storage organs. Moreover, monocot seeds are usually rich in starch, whereas, eudicot seeds are rich in proteins and/or lipids. All these energy-rich storage compounds are initially derived from sugars, the primary products of photosynthesis (Fig. 1). In addition, the maternal pericarp enclosing the seed in monocots functions as a transitory storage organ during early caryopsis (grain fruit) development and the pericarp starch is remobilized into the rapidly expanding endosperm when the grains reach maximum lengths2. In contrast, in eudicots such as fruit trees, the pericarp serves as the main storage organ and accumulates large amounts of sugars. Therefore, sugar metabolism in seed, along with the surrounding pericarp, is significantly different between monocots and eudicots. Sugars are the major components of edible horticultural crops such as fruits and vegetables and they have a strong impact on the overall organoleptic quality of fruits and vegetables. Thus, understanding the mechanism(s) that drive sugar accumulation in plants will lead to novel ideas for increasing sugar yields, not only for fruit and vegetable crops, but also for bioenergy crops as well.
ADP-Glc: ADP-glucose; Glc: glucose; triose-P: triose-phosphate; G6P: Glc-6-phosphate; G1P: Glc-1-phosphate; F6P: fructose-6-phosphate; UDPG: uridine diphosphate glucose; SE/CC: sieve element/companion cell; Suc-P: Suc-phosphate; SPS: Suc-phosphate synthase; SPP: Suc-phosphate phosphatase; PGI: phosphoglucose isomerase; NI: neutral invertase; SUSY: sucrose synthase; HK: hexokinase; FK: fructokinase; PGM: phosphoglucomutase; UDPase: UDP-glucose pyrophosphorylase; SS: starch synthase; SBE: starch branching enzyme; SDE: starch debranching enzyme; AMY: amylase; A: sucrose transporter; and B: sugar transport protein.
Sugars, the primary products of photosynthesis, function as energy-storage and carbon transport molecules and substrates for starch, protein, and lipid metabolism. Sugars also function as osmotica and hormone-like primary messengers in signal transduction. Therefore, sugars perform a variety of roles in plants3. Sugar metabolism in plants comprises a complex network of multiple pathways, and involves in cytosol and several organelles, such as vacuoles and amyloplasts (Fig. 1). In sink cells, sucrose is converted to fructose and glucose by neutral invertase (NI). Fructose and glucose are further converted to fructose-6-phosphate (F6P) and Glc-6-phosphate (G6P) by fructokinase (FK) and hexokinase (HK), respectively. Moreover, sucrose can also be converted to G6P via three successive reactions catalyzed by sucrose synthase (SUSY), UDP-glucose pyrophosphorylase (UDPase), and phosphoglucomutase (PGM), respectively. Both G1P and G6P are used for starch synthesis, whereas, F6P can be combined with UDP-glucose (UDPG) to re-synthesize sucrose via suc-phosphate synthase (SPS) and suc-phosphate phosphatase (SPP). In addition, G6P and F6P can interconvert into each other via phosphoglucose isomerase (PGI). Soluble sugars are transported into vacuole by transporter proteins located on the vacuole membrane, such as sucrose transporter (SUT) and monosaccharide transporter (MST)4. The MST family is diverse and contains seven distinct subfamilies, sucrose transporter (SUT), tonoplastic monosaccharide transporter (TMT), vacuolar glucose transporter (VGT), polyol/monosaccharide transporter (PMT), sugar transporter protein (STP), inositol transporter (INT), plastidic glucose transporter (pGlcT), and early-responsive to dehydration (ERD6)-like transporter5. Genes involved in sugar metabolism and transport between cytosol and vacuole have been identified in numerous plants, and their functions have been found remarkably conserved across all eudicots and grasses3,6.
In plants, sugar accumulation is dependent upon two processes, biosynthesis and transport. To date, many studies have been conducted to investigate regulatory mechanisms of sugar accumulation7,8,9. Some studies have reported that genes involved in sugar biosynthesis, including NI, SUSY, HK, and SPS, play important roles in sugar accumulation10,11,12,13,14, while other studies have argued that genes involved in sugar transport, such as SUT and MST families, are important for sugar accumulation15,16,17. Therefore, additional studies are needed to clarify whether it is sugar biosynthesis or transport that is more important in sugar accumulation.
More recently, several studies reveal evolutionary patterns of genes involved in starch biosynthesis in plants. For example, Yu et al.18 reported the strong evidence of positive selection at AGPL, GBSSI and SBEII in rice wild ancestor18. Li et al.19 found that divergent evolutionary pattern of genes involved in starch biosynthetic pathway between grasses and eudicots contributes to the observed differences in starch biosynthesis in seeds19. However, there are few reports on the mechanisms underlying the observed sugar accumulation difference in seeds and their surrounding pericarps between grasses and eudicots. In this study, we compared the evolutionary rates, gene duplication, and selective patterns of genes involved in sugar accumulation between grasses and eudicot plants, including four above-mentioned key gene families involved in sugar metabolism, HK, NI, SPS, and SUSY, and two sugar transporter gene families, SUT and MST. It is worth noting that a newly identified sugar transporter family called SWEET is involved in sugar transport in plants20. However, most SWEET genes play an important roles in phloem transport and are related to biotic and abiotic stress21,22,23,24,25. Since phloem transport is not a limited step for sugar accumulation in storage organs such as seeds or fruits26, the SWEET family was not included in this study. Our aim is to determine whether genes involved in either sugar metabolism or transport are most critical for sugar accumulation in plants. Our study not only provides insights into the observed divergent evolutionary patterns of genes involved in sugar accumulation in plants, but will also contribute to understanding of the observed morphological and physiological differences between grass and eudicot plants.
Materials and Methods
Retrieval and Comparison of Sequences
Amino acid sequences of genes involved in sugar accumulation and transport were collected using the protocol described by Tatusov et al.27, but with slight modifications. Briefly, the main steps were as follows: (1) Protein-coding transcripts of Brachypodium distachyon (v2.1 release)28, Oryza sativa (MSU Release 7.0)29, Setaria italic (v2.1 release)30, Sorghum bicolor (v2.1 release)31, Zea mays (6a release)32, Arabidopsis thaliana (TAIR release)33, Citrus sinensis (v1.1 release)34, Malus × domestica (v1.0 release)35, Glycine max (Glyma1.0, Soybean Genome Project)36, Ricinus communis (release v0.1)37, Solanum lycopersicum (iTAG2.3 release)38, Panicum virgatum (v1.1), and Populus trichocarpa (v3.0)39 were downloaded from either JGI (http://www.phytozome.net/) or the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) and a local BLAST database was constructed for each species using BLAST 2.2.24; (2) An all-against-all protein sequence comparison was carried out to identify homologous genes of Arabidopsis or Oryza sativa in eudicots and grasses, respectively; (3) In all the comparisons performed in the step 2, the orthologs were further identified using BLAST 2.2.24 with SUT2 in Arabidopsis or rice as a query, and obvious paralogs were collapsed; (4) All interspecies Best Hits (BeTs) of AtSUT2 or OsSUT2 and their paralogs were detected; (5) Steps 3 and 4 were repeated, with other SUT genes rather than SUT2 as BLASTp queries until all the SUT genes were identified; (6) Clusters of Orthologous Groups (COGs) were formed for the SUT gene family using the protocol previously reported by Tatusov et al.27; (7) COGs were constructed for other gene families using the same protocol as conducted for the SUT gene family.
Estimation of Gene Duplication and Gene Loss
Within each gene family, gene duplication and loss were analyzed by checking of each subfamily manually. Each gene family was assumed to have at least two subclades, grass and eudicot subfamilies, with at least six and seven members in grass and eudicot subfamilies, respectively. If there are two or more grass or dicot subclades, or if a subclade has two or more members from the same species, it was assumed to have one or more duplications. If a member was found to be missing from any species within the grass or eudicot subclade, it was concluded that there was a gene loss. Each gene loss was further validated by searching GenBank database using BLASTp.
Phylogenetic Analyses
Amino acid sequences were aligned using CLUSTAL X and manually adjusted using Seaview (Galtier et al.40. The resulting data matrix was used to construct a phylogenetic tree using the MEGA software (v6.06)41 with both Neighbor Joining (NJ) and Maximum Likelihood (ML) methods. For the NJ method, the parameters were as follows: model, p-distance; bootstrap, 1,000 replicates; and gaps/missing data treatment, pairwise deletion. For the ML method, the parameter setups were as follows: model, Jones-Taylor-Thornton (JTT); bootstrap, 1,000 replicates; gaps/missing data treatment, partial deletion; and branch swap filter, very strong.
Estimation of dN:dS Ratio
The codon sequences were aligned using MEGA v7.0 software with muscle codons alignment model, and termination codons were manually deleted. The alignment result was used for estimation of the ω values using maximum likelihood method in Codeml from the PAML v4.9a42. The significance of variation in the ω value among different branches in gene trees was tested using Codeml, a branch-specific model. This branch-specific model can be compared with one-ratio model that assumes a constant ω value across all branches using the likelihood ratio test (LRT).
Coding DNA sequences were aligned using CLUSTAL X and adjusted manually, as deemed necessary. The resulting data matrix was used to estimate ω value, the ratio of nonsynonymous substitutions per nonsynonymous site (dN) to synonymous substitutions per synonymous site (dS) of homologous gene pairs. The estimation of ω value was performed with the maximum likelihood method using a KaKs_Calculator package (v1.2)43,44.
The variation of the ω ratio among different branches in gene trees was tested using the pairwise comparison approach19. Briefly, the ω ratios between two randomly selected sequences within the grass or eudicot subclade were calculated using the KaKs_Calculator package. The difference of the ω ratios were compared between grass and eudicot subclades with the paired t-test using SPSS, and the comparison was only conducted between orthologous subclades. Significant difference was set as P < 0.05.
Identification of Positively Selected Sites
The codon sequences from grasses or eudicots were aligned using the online program MAFFT (http://mafft.cbrc.jp/alignment/server/), and the results were saved in PHYLIP format files. The aligned sequences were used to construct phylogenetic trees using the MEGA software (v6.06)41 with Neighbor Joining (NJ), and the results were stored in tree files. The PHYLIP format files, along with the trees file, were used for estimating dN:dS ratios with maximum likelihood method using the program Fitmodel (version: 0.5.3)45. The parameter setups were as follows: data type, DNA; DNA substitution model applies, codons; genetic code, universal; model of codon substitution, M3; model of swtiches between selection reigns, S1 or not; start with default values for switching rates, yes; start with default dn/ds ration parameters, yes; Ts/tv ratio, fixed; codon frequency, F3 × 4; analyze multiple data sets, no; optimis substitution parameters, yes. The dN:dS ratios were subsequently used to estimate positively selected sites in grasses or eudicots. Moreover, switching parameters were also estimated using the models M3 and M3 + 1, which were described by Yang and Nielsen46 and Guindon et al.45.
Detection of Functional Divergence after Gene Duplication
Estimation of gene functional divergence after gene duplication was conducted using the Type II model in DIVERGE v3.0 software47. The amino acid sequences from grasses and eudicots were aligned using MEGA v7.0 and the results were saved as FASTA format. NJ phylogenetic tree was constructed using PHYLIP v3.6 software following the instructions of the DIVERGE v3.0 software. In addition, each gene subfamily within the same phylogenetic tree was treated as one cluster.
Results
Phylogenetic Analyses
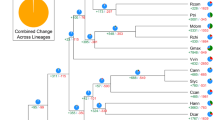
Twelve COGs were obtained and are shown in Fig. 2 and supplementary Fig. S1. Homologs of twelve gene families, HK, NI, SPS, SUSY, SUT, TMT, VGT, PMT, STP, ERD6-like, pGlcT, and INT, were included in a single COG (Fig. 2 and supplementary Fig. S1). Phylogenetic trees showed that both NJ and ML methods yielded identical topologies (Fig. 3 and supplementary Fig. S2). Each gene family consisted of multiple subfamily clades, which were supported by high levels of confidence (mostly > 90% bootstrap value) in all phylogenetic trees. For example, the SUT gene family was composed of three eudicot subfamilies and five grass subfamilies (Fig. 3), and all these eight subfamilies were clustered into three clades (SUT1, SUT2, and SUT4). This is consistent with earlier finding that the SUT gene family in A. thaliana is composed of three types48. However, all SUT genes in grasses were grouped into the SUT2 and SUT4 types, with none belonging to the SUT1 type (Fig. 3). As for the SUT1 type, gene duplication occurred frequently in most eudicot species such as A. thaliana following the monocot-eudicot split. For the SUT2 type, gene duplication occurred twice in grasses during the process of speciation. In addition, both grasses and eudicots contain SUT2 and SUT4 types, which suggested that the duplication of SUT genes have also occurred prior to the monocot-eudicot split. In short, gene duplication events in the SUT gene family have occurred at various time points during the process of plant evolution, covering both before and after the monocot-eudicot split.
Similar to the SUT gene family, the MST gene families, TMT, VGT, PMT, STP, ERD6-like, pGlcT, and INT, are also composed of multiple subfamilies (supplementary Fig. S2). The STP gene family contained the most abundant subfamilies, including nine grass subfamilies and eight eudicot subfamilies, while the VGT gene family had the lowest number of subfamilies, including two grass subfamilies and two eudicot subfamilies. The rest five transporter gene families, TMT, PMT, ERD6-like, pGlcT, and INT, contained 3–5 grass subfamilies and 3–4 eudicot subfamilies. Most duplication events of the MST gene families have primarily occurred following the monocot-eudicot split. Gene loss event was observed in all the MST gene families except the VGT family.
For sugar metabolism gene families, both HK and NI families contained eight grass subfamilies and four eudicot subfamilies, while SPS and SUSY families consisted of 5–6 grass subfamilies and 4–5 eudicot subfamilies. Similar to SUT genes, duplication events of these sugar metabolism genes have occurred both before and after the monocot-eudicot split.
Differences in Gene Duplication and Loss Patterns between Grasses and Eudicots
It has been reported that gene duplication can be classified into either recent or old duplications according to the time of occurrence49. In this study, both grass-specific (duplication prior to radiation of grasses) and eudicot-specific (duplication prior to radiation of eudicots) duplications were designated as old duplications, whereas, recent duplications were deemed as species-specific duplications (duplication within a species). Both recent and old duplications were investigated for each gene family (Table 1). In grasses, old duplication occurred approximately 26 times in ten gene families (PMT, HK, SUT, SUSY, PMT, TMT, NI, SPS, ERD6-like, and INT), but only five times in five gene families (SPS, HK, SUSY, STP and TMT) in eudicots. In addition, recent duplication in grasses occurred 17 times in six gene families (NI, SPS, HK, TMT, STP, and SUSY), whereas, recent duplication occurred 144 times in all the tested gene families in eudicots. The STP genes showed the highest frequency of recent duplication in eudicots (34 times). In short, old duplication frequently occurred in grasses, while recent duplication frequently occurred in eudicots. In addition, it is interesting to note that the frequencies of recent duplications showed significant variations among tested eudicot species, with most recent duplications observed in the following two species, M. ×domestica and G. max.
Recent gene loss in grasses was observed in ten gene families, including HK, TMT, NI, STP, SPS, PMT, SUT, pGlcT, ERD6-like, and INT (Table 1). Similarly, recent gene loss in eudicots was also observed in ten gene families, including HK, TMT, NI, STP, SPS, PMT, SUT, pGlcT, ERD6-like, and SPS. Moreover, old gene loss was only observed in the SUT1 clade in grasses (Table 1, Fig. 3). Overall, recent gene loss events occurred frequently in both grasses and eudicots, while old gene loss event rarely occurred in both grassed and eudicots (Table 1, Fig. 3, and supplementary Fig. S2).
Most Genes Involved in Sugar Metabolism and Transport Have Rapidly Evolved in Grasses
To determine whether sugar metabolism and transport gene families are under different evolutionary constraints in both grasses and eudicots, pairwise ω (dN/dS) values for all tested genes were calculated using the pairwise comparison model. The pairwise ω values within each subfamily were subsequently compared between grasses and eudicots. As a result, 10 subfamilies within the SUT, HK, VGT, PMT, STP and pGlcT gene families showed significantly higher mean ω values in grasses than in eudicots (Fig. 4, Supplementary Table S1), whereas, 13 subfamilies within the STP, TMT, INT, pGlcT, and ERD6-like families showed significantly higher mean ω values in eudicots than in grasses (Supplementary Table S1). This finding suggests that the majority of the genes involved in sugar transport are significantly different in evolutionary rates between grasses and eudicots.
Since pairwise comparison approach model cannot calculate the evolutionary rate of the most recent common ancestor of the subfamily, the branch-specific model was selected to check whether there are the changes in the evolutionary constraints across different subfamilies. As a result, all the metabolic gene families had p-values larger than 0.05, whilst almost all sugar transport gene families except VGT had p-values smaller than 0.05 (Table 2). This indicates that the evolutionary rate of sugar transport gene families is different between grasses and eudicots.
In addition, we compared the ω values between members within the SUT1 clade. Interestingly, all the ω values in three fruit crops, S. lycopersicum, C. sinensis, Malus × domestica, were higher than 1.0 (Fig. 5). This suggests that SUT1 in these three fruit crops has higher evolutionary rates compared with other eudicots investigated in this study.
A Divergent Selection of Sugar Transporter Gene Families Observed in Grasses versus Eudicots
To assess the potential for selection of genes involved in sugar metabolism and transport, the Fitmodel software was used to conduct selection analysis with both M3 and M3 + S1 models45, and the sites under positive selection for each gene family were compared between grasses and eudicots. Parameter estimates for selection analysis were found to be similar between M3 and M3 + S1 models (Table 3), thus, only results from M3 + S1 model analysis was presented.
Overall, more than 85% of sites (p1 and p2) in all tested genes, except for NI, were identified to be under purifying selection with ω1 ≤ 0.18 and ω2 ≤ 0.87. Moreover, 1% to 15% of identified sites (p3) within ten sugar transporter genes (SUT1, SUT2, SUT4, STP, VGT, TMT, PMT, pGlcT, ERD6-like, and INT) and one sugar metabolism gene NI were identified to be under positive selection with an ω3 value > 1.0. Among these positive selection sites, the number of sites for genes in SUT1, SUT2, TMT, PMT, VGT, pGlcT, ERD6-like, and INT gene families were significantly asymmetric across both grasses and eudicots (Table 4). This suggests that selection is mainly located either in the grass or eudicot subclade of the phylogenetic tree, thus providing a key evidence of the incidence of divergent selection between grasses and eudicots.
Type II Functional Divergence after Gene Duplication
The coefficient of type II functional divergence (θ) between any two clusters was calculated using DIVERGE v3.0. For sugar transport gene families, non-orthologous clusters had θ-II values that were significantly larger than 0, but not for orthologous clusters (Table 5, Supplementary Table S2). However, such a pattern was not observed between any two clusters of sugar metabolic gene families (Supplementary Table S2). This suggests that type II functional divergence may occur between sugar transport gene subfamilies.
Discussion
In this study, evolutionary features such as duplication patterns, evolutionary rates, and positive selection have been investigated for twelve gene families involved in sugar metabolism and transport in plants, including HK, NI, SPS, SUSY, PMT, STP, SUT, TMT, VGT, pGlcT, ERD6-like, and INT. Overall, sugar transporter gene families have demonstrated divergent evolutionary pattern between grasses and eudicots, whereas, sugar metabolism gene families have revealed similar evolutionary patterns between grasses and eudicots. For example, the SUT1 clade is found to be exclusively present in eudicots, and SUT2 genes show high frequencies of old gene duplication in grasses than in eudicots. Moreover, STP genes exhibit higher frequency of recent duplication in eudicots than in grasses, whereas, they show higher frequency of old duplication in grasses than in eudicots. In addition, both SUT2 and SUT4 gene families in grasses have faster evolutionary rates than in eudicots. Given the fact that sugar accumulation in seeds and/or pericarps is significantly different between grasses and eudicots, we hypothesize that this observed difference is associated with divergent evolutionary patterns of sugar transporter gene families.
Hypothesis 1 - Gene Duplication of Sugar Transporters Contributes to Sugar Accumulation in Eudicots
In this study, almost all analyzed transporter genes in eudicots, including PMT, STP, SUT, TMT, VGT, pGlcT, ERD6-like, and INT, have undergone one or more rounds of duplication in eudicots (Table 1, Fig. 3, and supplementary Fig. S2). The SUT genes serve as a typical example of this finding as they have undergone duplication eighteen times in eudicots.
In plants, the SUT gene family encodes sucrose/H+ symporters, and it consists of three types, termed SUT1, SUT2 and SUT450,51,52,53. In this study, the SUT1 subfamily is found to be present exclusively in eudicots. Previous studies show that both SUT1 and SUT2 proteins are localized to plasma membrane54,55, while SUT4 protein is localized to vacuolar membrane56,57. The SUT1 genes are expressed in both source and sink tissues and play essential functions such as those of phloem loading and increasing sucrose uptake into sink storage tissues58,59,60,61,62. On the other hand, SUT2 genes are mainly expressed in sink tissues and function in phloem loading63,64,65,66. Whereas, SUT4 genes show variable patterns of expression in different plant species and function in sucrose uptake from the vacuole into the cytoplasm61,62,63,64,65,66,67,68,69. In higher plants, it is commonly known that sucrose is the main mobile carbohydrate. SUT1 genes exhibit very high affinity and transport ability with sucrose70,71,72,73, while SUT2 and SUT4 genes have no or low affinity for sucrose55,74,75,76,77. Moreover, both SUT2 and SUT4 genes are not reported to increase sucrose uptake into the sink storage tissues. Thus, the SUT1 subfamily is deemed critical for sugar transport and its duplication is likely to be associated with observed high levels of sugar accumulation in eudicots. In turn, loss of the SUT1 subfamily in grasses is probably one of the main causes leading to observed differences of sugar accumulation in storage tissues between grasses and eudicots.
In this study, it is found that SUT2 genes, unlike SUT1 genes, show high frequencies of old duplication in grasses, but no duplication detected in eudicots. SUT2 genes are reported as potential candidates for starch accumulation55. Thus, duplication of SUT2 genes may contribute to starch accumulation in grasses.
Besides the SUT gene family, the STP gene family also functions as sugar transporters69,78,79,80. The STP gene family, also known as hexose transporters (HTs), is located in the plasma membrane, functions as monosaccharide/proton symporter, and plays an important role in monosaccharide import into sink cells80. In fruits, several studies have demonstrated that the STP gene family plays important roles in sugar accumulation81,82,83. In this study, the STP gene family is found to show differences in duplication patterns between eudicots and grasses. In grasses, the STP gene family has undergone four duplications in ancient times, but six duplications in recent times. In eudicots, the STP gene family has undergone only one duplication in ancient times, but has also undergone 34 recent duplications as well. Interestingly, the duplication of the STP gene family in recent times has reached as high as 24 times in three fruit crops, apple, tomato, and orange. This suggests that gene duplication of the STP subfamily may also contribute to sugar accumulation in eudicots. In addition, the rest six transporter gene families, PMT, TMT, VGT, pGlcT, ERD6-like, and INT, show similar duplication patters to SUT and STP gene families, which suggests that they are may also play important roles in sugar accumulation. However, sugar metabolism genes, unlike sugar transporter genes, show no significant differences in gene duplication patterns between grasses and eudicots (Fig. S2), thus indicating that they are unlikely responsible for observed differences in sugar accumulation between eudicots and grasses.
Taken together, all these findings support our hypothesis that sugar transporter gene duplication plays an important role in sugar accumulation in eudicots. In addition, our study reveals that functional divergence may occur after gene duplication between sugar transport gene subfamilies. More experiments are needed to clarify the roles of sugar transport gene subfamilies in sugar accumulation.
Hypothesis 2 - Evolutionary Rate Patterns of Sugar Transporters are Associated with Differences in Sugar Accumulation between Eudicots and Grasses
The ω value is an important index of genetic differentiation. In this study, both one-ratio model and a branch model were conducted to estimate the ω value, and the likelihood of these two models was compared using LRT statistic. The result reveals that evolutionary rates of most sugar transporter gene families are significantly different between grasses and eudicots. In contrast, no significant difference is observed for most sugar metabolism gene families between grasses and eudicots (Table 2). Among all tested genes in this study, those with ω > 1 are exclusively found in the SUT1 subfamily. Interestingly, all SUT1 genes with ω > 1 are derived from fruit crops, including apple, orange, and tomato (Fig. 5). This indicates that SUT1 genes in fruit crops have faster evolutionary rates than those in other eudicot species. As fruits are rich in sugars, we hypothesize that faster evolutionary rates of SUT1 genes contribute to sugar accumulation in eudicots.
In grasses, the SUT2 clade consist of four subfamilies. In addition, grass subfamily 2 has an orthologous relationship with the eudicot subfamily 3. The ω value for grass subfamily 2 is significantly higher than that for the eudicot subfamily 3. Similarly, SUT4 genes have significantly higher ω values in grasses than those found in eudicots. These results suggest that both SUT2 and SUT4 genes have significantly different evolutionary rates between grasses and eudicots. Several studies have reported that SUT2 is a sucrose sensor that can interact with SUT1 and/or SUT4 to regulate their relative activities55,75,84. Thus, the evolutionary rates of both SUT2 and SUT4 genes are also likely associated with sugar accumulation in plants.
As with SUT genes, other sugar transporter genes, such as STP, VGT, PMT, TMT, pGlcT, ERD6-like, and INT, have shown significant differences in ω values between grasses and eudicots. In a recent study, it is reported that rapid evolution of starch pathway genes contributes to starch accumulation in grasses20. Therefore, it is most likely that the evolutionary rate of sugar transporter gene families has influences on sugar accumulation in plants.
Unlike sugar transporter gene families, almost all gene families involved in sugar metabolism, except for HK4, show no significant differences in ω values between grasses and eudicots. The HK gene is responsible for hexose phosphorylation, and plays a vital role in plant sugar sense and sugar signal transduction85,86,87. The HK protein is localized in different organelles, including cell cytosol, chloroplastid, chondriosome, plasmid, and golgiosome88,89. In an earlier study, it has been demonstrated that the HK4 gene is incapable of catalyzing phosphorylation of hexoses, such as glucose or fructose87. Thus, it is clear that the patterns of evolutionary rates of HK genes involved in hexose phosphorylation are not associated with observed differences in sugar accumulation between grasses and eudicots. Such a phenomenon is quite commonly observed in genes involved in sugar metabolism (supplementary Table S1).
In this study, positive selection analysis has revealed that sites with ω3 values higher than one have been identified for all analyzed transport genes. However, positive selection sites are present exclusively in sugar transporter gene families in eudicots. This finding also demonstrates that sugar transporter genes are likely associated with high levels of sugar accumulation in eudicots.
In conclusion, sugar transporter genes rather than sugar metabolism genes play important roles in sugar accumulation in plants, and differences in sugar accumulation in storage tissues between grasses and eudicots may be attributed to at least three reasons as follows: 1) sugar transporter genes have higher frequencies of recent duplications in eudicots rather than in grasses; 2) patterns of evolutionary rates of sugar transporter genes are different between grasses and eudicots; and 3) sugar transporter genes have undergone positive selection in eudicots, but not in grasses. In short, divergent evolutionary patterns of sugar transporter genes are responsible for differences in sugar accumulation between grasses and eudicots. However, molecular mechanisms underlying the evolutionary divergence of sugar transporter genes between grasses and eudicots remain unclear.
As mentioned above, the SUT gene family highly influences sugar accumulation in plants, especially in fruit crops. During fruit development, sucrose is imported into the fruit, and it is subsequently cleaved into glucose and fructose. These latter sugar components not only contribute to fruit sweetness, but they can also be used as sources of energy for fruit growth. By increasing the uptake of sucrose into developing fruits, this may contribute to increased levels of sugar content in mature fruits. Therefore, manipulation of SUT genes may lead to enhanced sugar content in fruit crops.
Additional Information
How to cite this article: Wang, W. et al. Divergent Evolutionary Pattern of Sugar Transporter Genes is Associated with the Difference in Sugar Accumulation between Grasses and Eudicots. Sci. Rep. 6, 29153; doi: 10.1038/srep29153 (2016).
References
Sreenivasulu, N. & Wobus, U. Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu Rev Plant Biol. 64, 189–217 (2013).
Sreenivasulu, N. et al. Transcript profiles and deduced changes of metabolic pathways in maternal and filial tissues of developing barley grains. Plant J. 37, 539–553 (2004).
Halford, N. G., Curtis, T. Y., Muttucumaru, N., Postles, J. & Mottram, D. S. Sugars in crop plants. Ann. Appl. Biol. 158, 1–25 (2011)
Martinoia, E., Meyer, S., De Angeli A. & Nagy R. Vacuolar transporters in their physiological context. Annu. Rev. Plant Biol. 63, 183–213 (2012).
Büttner, M. The monosaccharide transporter(-like) gene family in Arabidopsis . FEBS Lett. 581, 2318–2324 (2007).
Ngampanya, B., Takeda, T., Narangajavana, J., Ikeda, A. & Yamaguchi, J. Sugar transporters involved in flowering and grain development of rice. J Appl Glycosci. 50, 237–240 (2003).
Granot, D., David-Schwartz, R. & Kelly-Frontiers, G. Hexose kinases and their role in sugar-sensing and plant development. Front. Plant Sci 44, 1–17 (2013).
Siswoyo, T. A., Oktavianawatia, I., Djenala, Murdiyantob, U. & Sugihartoa, B. Changes of sucrose content and invertase activity during sugarcane stem storage. J. Agr. Sci. 8, 75–81 (2013).
Tognetti, J. A., Pontis, H. G. & Martínez-Noël, G. M. A. Sucrose signaling in plants. Plant Signal. Behav. 8, e23316 (2013).
Dale, E. M. D. & Housley, T. L. Sucrose synthase activity in developing wheat endosperms differing in maximum weight. Plant Physiol. 82, 7–10 (1986).
Sun, J., Loboda, T., Sung, S. J. S. & Black, C. C. Sucrose synthase in wild tomato, Lycopersicon chmielewskii, and tomato fruit sink strength. Plant Physiol. 98, 1163–1169 (1992).
Klann, E. M., Hall, B. & Bennett, A. B. Antisense acid invertase (TW7) gene alters soluble sugar composition and size in transgenic tomato fruit. Plant Physiol. 112, 1321–1330 (1996).
D’Aoust, M. A., Yelle, S. & Quoc, B. N. Antisense inhibition of tomato fruit sucrose synthase decrease fruit setting and the sucrose unloading capacity of young fruit. Plant Cell 11, 2407–2418 (1999).
Baroja-Fernandez, E. et al. Enhancing sucrose synthase activity in transgenic potato (Solanum tuberosum L.) tubers results in increased levels of starch, ADP glucose and UDP glucose and total yield. Plant Cell Physiol. 50, 1651–1662 (2009).
Davies, C., Wolf, T. & Robinson, S. P. Three putative sucrose transporters are differentially expressed in grapevine tissues. Plant Sci. 147, 93–100 (1999).
Martinoia, E., Meyer, S., De Angeli, A. & Nagy, R. Vacuolar transporters in their physiological context. Annu. Rev. Plant Biol. 63, 183–213 (2012).
Lecourieux, F. et al. An update on sugar transport and signalling in grapevine. J. Exp. Bot. 65, 821–832 (2014).
Yu, G. Q., Olsen, K. M. & Schaal, B. A. Molecular evolution of the endosperm starch synthesis pathway genes in rice (Oryza sativa L.) and its wild ancestor, O. rufipogon L. Mol. Biol. Evol. 28, 659–671 (2011).
Li, C., Li, Q. G., Dunwell, J. M. & Zhang, Y. M. Divergent evolutionary pattern of starch biosynthetic pathway genes in grasses and dicots. Mol. Biol. Evol. 29, 3227–3236 (2012).
Eom, J. S. et al. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 25, 53–62 (2015).
Chen, L. Q. et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211 (2012).
Chong, J. et al. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea . J. Exp. Bot. 65, 6589–6601 (2014).
Chen, H. Y. et al. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 83, 1046–1058 (2015).
Hutin, M., Sabot, F., Ghesquière, A., Koebnik, R. & Szurek, B. A knowledge-based molecular screen uncovers a broad-spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703 (2015).
Chen, L. Q. et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532 (2010).
Ruan, Y. & Patrick, J. W. The cellular pathway of postphloem sugar transport in developing tomato fruit. Planta 196, 434–444 (1995).
Tatusov, R. L., Galperin, M. Y., Natale, D. A. & Koonin, E. V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic. Acids Res. 28, 33–36 (2000).
International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature 463, 763–768 (2010).
Ouyang, S. et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic. Acids Res. 35, D883–D887 (2007).
Bennetzen, J. L. et al. Reference genome sequence of the model plant Setaria . Nature Biotechnol. 30, 555–561 (2012).
Paterson, A. H. et al. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009).
Schnable, P. S. et al. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 (2009).
Lamesch, P. et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic. Acids Research 40, 1202–1210 (2013).
Wu, G. A. et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nature Biotechnol. 32, 656–662 (2014).
Velasco, R. et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42, 833–839 (2010).
Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature. 463, 178–183 (2010).
Chan, A. P. et al. Draft genome sequence of the oilseed species Ricinus communis. Nature Biotechnol. 28, 951–956 (2010).
Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
Tuskan, G. A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (2006).
Galtier, N., Gouy, M. & Gautier, C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12, 543–548 (1996).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24, 1586–1591 (2007).
Zhang, Z. & Li, J. & Yu, J. Computing Ka and Ks with a consideration of unequal transitional substitutions. BMC Evol. Biol. 6, 44 (2006).
Zhang, Z. & Yu, J. Evaluation of six methods for estimating synonymous and nonsynonymous substitution rates. Genomics Proteomics Bioinformatics 4, 173–181 (2006).
Guindon, S., Rodrigo, A. G., Dyer, K. A. & Huelsenbeck, J. P. Modeling the site-specific variation of selection patterns along lineages. Proc. Natl. Acad. Sci. 101, 12957–12962 (2004).
Yang, Z. & Nielsen, R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917 (2002).
Gu, X. Maximum likelihood approach for gene family evolution under functional divergence. Mol. Biol. Evol. 18, 453–464 (2001).
Sauer, N. et al. AtSUC8 and AtSUC9 encode functional sucrose transporters but the closely related AtSUC6 and At-SUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J. 40, 120–130 (2004).
Taylor, J. S. & Raes, J. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 38, 615–643 (2004).
Kühn, C. et al. The sucrose transporter StSUT1 localizes to sieve elements in potato tuber phloem and influences tuber physiology and development. Plant Physiol. 131, 102–113 (2003).
Sauer, N. Molecular physiology of higher plant sucrose transporters. FEBS. Lett. 581, 2309–2317 (2007).
Shiratake, K. Genetics of sugar transporter in plants. G3: Genes Genomes Genomics. 1, 73–80 (2007).
Kühn, C. & Grof, C. P. L. Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 13, 287–297 (2010).
Kühn, C., Franceschi, V. R., Schulz, A., Lemoine, R. & Frommer, W. B. Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275, 1298–1300 (1997).
Barker, L. et al. SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12, 1153–1164 (2000).
Payyavula, R. S., Tay, K. H., Tsai, C. J. & Harding, S. A. The sucrose transporter family in Populus: the importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant J. 65, 757–770 (2011).
Schneider, S. et al. Vacuoles release sucrose via tonoplast-localised SUC4-typetransporters. Plant Biol. 14, 325–336 (2012).
Riesmeier, J. W., Willmitzer, L. & Frommer, W. B. Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 13, 1–7 (1994).
Rosche, E. et al. Seed-specific overexpression of a potato sucrose transporter increases sucrose uptake and growth rates of developing pea cotyledons. Plant J. 30, 165–175 (2002).
Leggewie, G. et al. Overexpression of the sucrose transporter SoSUT1 in potato results in alterations in leaf carbon partitioning and in tuber metabolism but has little impact on tuber morphology. Planta. 217, 158–167 (2003).
Geiger, D. Plant sucrose transporters from a biophysical point of view. Mol. Plant 4, 395–406 (2011).
Reinders, A. et al. Protein-protein interactions between sucrose transporters of different affinities colocalized in the same enucleate sieve element. Plant Cell 14, 1567–1577 (2002).
Shakya, R. & Sturm, A. Characterization of source- and sink-specific sucrose/H+ symporters from carrot. Plant Physiol. 118, 1473–1480 (1998).
Meyer, S. et al. AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular tissue and in a carpel cell layer. Plant J. 24, 869–882 (2000).
Hackel, A. et al. Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant J. 45, 180–192 (2006).
Slewinski, T. L., Meeley, R. & Braun, D. M. Sucrose transporter 1 functions in phloem loading in maize leaves. J. Exp. Bot. 60, 881–892 (2009).
Endler, A. et al. Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 141, 196–207 (2006).
Reinders, A., Sivitz, A. B., Starker, C. G., Gantt, J. S. & Ward, J. M. Functional analysis of LjSUT4, a vacuolar sucrose transporter from Lotus japonicus. Plant Mol. Biol. 68, 289–299 (2008).
Schulz, A. et al. Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 68, 129–136 (2011).
Bock, K. W. et al. Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol. 140, 1151–1168 (2006).
Sivitz, A. B. et al. Arabidopsis sucrose transporter AtSUC9, high-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiol. 143, 188–198 (2007).
Sivitz, A. B., Reinders, A. & Ward, J. M. Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol. 147, 92–100 (2008).
Srivastava, A. C., Ganesan, S., Ismail, I. O. & Ayre, B. G. Functional characterization of the Arabidopsis AtSUC2 sucrose/H1 symporter by tissue-specific complementation reveals an essential role in phloem loading but not in longdistance transport. Plant Physiol. 148, 200–211 (2008).
Schulze, W., Weise, A., Frommer, W. B. & Ward, J. M. Function of the cytosolic N-terminus of sucrose transporter AtSUT2 in substrate affinity. FEBS Lett. 485, 189–194 (2000).
Weise, A. et al. A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12, 1345–1356 (2000).
Reinders, A., Sivitz, A. B. & Ward, J. M. Evolution of plant sucrose uptake transporters. Front. Plant Sci. 3, 22 (2012).
Barth, I., Meyer, S. & Sauer, N. PmSUC3: characterization of a SUT2/SUC3-type sucrose transporter from Plantago major. Plant Cell 15, 1375–1385 (2003).
Truernit, E., Schmid, J., Epple, P., Illig, J. & Sauer, N. The sink-specific and stress regulated Arabidopsis gene: enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 8, 2169–2182 (1996).
Nørholm, M. H. H. et al. Expression of the Arabidopsis high-affinity hexose transporter STP13 correlates with programmed cell death. FEBS letters. 9, 2381–2387 (2006).
Büttner, M. The Arabidopsis sugar transporter (AtSTP) family: an update. Plant Biol. 12, 35–41 (2010).
Hayes, M. A., Davies, C. & Dry, I. B. Isolation, functional characterization, and expression analysis of grapevine (Vitis vinifera L.) hexose transporters: differential roles in sink and source tissues. J. Exp. Bot. 58, 1985–1997 (2007).
Vignault, C. et al. VvHT1 encodes a monosaccharide transporter expressed in the conducting complex of the grape berry phloem. J. Exp. Bot. 56, 1409–1418 (2005).
McCurdy, D. W., Dibley, S., Cahyanegara, R., Martin, A. & Patrick, J. W. Functional characterization and RNAi-mediated suppression reveals roles for hexose transporters in sugar accumulation by tomato fruit. Mol. Plant 3, 1049–1063 (2010).
Riesmeier, J. W., Willmitzer, L. & Frommer, W. B. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 11, 4705–4713 (1992).
Halford, N. G., Purcell, P. C. & Hardie, D. G. Is hexokinase really a sugar sensor in plants? Trends Plant Sci. 4, 117–120 (1999).
Loreti, E., Bellis, L. D., Alpi, A. & Perata, P. Why and how do plant cells sense sugars? Ann. Bot. 88, 803–812 (2001).
Cho, J. I. et al. Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 149, 745–759 (2009).
Balasubramanian, R., Karve, A., Kandasamy, M., Meagher, R. B. & Moore, B. A role for F-action in hexokinase-mediated glucose signaling. Plant Physiol. 145, 1423–1434 (2007).
Karve, A. et al. Expression and evolutionary features of the hexokinase gene family in Arabidopsis . Planta. 228, 411–425 (2008).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 31420103914) and the Overseas Construction Plan for Science and Education Base, China-Africa Center for Research and Education, Chinese Academy of Sciences (Grant No. SAJC201327).
Author information
Authors and Affiliations
Contributions
Y.H. and W.W. conceived and designed the experiments. W.W., H.Z. and B.M. performed the experiments. Y.H. and W.W. wrote the paper. S.S.K. and A.O. revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, W., Zhou, H., Ma, B. et al. Divergent Evolutionary Pattern of Sugar Transporter Genes is Associated with the Difference in Sugar Accumulation between Grasses and Eudicots. Sci Rep 6, 29153 (2016). https://doi.org/10.1038/srep29153
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep29153
This article is cited by
-
Comparative genomics reveals probable adaptations for xylose use in Thermoanaerobacterium saccharolyticum
Extremophiles (2024)
-
Exogenous Sorbitol Modifies Sugar Metabolism, Transport and Signalling in In Vitro-Grown ‘Myrobalan 29C’ According to Tissue
Journal of Plant Growth Regulation (2023)
-
Physiological responses and expression of sugar associated genes in faba bean (Vicia faba L.) exposed to osmotic stress
Physiology and Molecular Biology of Plants (2021)
-
The sucrose transporter MdSUT4.1 participates in the regulation of fruit sugar accumulation in apple
BMC Plant Biology (2020)
-
Root lodging is a physical stress that changes gene expression from sucrose accumulation to degradation in sorghum
BMC Plant Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.