Abstract
Mustard aphid, also known as turnip aphid (Lipaphis erysimi) is a major insect pest of rapeseed-mustard group of crops. Tremendous economic significance has led to substantial basic research involving gene-expression studies in this insect species. In qRT-PCR analysis of gene-expression, normalization of data against RNA variation by using appropriate reference gene is fundamental. However, appropriate reference genes are not known in case of L. erysimi. We evaluated 11 candidate reference genes for their expression stability in 21 samples of L. erysimi subjected to various regimes of experimental treatments. Unlike other studies, we validated true effects of the treatments on the samples either by gene-expression study of an associated marker gene or by biochemical tests. In the validated samples, expression stability of the reference genes was analysed by employing four different statistical softwares geNorm, NormFinder, BestKeeper and deltaCt. Drawing consensus on the results from different softwares, we recommend three best reference genes 16S, RPS18 and RPL13 for normalization of qRT-PCR data in L. erysimi. This study provides for the first time a comprehensive list of suitable reference genes for mustard aphid and demonstrates the advantage of using more than one reference gene in combination for certain experimental conditions.
Similar content being viewed by others
Introduction
Mustard aphid, also known as turnip aphid (Lipaphis erysimi Kaltenbach), inflicts heavy damage in several oilseed and vegetable Brassicas1. Particularly in rapeseed-mustard it has evolved as the most dreaded insect-pest in tropical and subtropical regions including India1,2. Genetic resistance against aphids is either unavailable or limited to a few wild accessions of Brassica coenospecies. Therefore, extensive basic research on physiological aspects of aphid biology with plausible implication in its integrated management strategy is being perused worldwide3,4. In this context, gene-expression studies of important aphid genes, involved in parasitic lifecycle, have assumed significance in entomological research. An immediate application of such studies, for example, has led to several attempts for developing RNAi mediated host-resistance by targeted silencing of aphid genes5,6,7,8,9.
qRT-PCR has rapidly gained importance as a robust method for studying gene-expression because of its high sensitivity and convenience in large throughput. However, certain limitations such as, batch to batch variation in output, variable efficiency of reverse transcription and PCR reaction which influences the threshold (Ct) values have rendered this technique skill-intensive10. Consequently, the technical challenges of attaining precision in qRT-PCR based analysis depend on appropriate selection of the reference gene(s) in the experimental set up. Commonly, housekeeping genes are used as the reference genes with the fundamental assumption that their expressions remain constitutive and unaffected irrespective of the treatments or changes in physiological condition of the samples during the course of the experiments11,12,13. Accordingly, any sample to sample variation indicated by change in expression data of the reference gene is considered as art-effect and the same is nullified in the output data through the process of ‘normalization’. Therefore, much credibility of the qRT-PCR data relies on true constitutive expression of the reference gene across the samples under study.
For qRT-PCR based gene-expression studies in insects including a few aphid species, housekeeping genes such as 18S rRNA, ACT1, EF-1a, GAPDH, UBI, RPSs, RPLs and TUB have been commonly used as reference genes6,12,14,15,16,17,18. However, several recent reports on expression inconsistency of these genes question their suitability as reference genes in qRT-PCR studies (reviewed by Gutierrez et al.19; Kozera and Rapacz20). In fact, it has now been shown that most of the house keeping genes demonstrate variable expressions depending on the organism, its developmental stage and also the treatment conditions21,22,23,24. Therefore, none of these genes can be universally used as reference gene; rather a validation of their suitability as reference genes in the target species is a prerequisite.
Owing to the importance of aphids as the most significant insect-pest of rapeseed-mustard there have been an increasing number of scientific reports on functional genomics of its biology as well as annotated gene sequences25,26. In most of the aphid species including L. erysimi, qRT-PCR studies are carried out with non-validated reference genes commonly based on their use in other organisms. Report on validation of references genes for qRT-PCR, though rapidly being available in other organisms, is lacking for this aphid species. As a result, the impulse for appropriate reference gene for L. erysimi system is felt in many of the research studies that involve qRT-PCR. In this context, the current study was undertaken for validating 11 housekeeping genes, such as, 16S ribosomal RNA (16S), succinate dehydrogenase B (SDHB)27, actin (ACT)28, elongation factor 1a (EF1A)12,16, ribosomal protein L13a (RPL13)29, ribosomal protein S18 (RPS18)15, ribosomal protein L27 (RPL27)28, ribosomal protein L29 (RPL29)12, β-tubulin (TUB)30, glyceraldehydes 3-phosphate dehydrogenase (GAPDH)18, arginine kinase (AK)30, as reference gene for qRT-PCR in L. erysimi. The expression stability of these genes across the regime of different experimental conditions was evaluated by using four statistical algorithms, geNorm11, NormFinder31, BestKeeper32 and deltaCt method33. To the best of our knowledge it is the first report on comprehensive evaluation of the reference genes in L. erysimi. The outcome of the study will not only benefit those experiments which involve gene-expression study in this aphid species but will immediately find translational application in other closely related aphids.
Results
PCR amplification of candidate reference genes
The sequence information of the reference genes in L. erysimi was not available in the genbank database. Therefore, the primers were initially designed based on the A. pisum sequences (Table S1). The specificity of PCR amplification on L. erysimi cDNA was validated by single amplification of expected size in each case. The amplicons were sequenced and the sequence data were validated by homology to their heterologous counterparts (Table S2). Based on L. erysimi specific sequences of the reference genes, the qRT-PCR primers were designed. The specificity of primer binding in qRT-PCR was further confirmed by the desired amplicon, visualized on agarose gel and melt curve analysis (Fig. 1). Linear regression coefficient R2 of all the primer-pairs were ranged between 0.987–1.000 and the PCR efficiency determined by the standard curve ranged from 91.2–102.4% (Table S3; Fig. S1).
Validation of treatment-effects by gene-expression and biochemical test
Treatment-effects on the L. erysimi samples were validated by assaying expression level of the marker genes in each case. In the samples of different developmental stages, a gradual increase in AP-1 transcript level confirmed the stage-difference among the samples (Fig. 2A). For imposing antibiosis, the aphids were fed on diet supplemented with variable levels of sinigrin and for variable duration. Ingestion of sinigirin led to enhanced expression of the myrosinase gene in the treated aphids (Fig. 2B,C). Similarly, the effect of temperature stress was verified by multifold induction of Hsp83 transcripts at 37 °C compared to lower (10 °C) and optimal (22 °C) temperatures (Fig. 2D). Feeding of the aphids on artificial diet was validated by honeydew deposition detected through ninhydrin staining. A gradual increase in staining intensity was observed on the whatman paper with increasing feeding duration (Fig. 2E). As expected, in case of starvation no stain was detected (Fig. 2F).
Validation of treatments by molecular markers and biochemical tests.
(A) AP-1 expression in four developmental stages; (B,C) Transcriptional activation of MYR in aphids due to sinigrin ingestion at different doses and for different duration; (D) Transcriptional activation of Hsp83 by temperature stress; (E) Ninhydrin assay of honeydew secreted by aphids on artificial diet; (F) Ninhydrin assay for honeydews in starved aphids.
Expression profile of the candidate reference genes in L. erysimi
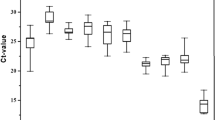
In qRT-PCR, a variable Ct value of all the reference genes across the 5 treatments indicated that their gene-expression levels are affected by the treatments albeit at differential extents (Fig. 3). The expression of SDHB, RPL29 and AK were maximally affected by the developmental stages and temperature stress. Similarly, the expression of EF1A, GAPDH, AK and SDHB varied significantly among the samples collected from aphids either fed on artificial diet for different durations or treated with sinigrin in the diet. In a comparative assessment, a narrow range of variable Ct values in case of 16S, RPS18 and RPL13 across the type and time course of the treatments empirically suggested their relatively more stability of expression. In contrary, RPL29, EF1A and GAPDH displayed highest variations in Ct values when all the experimental treatments were taken into account. Irrespective of the treatments, 16S rRNA was found to be most abundant, with the lowest mean Ct value 9.23, compared to the transcript levels of other protein coding genes (Fig. 3).
Expression stability and ranking of the candidate reference genes
The candidate reference genes were ranked based on their gene-expression stability over the treatments by employing four statistical algorithms. In analysis by different softwares, the rank order of the reference genes for individual treatment showed similar trends with subtle variation, which may be attributed to the differences in algorithms. For the purpose of more clarity in depiction the ranking of the reference genes generated by each statistical method has been dealt separately.
geNorm
In geNorm analysis, all the reference genes significantly varied in their expression stability across the treatment conditions. More importantly, geNorm ranking unambiguously showed 16S as the most stable reference gene across the treatments of developmental stages, starvation and antibiosis whereas RPL13 was the best for the treatments of temperature stress and artificial diet (Fig. 4). However, taking into account all the treatments together, expression stability of ACT and TUB also scored below the threshold value along with RPL13 and 16S. Further, the optimal number of reference genes required for normalization of samples involving a specific treatment was calculated. If the pairwise variation (Vn/Vn + 1) between sequential normalization factors, NFn and NFn + 1 is >0.15, it is not necessary to use ≥n + 1 as an internal control for particular experimental conditions11. The pairwise variation V2/3 value was less than 0.15 for most of the conditions except in the case of developmental stages and starvation (Fig. 5). This indicated that combined use of two stable reference genes would be mandatory for normalization purpose in studies involving variable regimes of temperature, glucosinolate and artificial diet conditions. In developmental stages, the V3/4 value was less than 0.15, suggesting that the optimal number of reference genes for normalization would be three, namely 16S, GAPDH and RPS18. However, in the case of starvation and all the treatments together, the pairwise variation values were above the cut-off value. The geNorm manual suggests use of minimum three most stable reference genes in such situations (Fig. 5).
Expression stability and relative ranking of the 11 reference genes predicted by the geNorm.
The mean expression stability (M) was calculated by stepwise exclusion of the least stable gene across all the samples within a particular group set. The mean stability of different genes is plotted; the least stable genes are represented on the left and the most stable on the right side of the plot. The dotted line indicates the threshold value of mean expression stability. The genes with M ≤ 1.5 are considered significant with stable expression.
Optimal number of reference genes for accurate normalization calculated by geNorm analysis.
Average pairwise variations (V) were calculated between the normalization factors NFn and NFn+1 by geNorm software to indicate the optimum number of reference genes required for qRT-PCR data normalization. A threshold value of V ≤ 0.15 was suggested for significant normalization, the value below 0.15 indicated that the additional reference gene has no significant improvement on normalization in qRT-PCR data. Arrow heads indicate the optimal number of genes for normalization for that particular condition.
BestKeeper
BestKeeper software tool calculates the standard deviation (SD) which is inversely proportional to the stability of expression. The rank order of the reference genes across the treatments suggested that no single reference gene can be used ideally for all the treatments (Table S4). The reference gene 16S was most stably expressed and ranked the best in both starvation and temperature-stress conditions followed by RPS18 and RPL13, respectively. Interestingly, gene-expression of RPL13 and RPS18 was least affected in antibiosis treatment compared to other treatments and ranked ahead of 16S. In developmental stages, GAPDH showed more stable expression compared to 16S and ranked as the best reference gene. Analysis of the aphid samples collected from the field validated similar trend in which SDBH and GAPDH were ranked as the best reference genes followed by 16S. When the aphids were fed on artificial diet, expression of RPL27 followed by RPL29 was least affected among all the reference genes. In contrary, RPL29 expression showed significant variation, in fact highest SD value, in samples of different developmental stages, starvation and temperature stress.
NormFinder
Based on the rank order assigned by NormFinder algorithm either EF1A or RPS18 were found to be most consistent in expression-level within and across the treatments, except starvation (Table S5). Under starvation 16S was more stable and ranked higher than RPS18.
DeltaCt method
Delta Ct method identified 16S as the most stable reference genes across the treatments of starvation, glucosinolate and temperature stress (Table S6). In developmental stage and artificial diet conditions EF1A and ACT were ranked as most stable, respectively.
Validation of the reference genes
It was apparent that the best ranked reference gene for one treatment was not applicable to the other treatments. Therefore, for experimental convenience it was imperative to identify the reference genes that show acceptable, if not the highest, stability in expression within and across the treatments. Analysis of consolidated data indicated 16S, RPS18 and RPL13 as the top ranked reference genes in terms of stability in gene-expression for most of the treatment conditions. In contrary, RPL29 was found to be the least stable. Applicability of these reference genes in normalization, either as single or in combination, was tested. This was carried out by assessing the transcript level of AP-1, MYR and Hsp83 in the treatments of developmental stage, sinigrin and heat stress, respectively (Fig. 6). In case of developmental stage, either of RPS18 and RPL13 or combination of them as normalizer showed higher consistency in qRT-PCR data compared to normalization by 16S and its combinations (Fig. 6A). However, for sinigrin treatment and heat stress normalization by a single recommended gene produced consistent and comparable result (Fig. 6B,C). In contrary, RPL29 being the least stable reference gene when used as normalizer showed significant variation in the results (Fig. 6).
Variation in AP-1, MYR and Hsp83 gene-expression data normalized by different reference genes and their combinations.
Fold change in AP-1, MYR and Hsp83 transcripts in aphids subjected to treatments of developmental stage (A), sinigrin (B) and temperature stress (C), respectively. Stable reference genes 16S, RPS18, RPL13, their combinations and least stable reference gene RPL29 were independently used as normalizer. Values represent mean ± SE (3n = 9) and comparison of means was carried out by student’s t test (P < 0.05). Different letters indicate significantly different values.
Discussion
Most of the qRT-PCR studies in L. erysimi, have used reference genes from heterologous systems without prior validation in this species5,6,7,8,9. This becomes a serious impediment in accurate data interpretation. Such concern necessitates a comprehensive validation of the reference genes in L. erysimi under varied experimental conditions. In this study, 11 housekeeping genes frequently used as reference genes in other sap-sucking insects were analysed for their expression-stability in this insect-pests across the various treatment conditions12,16,23,24,27,30. The treatments per se as well as different intensity of treatments in terms of either dose or duration were set as the variables for assessing expression-stability of the reference genes. Perception of the treatments in the aphid samples was validated by anticipated change in marker gene-expression. In pea aphid, expression of AP-1gene is regulated by metamorphotic transition and increases gradually as the individual nymphs mature to alate (winged) adults34. Therefore, differential expression of AP-1 validated true stage-difference of the aphids from different developmental stages (Fig. 2A). In specialist aphids like L. eysimi and Brevicoryne brassicae, myrosinase sequesters plant glucosinolates into crystalline microbodies and its expression is activated by glucosinolates in a dose dependent manner35,36,37. A commensurate increase in myrosinase expression in L. erysimi with increasing dose and duration of sinigrin treatment confirmed true treatment-effect on the test samples (Fig. 2B,C). High temperature rapidly induces the heat shock protein Hsp83 in the larvae and pupa of Aedes aegypti, Tribolium castaneum and cotton aphid Aphis gossypii12,38,39. Therefore, perception of high temperature stress at the gene-expression level was verified by transcriptional activation of Hsp83 in the treated samples (Fig. 2D). Aphid feeds in continuous fashion and secretes honeydews which are rich in free amino acids and can be quantitatively assayed by ninhydrin dyes. The volume of secreted honeydew depends on the feeding efficiency. In bioassay, deterrence in aphid-feeding by potential insecticidal compounds has been quantitatively measured by decrease in honeydew deposition40,41,42. In our experiments, prolonged aphid feeding on artificial diet was monitored by continuous increase in honeydew deposition in a time course manner (Fig. 2E). Similarly, significant decline in honeydew secretion was an appropriate starvation response of the aphids (Fig. 2F). Validation of treatment-effect on the gene-expression of the samples was imperative because escapes of the treatment, if any, were likely to erroneously demonstrate null-effect of the treatment on the reference gene in the statistical analysis. Surprisingly, such authentication of treatment-effects on the test-samples has not been considered in any of the earlier studies on expression of the reference genes in multiple treatments12,16,23,24,27,30. Our study for the first time explicitly highlights the importance of validating the treatment-effects in the test-samples.
Interestingly, the raw gene-expression data clearly demonstrated profound effect of all the treatments on the reference genes; though, all of them were not affected by each treatment. Collectively, the results re-ascertained the earlier reports that no single reference gene is consistent across all the treatments22,23,43. The most suitable reference gene(s) for each treatment was indicated by rank order of the reference genes within the treatment. Though, rank order of the reference genes was not identical in the output of the four independent statistical analysis, an empirical consensus clearly indicated the top order reference genes for each treatment conditions. For instance, GAPDH was ranked most stable reference gene by geNorm as well as BestKeeper for studying gene expression regulated by developmental stages; where as NormFinder and deltaCt method ranked EF1A as the best. The subtle differences in ranking among the top order reference genes could be attributed to difference in algorithms of the employed softwares and sensitivities towards the co-regulated reference genes. Similar variability in stability-ranking was also evident among the probable reference genes identified in other insect species13,30,44,45. Analysis of gene-expression data across the treatments and using all the four statistical methods identified 16S, RPL13 and RPS18 as the most stable reference genes based on their consistent top order ranking (Tables S4–S6).
There has been a long debate about the optimal number of reference genes required for qRT-PCR analysis11,24. For avoiding errors in normalization, researchers have demonstrated use of multiple reference genes instead of one28,46,47. In our analysis geNorm pair-wise values advocate use of two reference genes for attaining better accuracy in normalization for most of the experimental conditions such as, high temperature, antibiosis and artificial diet. However, conditions like change in developmental stage and starvation would require normalization by three or more reference genes, respectively16,24. Thus, we suggest the use of more than one reference gene in combinations for studying gene-expression by qRT-PCR in L. erysimi, under different experimental conditions.
In conclusion, keeping in view the economic importance of L. erysimi as agricultural pest, functional genomics and gene-expression study would continue to constitute an important part of basic research in this insect-pest. Hence, establishing a standardized qRT-PCR in L. erysimi will benefit the peers in this research area. Our study empirically demonstrates that the commonly known reference genes may not be suitable for normalization in qRT-PCR without prior validation across the regime of the experimental treatments. Nevertheless, we recommend the use of 16S, RPL13 and RPS18, preferably in combination for better normalization in qRT-PCR for most of the treatment conditions. This study would be a way forward to the qRT-PCR study in L. erysimi that are undertaken in diverse research projects worldwide owing to the growing economic importance of this insect-pest.
Materials and Methods
Insect rearing
Apterae adults of mustard aphids (L. erysimi) were collected from mustard (B. juncea) plants grown in the field at Indian Agricultural Research Institute, New Delhi, India during the mustard growing season, January 2015. The adults were reared on one month old mustard plants and allowed to breed repeatedly producing large number of nymphal colonies, in a growth chamber under 16 h light (140 μmol m−2 s−1), 8 h dark cycles at 22 ± 2 °C and 62–72% relative humidity.
Experimental treatments
Aphid developmental stages
Aphids of four different developmental stages, first instar nymphs, third instar nymphs, wingless and winged adults were collected from the plants grown either in the growth chamber or in the field. Twenty female aphids were released on B. juncea plants and allowed to lay offspring. After 24 h the first instar nymphs and after five days the third instar nymphs were collected at least from three different clonal progeny. The wingless and winged adults were collected in between 7–12 days of aphid colonies. At least 50 aphids constituting one biological replicate for each sample were flash frozen in liquid nitrogen and stored at −80 °C until used. The samples were collected for at least three biological replicates.
Temperature
The wingless adults were subjected to three different temperature regime, 10 °C, 22 °C and 37 °C (BOD incubator, Kuhner shaker LT-X, Switzerland) for 1 hour in Petri-dishes and harvested.
Starvation
For creating starvation stress, a group of 20 adult aphids were starved for 12 h and 24 h in Petri-dishes at 22 ± 2 °C in a BOD incubator. Ninhydrin test of the aphid honeydews was performed as described by Kanrar et al..
Feeding on artificial diet
A group of 20 wingless adults were maintained on artificial diet at 22 ± 2 °C in BOD incubator and harvested at different time intervals ranging from 24, 48 and 72 h.
Glucosinolate treatment
For the treatment of glucosinolates sinigrin at 0, 0.5, 1, 2 mg/L was supplemented in the artificial diet and a group of wingless adults were released. The aphids were collected in a time course manner at 24, 48, 72 h.
Marker genes and biochemical test for validating treatment effects
For assessing treatment-effects on the gene expression of the treated samples either expression of a marker gene explicitly responsive to the treatment was monitored or biochemical test was performed on the samples. For aphid developmental stages, temperature and sinigrin treatment the expression of Apterous-1 (AP-1), heat shock protein (Hsp83) and myrosinase (MYR), respectively was evaluated12,34,35,36,37. For treatment of feeding on artificial diet and starvation, ninhydrin assay of the honeydews was performed48.
Candidate reference genes and primer design
The candidate reference genes, chosen for the present study is shown in Table S1. Primers were designed (http://www.idtdna.com/scitools/applications/primerquest/) based on their sequence in Acyrthosiphon pisum. Homologous sequences from L. erysimi were PCR amplified from cDNA and sequenced. Low quality sequence ends were trimmed by using NCBI Vecscreen (http://www.ncbi.nlm.nih.gov/tools/vecscreen/). The sequences were validated for homology using BlastN programme (Table S2). For all the qRT-PCR studies in L. erysimi specific primers were designed based on the obtained sequences (Table S3).
RNA isolation and cDNA synthesis
Total RNA was isolated by using RNAiso Plus reagent (Takara Bio Inc. Tokyo, Japan) according to manufacturer’s protocol. The residual DNA was removed by using TURBO DNA free kit (Ambion Inc., UK). Yield and purity of RNA was determined by NanoDrop ND-1000 spectrophotometer (Nanodrop technologies, USA). RNA samples with an absorbance ratio OD 260/280 between 1.9–2.2 and OD 260/230 greater than 2.0, were used for further analysis. RNA-integrity was verified by resolving the samples on 1.8% agarose gel in 1X TAE. First stand cDNA was synthesized from 2 μg of total RNA in a 20-μl reaction volume using Superscript III First-Strand cDNA Synthesis Kit (Invitrogen, USA) as per manufacturer’s instruction. cDNA was stored at –20 °C for further use. For qRT-PCR analysis, each cDNA sample was diluted 20 times with nuclease free water.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
All the qRT-PCR reactions were performed using SYBR green detection chemistry, in a StepOne plus real time PCR machine (Applied Biosystems, USA). A reaction cocktail of 20 μl was constituted of 2 μl diluted cDNA, 10 μl 2X SYBR Premix Ex Taq II (Takara Bio Inc, Japan), 0.4 μl of ROX reference dye and 0.4 μl each of the forward and reverse primers. PCR cycling was carried out at an initial denaturation for 30 sec at 95 °C, followed by 40 repeated cycles, each consisting of 95 °C for 10 s, 60 °C for 30 s and 72 °C for 30 s. A serial 10- fold dilution (1–1000) of cDNA was used to generate standard curve for determining the gene specific PCR efficiency (E) of the primer pairs in qRT-PCR by using linear regression model. The amplification efficiency (E) for each gene specific primers was calculated according to the equation: E (%) = (10−1/slope−1) × 100%.
Data mining and statistical analysis
The stability of the candidate reference genes was ranked by using Microsoft Excel based software tools, geNorm11, NormFinder31, BestKeeper32 and deltaCt method33. BestKeeper uses raw data (Ct values) and PCR efficiency (E) to compute best-suited standards and combines them into an index. The geNorm program calculates an expression stability value (M) and ranks the genes in an order for given set of samples. The lower the M value, the higher is the expression stability and the M value less than 1.5 is recommended to identify stably expressed gene. It also compares the pair wise variation (V) of these genes with others. The pair wise variation value of Vn/Vn+1 between two sequential normalization factors was used to determine the optimal number of reference genes required for better normalization. A threshold value below 0.15 suggests the requirement of no additional reference gene for normalization11. NormFinder calculates gene expression stability for all the samples in any number of groups based on intra- and inter-group variations and combines these values to provide gene rank order depending on the variation in gene expression. Genes with the lowest rank are considered to be most stably expressed and are ideal to select as reference gene(s) for that particular experimental conditions. In Delta Ct method, rank order is determined based on pair-wise comparisons of gene-sets using mean delta Ct values within a particular treatment. Therefore, the average standard deviation of each gene-set is inversely proportional to the gene-expression stability.
Additional Information
How to cite this article: Koramutla, M. K. et al. Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt.). Sci. Rep. 6, 25883; doi: 10.1038/srep25883 (2016).
References
Blackman, R. L. & Eastop, V. F. Aphids on the World’s Crops: An Identification and Information Guide 2nd edn (John Wiley & Sons, 2000).
Bakhetia, D. R. C. & Ghorbandi, A. W. Relationship between the parameters of aphid population per plant and percentage of plants infested by Lipaphis erysimi (Kaltenbach) in Indian mustard crop. J. Aphidology. 3, 119–124 (1989).
Jaouannet, M. et al. Plant immunity in plant-aphid interactions. Front. Plant Sci. 5, 663 (2014).
Furch, A. C., van Bel, A. J. & Will, T. Aphid salivary proteases are capable of degrading sieve-tube proteins. J. Exp. Bot. 66, 533–539 (2015).
Pitino, M., Coleman, A. D., Maffei, M. E., Ridout, C. J. & Hogenhout, S. A. Silencing of aphid genes by dsRNA feeding from plants. PLoS One. 6, e25709 (2011).
Bhatia, V., Bhattacharya, R. C., Uniyal, P. L., Singh, R. & Niranjan, R. S. Host Generated siRNAs attenuate expression of serine protease gene in Myzus persicae. PLoS One. 7, e46343 (2012).
Bonning, B. C. et al. Toxin delivery by the coat protein of an aphid-vectored plant virus provides plant resistance to aphids. Nature Biotechnol. 32, 102–105 (2014).
Mao, J. & Zeng, F. Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae. Transgenic Res. 23, 145–152 (2014).
Zhang, J. et al. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science. 347, 991–994 (2015).
Bustin, S. A., Benes, V., Nolan, T. & Pfaffl, M. W. Quantitative real-time RT-PCR-a perspective. J. Mol. Endocrinol. 34, 597–601 (2005).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 0034 (2002).
Li, R. M. et al. Reference gene selection for qRT-PCR analysis in the sweet potato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS One 8, e53006 (2013).
Zhu, X. et al. Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hu¨bner) (Lepidoptera: Noctuidae). PLoS One. 9, e84730 (2014).
Puinean, A. M. et al. Amplification of a cytochrome P450 gene is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. Plos Genetics 6, e1000999 (2010).
Maroniche, G. A., Sagadin, M., Mongelli, V. C., Truol, G. A. & del Vas, M. Reference gene selection for gene expression studies using RT-qPCR in virus-infected planthoppers. Virol. J. 8, 308 (2011).
Bansal, R., Mian M. A. R., Mittapalli, O. & Michel, A. P. Characterization of a chitin synthase encoding gene and effect of diflubenzuron in soybean aphid. Aphis Glycines. Int. J. boil. Sci. 8, 1323–1334 (2012).
Zhang, M. et al. Identifying potential RNAi targets in grain aphid (Sitobion avenae F.) based on transcriptome profiling of its alimentary canal after feeding on wheat plants. BMC Genomics. 14, 560 (2013).
Lu, Y. et al. Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS One 8, e68059 (2013).
Gutierrez, L., Mauriat, M., Pelloux, J., Bellini, C. & Wuytswinkel, V. O. Towards a systematic validation of references in real-time RT-PCR. Plant Cell. 20, 1734–1735 (2008).
Kozera, B. & Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 54, 391–406 (2013).
Shi, X. Q. et al. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Res Notes. 6, 93 (2013).
Fu, W. et al. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 9, 792 (2013).
Yuan, M. et al. Selection and evaluation of potential reference genes for gene expression analysis in the Brown Planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS One. 9, e86503 (2014).
Yang, C. et al. Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 5, 18201 (2015).
Chilana, P., Sharma, A. & Rai, A. Insect genomic resources: status, availability and future. Curr. Sci. 102, 571–580 (2012).
Yin, M., Ma, Z., Cai, Z., Lin, G. & Zhou, J. Genome sequence analysis reveals evidence of quorum-sensing genes present in Aeromonas hydrophila strain KOR1, isolated from a mangrove plant (Kandelia obovata). Genome Announc. 3, genomeA. e1461–15 (2015).
Yang, C., Pan, H., Liu, Y. & Zhou, X. Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLoS One. 9, e110454 (2014).
Sinha, D. K. & Smith, M. Selection of reference genes for expression analysis in Diuraphis noxia (Hemiptera: Aphididae) fed on resistant and susceptible wheat plants. Sci. Rep. 4, 5059 (2014).
Scharlaken, B. et al. Reference gene selection for insect expression studies using quantitative real-time PCR: The head of the honeybee, Apis mellifera, after a bacterial challenge. J. Insect Sci. 8, 33 (2008).
Zhai, Y. et al. Identification and validation of reference genes for quantitative real-time PCR in Drosophila suzukii (Diptera: Drosophilidae). PLoS One 9, e106800 (2014).
Andersen, C. L., Jensen, J. L. & Orntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 (2004).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotech. Lett. 26, 509–515 (2004).
Silver, N., Best, S., Jiang, J. & Thein, S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7, 33 (2006).
Brisson, N. et al. Why are wheat yields stagnating in Europe? A comprehensive data analysis for France. Field Crops Res. 119, 201–212 (2010).
Mainguet, A. M., Louveaux, A., Sayed G. E. & Rollin, P. Ability of a generalist insect, Schistocerca gregaria, to overcome thioglucoside defense in desert plants: tolerance or adaptation? Entomol. Exp. Appl. 94, 309–317 (2000).
Bridges, M. et al. Spatial organisation of the glucosinolate-myrosinase system in Brassica specialist aphids is similar to that of the host plant. Proc. Royal Soc. Biol. Sci. 269, 187–191 (2002).
Kazana, E. et al. The cabbage aphid: a walking mustard oil bomb. Proc. R. Soc. Lond. 274, 2271–2277 (2007).
Xu, J., Shu, J. & Zhang, Q. Expression of the Tribolium castaneum (Coleoptera: Tenebrionidae) hsp83 gene and its relation to oogenesis during ovarian maturation. J. Genet. Genomics. 37, 513–522, (2010).
Zhao, L., Becnel, J. J., Clark, G. G. & Linthicum, K. Expression of AeaHsp26 and AeaHsp83 in Aedes aegypti (Diptera: Culicidae) larvae and pupae in response to heat shock stress. J. Med. Entomol. 47, 367–375 (2010).
Sadeghi, A., Van Damme, E. J. M. & Smagghe, G. Evaluation of the susceptibility of the pea aphid, Acyrthosiphon pisum, to a selection of novel biorational insecticides using an artificial diet. J. Insect Sci. 9, 65 (2009).
Smagghe, G., Mahdian, K., Zubrzak, P. & Nachman, R. J. Antifeedant activity and high mortality in the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidae) induced by biostable insect kinin analogs. Peptides. 31, 498–505 (2010).
Li, J. et al. A simplified procedure for the determination of organochlorine pesticides and polychlorobiphenyls in edible vegetable oils. Food Chem. 151, 47–52 (2014).
Ponton, F., Chapuis, M. P., Pernice, M., Sword, G. A. & Simpson, S. J. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57, 840–850 (2011).
Velada, I., Ragonezi, C., Arnholdt-Schmitt, B. & Cardoso, H. Reference genes selection and normalization of oxidative stress responsive genes upon different temperature stress conditions in Hypericum perforatum L. PLoS One 9, e115206 (2014).
Petriccione, M., Mastrobuoni, F., Zampella, L. & Scortichini, M. Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 5, 16961 (2015).
Chandna, R., Augustine, R. & Bisht, N. C. Evaluation of candidate reference genes for gene expression normalization in brassica juncea using real time quantitative RT-PCR. PLoS One. 7, e36918 (2012).
Xiao, X. et al. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 5, 788 (2015).
Kanrar, S., Venkateswari, J., Kirti, P. B. & Chopra, V. L. Transgenic Indian mustard (Brassica juncea) with resistance to the mustard aphid (Lipaphis erysimi Kalt). Plant Cell Rep. 20, 976–981 (2002).
Acknowledgements
This work was supported by National Agricultural Science Fund, Indian Council of Agricultural Research and Department of Science and Technology, Govt. of India.
Author information
Authors and Affiliations
Contributions
R.B. and M.K.K. conceived and designed research. M.K.K. and R.A. conducted experiments, M.K.K. analysed data. R.B., M.K.K. and R.A. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Koramutla, M., Aminedi, R. & Bhattacharya, R. Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt). Sci Rep 6, 25883 (2016). https://doi.org/10.1038/srep25883
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep25883
This article is cited by
-
Reference genes selection for expression studies in Maconellicoccus hirsutus (Green) (Pseudococcidae: Hemiptera) under specific experimental conditions
Molecular Biology Reports (2023)
-
Double-stranded RNA mediated knockdown of sucrase gene induced mortality and reduced offspring production in Aphis gossypii
Functional & Integrative Genomics (2023)
-
Identifying optimal reference genes for gene expression studies in Eurasian spruce bark beetle, Ips typographus (Coleoptera: Curculionidae: Scolytinae)
Scientific Reports (2022)
-
Identification of suitable reference genes for expression profiling studies using qRT-PCR in an important insect pest, Maruca vitrata
Molecular Biology Reports (2021)
-
Selection and validation of experimental condition-specific reference genes for qRT-PCR in Metopolophium dirhodum (Walker) (Hemiptera: Aphididae)
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.