Abstract
The diploid woodland strawberry (Fragaria vesca) is an important model for fruit crops because of several unique characteristics including the small genome size, an ethylene-independent fruit ripening process, and fruit flesh derived from receptacle tissues rather than the ovary wall which is more typical of fruiting plants. Histone methylation is an important factor in gene regulation in higher plants but little is known about its roles in fruit development. We have identified 45 SET methyltransferase, 22 JmjC demethylase and 4 LSD demethylase genes in F. vesca. The analysis of these histone modifiers in eight plant species supports the clustering of those genes into major classes consistent with their functions. We also provide evidence that whole genome duplication and dispersed duplications via retrotransposons may have played pivotal roles in the expansion of histone modifier genes in F. vesca. Furthermore, transcriptome data demonstrated that expression of some SET genes increase as the fruit develops and peaks at the turning stage. Meanwhile, we have observed that expression of those SET genes responds to cold and heat stresses. Our results indicate that regulation of histone methylation may play a critical role in fruit development as well as responses to abiotic stresses in strawberry.
Similar content being viewed by others
Introduction
Chromatin is a complex composed of histones, chromosomal proteins, DNAs and small RNAs. The nucleosome is the basic unit of chromatin, consisting of a histone octamer (two of each histone 2A, histone 2B, histone 3 and histone 4), surrounded by a 146–148 bp DNA wrapping. The post-transcriptional modifications of the N-terminal tails of core histones could affect the nucleosome spacing, higher-order nucleosome interaction, and greatly affect the accessibility of the transcriptional regulatory proteins. Thus, in eukaryotes, post-transcriptional histone modifications are a determinant of the active/silent state of the associated genes, and are of great importance to a variety of important biological processes1.
The covalent modifications of histones include methylation, acetylation, phosphorylation, ubiquitation and SUMOylation2. Methylation on the lysine residues is among the most variant and important histone modifications. The majority of methyl marks are located on lysine 4, 9, 27, 36, 79 on histone H3, and lysine 20 on histone H4. The different active/silent chromatin states are characterized by different combinatorial patterns of histone modifications. Generally speaking, methylations on H3K9, H3K27, H3K79 and H4K20 are silent marks, while methylations on H3K4 and H3K36 are active marks3,4. Although having some variations, the mechanisms determining the chromatin states are quite conserved in plants and animals.
Histone lysine methylation is dynamic during organ development, and determined by histone modifying proteins including histone lysine methyltransferases (HKMTases), histone demethylases (HDMases) and histone turnovers. The majority of HKMTases have a SET (Suppressor of variegation, Enhancer of zeste and Trithorax in Drosophila) domain mediating the methyltransferase catalytic activity5. The only known HKMTase that does not have a SET domain is Dot1/Dot1L, which is responsible for H3K79 methylation6,7.
Based on the amino acid sequence conservation of SET domains, there are seven classes of SET protein encoding genes in Arabidopsis, each of which have preferred targets5,8. Class I consists of three polycomb group genes homologous to the Drosophila ortholog E(Z) (Enhancer of Zeste), capable of transferring methyl groups to H3K27. Class II consists of five SET genes homologous to the Drosophila ortholog ASH1, responsible for methylations on H3K4 and/or H3K36. Class III is another group of SET genes responsible for the active mark H3K4me1/2/3, similar to the Drosophila ortholog TRX. Class IV is plant-specific, responsible for monomethylation on H3K27, a silent mark essential for transposon silencing9. Class V is the largest SET group consisting of 15 SET genes homologous to Drosophila SU(VAR)3–9. Similar to their orthologs in animals, class V SET genes in Arabidopsis play an essential role in the establishment of heterochromatic mark H3K9me1/2/3, as well as H4K20me and H3K27me2. Class VI and VII consist of genes having a SET domain with functions to be determined.
Histone HDMases consist of two major types of enzymes, the LSD (Lysine Specific demethylase) type and the JmjC domain-containing HDMases10. LSD HDMases can only demethylate mono- and di-methylated lysine residues7. LSD family in Arabidopsis has four members, being able to demethylate H3K4me1/2 and H3K9me1/2. While JmjC is a much larger gene family consisting of 21 genes in Arabidopsis, and is compatible with trimethylated residues11. Within the JmjC family, members with the activity to demethylate methyl groups on H3K4, H3K9, H3K27, H3K36 and H4K20 have been identified7. H3K79 methylation is mediated by the only non-SET domain HKMTase Dot1/Dot1L, thus it is tempting to speculate that another class of histone HDMase might be responsible for H3K79 demethylation.
The first plant SET genes identified are CURLY LEAF (CLF) and MEDEA (MEA) in Arabidopsis thaliana12,13. Since then, the identification and functional investigation of histone HKMTases and HDMases in plants have been the subject of numerous studies. These studies suggest that HKMTases and HDMases are pivotal in phase transitions between sporophyte and gametophyte, gametophyte and seed development, embryo-seedling transition, induction of flowering, and vernalization1. In addition, histone methylations determined by both HKMTases and HDMases play an important role in the memory mechanism in response to recurring stresses1,14. For example, the “memory genes” responded to recurrent dehydrations maintain the active mark H3K4me3 during the recovery phase when transcription is low, which serves as a mark of “transcriptional stress memory”14. The list of cellular processes known to involve HKMTases/HDMases is still growing, and these histone modifiers are believed to play essential roles in all aspects of regulations of plant development.
In spite of the essential roles of histone modifications in cellular processes, little is known about histone modifiers in strawberry. The cultivated strawberry (F. x ananassa) is a young crop species as a model plant considered to be non-climacteric. F. x ananassa has an extremely complex octaploid genome harboring 56 chromosomes (2n = 8x = 56) derived from 4 diploid ancestors. Thus, the sequenced diploid woodland strawberry Fragaria vesca with a small genome (240 Mb, 2n = 2x = 14) offers substantial advantages for genomic research15. In this study, we identified genes encoding the histone lysine methylation modifiers, both HKMTases and HDMases in F. vesca. Comprehensive studies about the phylogeny, evolutionary history, structure, expression patterns in different stages/organs and in response to abiotic stresses were performed to give an overview of this important group of genes in F. vesca. This study provides the first characterization of the full set of histone lysine methylation modifiers in strawberry, and should greatly facilitate the functional characterization of those epigenetic regulators in this economically important crop species.
Results
Identification of genes encoding putative histone HKMTases containing SET domains in F. vesca
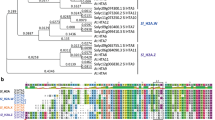
To identify histone HKMTases, the full alignment of SET domains downloaded from Pfam was searched against the F. vesca proteome by HMMER toolset (Methods for details). This sequence-based search identified 45 SET domain-containing genes in F. vesca (Fig. 1, Table 1 and Supplementary Table S1). To better understand the expansion and evolutionary history of SET genes in F. vesca, genes encoding SET-domain containing proteins were also identified in seven other species representing the major clades of terrestrial plants (Fig. 1). The basal angiosperm Amborella trichopoda is suggested to be the single living representative of the sister lineage to all other extant flowering plants16. A. trichopoda originated prior to the split of eudicots and monocots, and has not experienced any whole genome duplication (WGD) since then16; while the other seven angiosperms had several rounds of whole genome duplication/triplications after their split from A.trichopoda (Fig. 1, data from PGDD website, http://chibba.agtec.uga.edu/duplication/)17, which should have contributed to the evolution of SET genes. To standardize gene names, the Arabidopsis genes with known functions were named as published, following the standard gene symbol conventions with all capital letters; while genes in other species were named as SET1-62 following the species abbreviation.
The Taxonomy Common Tree was constructed online by Taxonomy Browser in NCBI (http://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/www.cmt.cgi).
Expansion and evolution of SET genes in F. vesca
In order to investigate the classification of F. vesca SET genes, phylogenetic trees were constructed using the Maximum Likelihood method (see Methods for details) based on amino acid sequences in the conserved SET domains on the SET genes identified in the agiosperms F. vesca, A. thaliana, Z. mays, O. sativa and A. trichopoda (Fig. 2). It is noted that the topologies of phylogenetic trees constructed by different methods are slightly different, and the results of the bootstrapping analysis for some nodes are lower than 60 (Fig. 2, Supplementary Fig. S1, S2–S6). Thus, domain composition for the whole proteins (Supplementary Fig. S1) and motif construct for the SET domains (Supplementary Fig. S2) were taken into consideration for the classification of SET genes as well. Accordingly, the SET genes could be grouped into seven classes (Fig. 2 and Table 1), which is consistent with previously reported in A. thaliana8. Class I–V consist of most of the canonical SET genes known to be involved in the catalysis of histone methylation (in Arabidopsis); while class VI and VII consist of relatively shorter genes with no known specificity (Table 1 and Fig. 2).
The phylogenetic tree was constructed based on the amino acids sequences of the SET domains with 1000 bootstrapping replicates. The F. vesca lineage-specific duplicated gene pairs (since its split form A. thaliana) are highlighted as yellow. The seven classes of SET genes are marked by different colors. Refer to Table 1 for more basic information of SET genes in F. vesca and A. thaliana.
The seven SET classes have different domain architectures and motif compositions (Supplementary Fig. S1–S6). In addition to the domain compositions identified by pfam, the 20 most common motifs embedded in the SET domains were identified by MEME for class I–V SET genes (Supplementary Fig. S2). Class I has few domains beside SET, but all the SET domains in this class consistently have the class I-specific motif 16 and 17. Although four SET genes in other classes have the motif 16 as well, the sequences are highly degenerate. Class II is characterized by AWS, SET and Post-SET domains, motif 1, 2, 3, 4, 9, and a class II-specific motif 20. Class III SET genes have PHD, zf-HC5HC2H_2, SET and Post-SET domains at high frequency, motif 1, 2, 3, 9 and class III-specific motif 7, 8, 12 and 15. Class IV is characterized by PHD and SET domains, and motif 1, 2 and class IV-specific motif 18. Class V SET genes have SAD_SRA, Pre-SET, SET and Post-SET domains, motif 1, 2, 3 and 4 at high frequency, and several class V-specific motif 5, 6, 11, 12, 14 and 15 (Supplementary Fig. S1,S2). While class VI and VII SET genes are relative short, and have few domains known to be essential for an HKMTase catalysis function.
To explore the detailed evolutionary history of the SET domain containing genes, the phylogenies of each class were investigated. In general, most of the SET genes reside in the sub-clades consisting of genes from all five species (Supplementary Fig. S3–S6), but there are some exceptions as well. For example, the AT-RE-PR sub-clade in class VI, and the AT-SETC3 sub-clade in class VII do not have an F. vesca member, indicating a lineage-specific loss in F. vesca (Supplementary Fig. S5,S6). On the other hand, in some sub-clades, two F. vesca genes clustered together with either a single Arabidopsis gene, or without any corresponding genes in Arabidopsis (Fig. 2, highlighted by yellow), indicating a lineage-specific duplication in F. vesca (duplications happened in F. vesca after its split from A. thaliana). In total, there are 7 pairs of such F. vesca SET genes, with 4 pairs in class V and the other 3 in other classes. To study the evolutionary constraints performed on the 7 recent duplicates in F. vesca, dn (nonsynonymous substitutions per site) and ds (synonymous substitutions per site) between each duplicate were calculated. dn of class V duplicated pairs are higher than other classes (Fig. 3B). Furthermore, dn/ds value of class V duplicates ranges from 0.62 to 0.93, significantly higher than the 3 duplicates from other classes (Fig. 3B), indicating a relaxation of negative selection on the class V duplicated pairs after duplication events happened18. In addition, some of those recent duplicates are coupled with domain gains and losses. For example, in duplicate pair FV-SET22 and FV-SET44 of class V, FV-SET22 obtained two domains at the N-terminal (Supplementary Fig. S3). Overall, recent duplicate pairs were more frequently originated or maintained in class V, and class V SET gene duplicate pairs evolved faster than other classes.
(A) Chromosomal locations of SET and JmjC genes on the seven chromosomes of F. vesca. The lineage-specific duplicated SET and JmjC pairs are connected by dashed lines. The scale on the left is in megabases. (B) The correlation between ds vs. dn, and ds vs. dn/ds for those duplicated gene pairs. Red circles denote duplicate genes pairs in class V, while blue diamonds denote those in other classes. For the FV-SET34/45/18/26 gene set, ds and dn were calculated for FV-SET34 vs.45, 34 vs.18 and 34 vs.26, according to the phylogenetic tree shown in Supplementary Fig. S5.
On the DNA level, WGD, large segmental duplication, or tandem duplication might lead to those duplicated pairs. To evaluate the relative contribution of those mechanisms in the expansion of the SET gene family in F. vesca, all SET genes were mapped to the seven chromosomes (Fig. 3A) and analyzed by MCscan19. The MCscan results suggest that in F. vesca, 6 out of the 45 SET genes were related to WGD, while the others resulted from dispersed duplications (Supplementary Table S2,S3). In class V, FV-SET34/45/18/26 cluster with a single Arabidopsis gene AT-SUVH4 (Fig. 2), indicating that more than one duplication event happened to the F. vesca orthologs of AT-SUVH4. The fact that both FV-SET18 and FV-SET 26 have a single exon suggests that retro-transposition may have contributed to the expansion of this gene set. In summary, our results suggest that most of the F. vesca SET genes originated before the split of eudicots and monocots, and that WGDs, dispersed duplications via retro-transpositions in some cases, have contributed to the evolution of SET genes in F. vesca as well.
Identification of genes encoding histone HDMases and investigation of their evolution in F. vesca
To investigate the histone HDMases in F. vesca, LSD HDMases and JmjC domain-containing HDMases were identified by sequence-based search using HMMER toolset. All the LSD HDMases characterized previously contain two domains, a SWIRM domain and an amino oxidase domain7. Thus, the proteins with both domains were identified as putative LSD HDMases.
In total, the sequence-based search identified 5, 4, 4, 4 and 4 genes encoding proteins with both the SWIRM and amino oxidase domains in A. trichopoda, O. sativa, Z. mays, A. thaliana and F. vesca, respectively (Fig. 4, Table 2 and Supplementary Table S4). The consistent number of LSD HDMases indicates that duplication events may not contribute much to the evolution of LSD HDMases in angiosperms.
Most likelihood phylogenetic trees and schematic diagrams for domain composition of JmjC (A) and LSD (B) genes in the species investigated. The phylogenetic tree was constructed based on the amino acids sequences of either the JmjC domain (A) or the whole LSD protein (B) with 1000 bootstrapping replicates, and the results of the bootstrapping analysis larger than 50% are shown.
In contrast to LSD HDMases, the number of JmjC HDMases varies in the five species. In total, A. trichopoda, O. sativa, Z. mays, A. thaliana and F. vesca have 17, 17, 26, 21 and 22 JmjC domain-containing genes respectively (Fig. 1 and Fig. 4). According to phylogenetic trees and domain constructs, JmjC HDMases are grouped into 9 classes (Fig. 4), which is consistent with previous reports20. Specifically, F. vesca lineage-specific duplications only happened in the KDM3 class, in which the sister group of AT-KDM3C (AT_JmjC10) in F. vesca has five members (FV_JmjC12, 13, 14, 20 and 21). Interestingly, among all the JmjC domain-containing genes, FV_jmjC12 and FV_jmjC21 are the only two genes having a single exon (Supplementary Fig. S7), suggesting that the ancestor of FV_JmjC12 and FV_JmjC21 resulted from a retrotransposition event where transcribed messenger RNA was inserted into the genome to form the ancestor of FV_JmjC12 and FV_JmjC21. In most classes, the F. vesca JmjC HDMases have not expanded, but in the KDM3 class, a series of duplication events occurred leading to the F. vesca lineage-specific expansion in this particular class.
In order to investigate which mechanisms might have contributed to those duplication events, JmjC genes were mapped to F. vesca chromosomes (Fig. 2) and analyzed by MCscan. MCScan results suggest that out of the 26 HDMase genes, 2 were WGD-related, and 24 resulted from dispersed duplications (Supplementary Table S2,S3). Therefore, similar to SET HKMTases, most of the F. vesca LSD and JmjC HDMases originated before the split of eudicots and monocots; and recent dispersed duplications and retro-transpositions might have played a pivotal role in the evolution of histone HDMases in F. vesca.
Expression profiles of histone HKMTases and HDMases in flower and fruit development in F. vesca
To investigate the expression profiles of individual histone HKMTase and HDMase genes in different organs and developmental stages, transcriptome data were investigated in flower development and early-stage fruit development21,22. One out of 45 F. vesca SET genes have no expression data available from the database, and were omitted from the following analysis.
The genes encoding HKMTases and HDMases have quite diversified expression patterns (Fig. 5A). Firstly, for the seven classes of SET genes, the members within a particular class show different tissue/stage-specific expressions. For example, in class I, CLF is moderately expressed in each tissue/developmental stage; while the mRNA of EZA1 is depleted in pollen and the early-stage embryo, and is more enriched in the developing pith and cortical tissues of strawberry flesh. Secondly, the respective members of the recently duplicated SET pairs have different tissue-specificity. Based on available transcription data, five out of the six duplicated F. vesca SET pairs with transcription data available have a similar expression pattern: one gene of the duplicated pair is more evenly and ubiquitously expressed, while the other gene is silent in the flesh (pith and cortex), the anther, and in some tissues in the seed (embryo, ghost, wall). It suggests that although highly conserved on amino acids sequence, those duplicated genes are differently regulated. Thirdly, LSD and different classes of JmjC HDMases show different expression profiles in different organs/stages as well; and the recent JmjC HDMase duplicates also express differentially (Fig. 4). Overall, the expression of histone HKMTase and HDMase genes has different organ/stage specificity, indicating a functional diversification coupled with the expansion of those gene families in F. vesca.
(A) The mRNA levels of histone modifiers in different tissues in flower and early-stage fruit development. The expression levels (RPKM) for the genes of interest were directly retrieved from http://bioinformatics.towson.edu/strawberry/) and plotted in log2 scale. (B) Expression profiles of SET genes in flesh (including pith and cortex, without seeds) during fruit ripening. (C) Expression profiles of SET genes in response to heat and cold stresses. For (B,C), the expression levels relative to GAPDH were measured by quantitative RT-PCR, and displayed in log2 scale. Three biological replicates and three technical replicates were done for each data point.
On the other hand, the different organs/stages have very different combinatorial expression patterns of HKMTase and HDMase genes (Fig. 5A). Firstly, most of the SET genes are expressed at extremely low level in pollen, with the majority of the class V and class I SET genes silent there. Secondly, the mRNAs of all the LSD HDMases and most of the JmjC HDMases are depleted in pollen as well, but interestingly, FV-JmjC5 and FV-JmjC16 show highest expression, indicating that those two JmjC HDMases might play a dominant role in histone demethylation in pollen. Thirdly, in the developing strawberry flesh (pith and cortex), both active and silent mark-related SET genes, LSD and JmjC genes show decreasing expression levels during early-stage fruit development (pollination to big green). Fourthly, overall, genes encoding both HKMTases and HDMases express higher in tissues of developing flowers (carpels, perianth, flowers, receptacles and microspores) than in other tissues. Thus, the different tissues in different developmental stages have diversified expression patterns of histone lysine methylation related genes, indicating the specific regulatory roles of those genes in cellular processes.
The expression profiles shown above reveal that duplicated SET pairs are quite distinct, with one silent in early-stage fruit development, while the other relatively ubiquitously expressed in all tissues, (Fig. 5A). To investigate how those duplicated SET genes express during strawberry fruit ripening, the expression levels of the more ubiquitously expressed genes were investigated in strawberry fleshy fruits (stripped with seeds, including pith and cortex only) at big green stage, big white stage (with red seeds and white flesh), turning stage (with red seeds and light white flesh) and red stage (2–3 days after turning stage) by quantitative RT-PCR assays (Fig. 5B). In addition, a subset of SET genes representing different classes was investigated as well. In contrast to the overall decreasing expression during early-stage fruit development, the mRNA levels of a substantial number of SET genes showed increasing expression levels during fruit ripening, and peaked at the turning stage (9 out of 14 genes investigated, Fig. 5B). The expression patterns of those SET genes in fruit development revealed that histone lysine HKMTase genes are dynamically expressed, and that the genomic histone lysine methylation patterns might undergo a dramatic change at the onset of fruit ripening.
Expression profiles of SET genes during heat/cold shock in F. vesca
Histone modifications are suggested to play an important role in the regulation of gene expression in response to abiotic stresses1,14. Strawberry plants are quite sensitive to extreme temperatures, and cold and heat shock are two recurrent stresses strawberry encounters in the natural environment. To study how HKMTase genes are regulated during heat and cold stresses, the expression patterns of a subset of SET genes were investigated in seedlings. The qRT-PCR results demonstrate that those HKMTase genes respond differentially to a particular abiotic stress (Fig. 5C). Three out of 13 investigated SET genes show increased expression levels upon cold shock at 3 h, while other 10 genes display no significant changes. Two SET genes show increased expressions upon heat shock at 4 h (3 h heat shock + 1 h recovery) as well. Interestingly, ATX3b and SUVH4a response to both cold and heat shock. Furthermore, the recent duplicated gene pairs response differently. For SUVH4a/b/c/d, the expression level of SUVH4a increases after cold and heat shock, while the other three are not responsive at all. In summary, some SET genes show dynamic expression patterns upon cold and heat shock, which indicates that these genes may be involved in F. vesca’s responses to temperature stresses.
Discussion
Sequence-based searching and phylogenetic analysis proved to be an effective way to identify histone modifiers in a sequenced genome20,23,24,25,26,27. In this study, we identified genes encoding SET HKMTases, LSD HDMases and JmjC HDMases in F. vesca plus seven other plant species representing the major clades of terrestrial plants. The extensive phylogenetic analysis revealed the evolutionary history of those histone modifiers in F. vesca and also in other angiosperms.
In total, 45 SET HKMTase genes grouped in seven classes were identified in F. vesca. These phylogenetic studies suggest that those identified SET genes were highly conserved in each class across a wide spectrum of plants, indicating their essential regulatory roles in the plant kingdom. Of the SET genes studied, the most intriguing observation was the expansion of class V in both eudicots and monocots, especially in Z. mays. Class V SET HKMTases are specific for methylations on H3K9, which is involved in transposon silencing and heterochromatin formation2,5. There is a vast expansion of transposable elements in Z. mays28, which might explain the maintenance of a large number of class V SET genes in Z. mays to protect the genome integrity. In addition, class V genes diverged faster than other classes (Fig. 3 and Table 2).
For each species, the numbers of genes grouped into each class is summarized in Table 2. Overall, there is no simple linear correlation between SET gene numbers and genome size, or total gene numbers (Table 3). Furthermore, there is no significant difference among the five species in class I, II, III, VI or VII, in terms of gene numbers. It suggests that those five classes did not experience any extensive expansion in angiosperms, and that most of the duplicated genes from the multiple whole genome duplication/triplication events were lost during evolution.
In contrast to the conserved number of SET genes in the five classes mentioned above, class IV and V show distinct evolutionary characteristics. Firstly, class IV SET genes are absent in A. trichopoda. Phylogenetic trees suggest that class IV genes are present in S. moellendorffii (Supplementary Fig. S7). Thus it is likely that the H3K27me1-specific HKMTase was lost in A. trichopoda. Secondly, there is an expansion of class V SET genes in both eudicots and monocots, especially in Z. mays. Overall, for the five species investigated, the number of SET genes in class I, II, III, VI and VII remains quite constant, while the number of genes in class IV and V fluctuates in angiosperms, indicating different evolutionary histories accompanied by rounds of WGDs and subsequent gene losses/gains by natural selection constrains.
We identified 26 histone HDMase genes in F. vesca. The number of LSD HDMase genes remained nearly the same in eight angiosperm species and the domain construct was highly conserved (Fig. 4B). The JmjC HDMase genes fall into 11 classes indicating that the genes coding for JmjC HDMases underwent rapid expansion and probably functional specification in plant genomes. This expansion and specification process was likely involved in the evolution of epigenetic regulatory mechanisms in plants with distinct biological features. Furthermore, JmjC genes in the KDM3 group were preferentially expanded in the strawberry genome compared to other species, implying that KDM3 group genes may have evolved in strawberry to meet some unique regulatory needs. It is noticeable that there was no H3K79me-specific Dot1/Dot1L HKMTase identified in the five angiosperms (Fig. 1). Considered that Dot1/Dot1L gene is present in S. moellendorffii (Fig. 1) and also in animals6, it is likely that angiosperms have lost the Dot1/Dot1L HKMTases in their common ancestor.
The phylogeny based on both sequence conservation and domain construct in this study suggests that WGDs and dispersed duplications contributed to the expansion of some histone lysine modifiers in angiosperms, which is consistent with that previously reported20,29. On the other hand, all the F. vesca lineage-specific duplications originated from dispersed duplications, particularly retro-transpositions in some cases. The fact that those recently duplicated gene pairs have greatly diverged in expression patterns suggests that they might have been retained in the F. vesca genome by selection to more precisely regulate the developmental processes which histone methylation plays a role.
All the SET, LSD and JmjC families have several genes, and their functions could be both redundant and/or complementary. Thus, the overall histone modifications in particular tissues are determined by the combinatorial expression profiles of histone modifiers. Indeed, the different organs/stages have distinct combinatorial expression patterns during flower and fruit development, and many of those histone modifier genes show abrupt up- or down-regulation in specific tissues or developmental stages. Anthers and carpels where sporogenesis and gametogenesis occur, appear to have more genes being up-regulated, irrespective of whether they code for HKMTases or HDMases, highlighting the potentially active and dynamic regulation of histone modifications in these tissues. However, pollen (considered to be in a division- and growth-quiescent state) showed the least number of genes with active expression, particularly the class V SET genes (Fig. 5A). The lack of expression of both H3K9me- and H2K27me2/3-specific SET genes (both for silencing chromatin) is consistent with the de-condensed chromatin states in the vegetative cells in pollen. Pollen grains have three cells, one large vegetative cell and two germ cells. It is known that in Arabidopsis, the vegetative cell lack H3K9me2 marks, resulting in the genome-wide activation of transposons. The small RNAs produced by transposon activation are delivered to the germ cells to silence the transposons and thus maintain DNA integrity30. The lack of expression of class V SET genes might reflect this compromised strategy of the large vegetative cell to protect the germ lines. A few genes coding for demethylase were sharply up-regulated in specific stages or tissues, for example FV_JmjC9 and FV_JmjC22 in pollen, which might serve critical regulatory function there.
Another interesting phenomenon is that the expression levels of the some active SET genes that we investigated in pith and cortical tissues decreased during early-stage fruit development (from pollination to big green stage), but gradually increased beginning at the white stage and reaching a peak at the turning stage (Fig. 5). It has been reported that epigenetic marks fluctuate during fruit development and ripening, e.g. overall DNA methylation levels decrease during tomato fruit ripening31. Global DNA methylation measurements revealed that DNA methylation varied in both peal and flesh of sweet orange32. Our results suggest that histone modifications might be dynamic during strawberry fruit development and ripening as well, and histone modifiers are probably involved in this regulatory process.
Epigenentic regulation of cellular processes during abiotic stresses has been the subject of several recent investigations that suggest that both DNA modifications and histone modifications play a pivotal role in plant responses to various stresses1,14,33,34. The RT-qPCR results revealed that the expression levels of a set of HKMTases were found to be elevated after cold/heat shock (Fig. 5C), indicating their possible participation in response to extreme temperatures in strawberry plants. Overall, expressions of histone modifers are dynamic in different tissues during different developmental stages, and in response to abiotic stresses as well, demonstrating their regulatory roles in various cellular processes in strawberry.
Compared to the well-studied model plants Arabidopsis and rice, strawberry has several distinct characteristics including that “fruits” develop from receptacle tissues and the adaptability of the species to different environments. As essential regulatory factors, how histone modifiers are involved in various cellular process is of great interest. The majority of previous studies about histone modifiers focused on Arabidopsis, which does not have fleshy fruits. Although some work has been published about histone modifiers in tomato, the data are limited. Our identification and characterization of histone modifiers in woodland strawberry is the first comprehensive analysis of HKMTases and HDMases in a non-climacteric fruit species. Our study provides an overview of the histone lysine methylation modifiers in strawberry, and should greatly facilitates molecular, biochemical and physiological characterizations of histone methylations in strawberry and other Rosaceae species.
Methods
Data retrieve
Eight plant genomes were analyzed, Fragiaria vesca, Arabidopsis thaliana, Vites vinifera, Nelumbo nucifera, Orazy sativa, Zea mays, Amborella trichopoda and Selaginella moellendorffii. The Fragaria vesca and Nelumbo nucifera complete protein sequences and corresponding annotation information were downloaded from NCBI and others were download from Phytozome (version 10.3; http://phytozome.jgi.doe.gov/pz/portal.html). See Supplementary Table S5 for versions and resources of the databases. In proteome datasets, if more than one protein are annotated for the same gene from alternative splicing, the longest form was used for further analysis.
Identification of genes with the domain(s) of interest
To identify the genes with the domain(s) of interest, the amino acid sequences of SET domain (PF00856), JmjC domain (PF02373), Amino_oxidase domain (PF05193) and SWIRM domain (PF04433) were downloaded from pfam database V27.035 and used as a query to find homologous sequences in proteome datasets, respectively. To verify the presence of those domains, the resulting sequences were verified using the Pfam database (http://pfam.xfam.org/search), Conserved Domain Database36 (CDD; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) available from NCBI, and the Simple Modular Architecture Research Tool database37 (SMART; http://smart.embl-heidelberg.de/), with a threshold of e-value < 1e−4. Proteins with both Amino_oxidase domain and SWIRM domain were identified as LSD genes.
Sequences alignment and phylogenetic analysis
Protein sequences were aligned using MUSCLE v3.8.31 with default parameters38. Phylogenetic trees were constructed by Raxml (version 8.1.16) with gamma distribution and 1000 bootstrapping replicates39. The construct of each phylogenetic tree was verified by MrBayes v3.2.440.
Domain and motif analysis, and identification of F. vesca lineage-specific duplicated pairs
All identified proteins were used to search against the PFAM, SMART and CDD databases to search for other known domains. All domains found by any of the three databases with e-value < 10−4 were kept. In addition, motif analyses were performed online by MEME (MEME, Version4.10.2, http://meme-suite.org/tools/meme)41. The number of motifs was set at no more than 20 with the length from 15–50 amino acids for each search. The lineage-specific duplicated gene pairs were identified based on both phylogenetic trees, domain composition of the whole proteins and motif composition of the SET domains. FV-SET13/35 was not identified as an F. vesca lineage-specific duplicated pair by the phylogenetic tree constructed for SET class V (Supplementary Fig. S4) and thus was omitted; while FV-SET33/40 and FV-SET10/12 were included based on either domain composition (Supplementary Fig. S2) or phylogenetic trees constructed for each SET class (Supplementary Fig. S6).
Plant growth conditions, stress treatments and material collection
A 7th generation inbred line of woodland Fragaria vesca, Ruegen F7-4 (Kindly provided by Janet Slovin) was used for all strawberry material collection. Strawberry flesh at different development stages was collected from plants grown in 10 cm x 10 cm pots in a controlled-environment growth chamber, set at 16 h light/8 h dark cycles, 22 °C, and 65% relative humidity. Strawberry fruits at 12-day old big green stage, big white stage (white flesh with red seeds), pink stage (slight pink flesh and red seeds), and red stage (2–3 days after the pink stage) were collected and immediately put into liquid nitrogen.
The strawberry seedlings for cold/heat shock were grown in MS media in a growth chamber set at 16 h light/8 h dark cycles, 22 °C. Four-week old seedlings were transferred to a growth chamber set at either 4 °C or 38 °C for cold/heat shock. Cold shocked seedlings were collected at 1 h, 3 h and 8 h; while heat shocked seedlings were collected at 1 h and 3 h, or at 4 h (3 h heat shock and 1 h recovery at 22 °C) and 8 h (3 h heat shock and 5 h recovery at 22 °C). The collected materials were immediately put into liquid nitrogen for RNA processing.
RNA extraction and expression analysis
Before RNA extraction, the achenes were stripped from the frozen strawberry fruit, and only the de-seeded flesh was processed for RNA isolation. RNA was isolated from either the deseeded flesh or seedlings by a modified CTAB method. After DNase I treatment, RNAs were used for cDNA synthesis by using the Primerscript RT reagent Kit with gDNA Erase (Takara). The cDNAs were used as templates for quantitative RT-PCR to measure the abundance of a certain transcript. Quantitative RT-PCR was performed using SYBR Premix Ex Tag (Takara) on a Bio-rad iQ5. Primers used are listed in Supplementary Table S6. Results were analyzed by using the ΔΔCT method42 using GAPDH as the control locus43. Three biological and three technical replicates were performed and analyzed.
Additional Information
How to cite this article: Gu, T. et al. Identification and characterization of histone lysine methylation modifiers in Fragaria vesca. Sci. Rep. 6, 23581; doi: 10.1038/srep23581 (2016).
References
Thorstensen, T., Grini, P. E. & Aalen, R. B. SET domain proteins in plant development. Biochim Biophys Acta 1809, 407–20 (2011).
Deal, R. B. & Henikoff, S. Histone variants and modifications in plant gene regulation. Curr Opin Plant Biol 14, 116–22 (2011).
Zhang, Y. & Reinberg, D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes & Development 15, 2343–2360 (2001).
Lachner, M. & Jenuwein, T. The many faces of histone lysine methylation. Curr Opin Cell Biol 14, 286–98 (2002).
Ng, D. W. et al. Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta 1769, 316–29 (2007).
Feng, Q. et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 12, 1052–8 (2002).
Kooistra, S. M. & Helin, K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13, 297–311 (2012).
Pontvianne, F., Blevins, T. & Pikaard, C. S. Arabidopsis Histone Lysine Methyltransferases. Adv Bot Res 53, 1–22 (2010).
Jacob, Y. & Michaels, S. D. H3K27me1 is E(z) in animals, but not in plants. Epigenetics 4, 266–269 (2009).
Shi, Y. G. & Tsukada, Y. The discovery of histone demethylases. Cold Spring Harb Perspect Biol 5 (2013).
Klose, R. J., Kallin, E. M. & Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7, 715–27 (2006).
Goodrich, J. et al. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis . Nature 386, 44–51 (1997).
Grossniklaus, U., Vielle-Calzada, J. P., Hoeppner, M. A. & Gagliano, W. B. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis . Science 280, 446–50 (1998).
Avramova, Z. Transcriptional ‘memory’ of a stress: transient chromatin and memory (epigenetic) marks at stress-response genes. Plant Journal 83, 149–159 (2015).
Shulaev, V. et al. The genome of woodland strawberry (Fragaria vesca). Nat Genet 43, 109–16 (2011).
Amborella Genome, P. The Amborella genome and the evolution of flowering plants. Science 342, 1241089 (2013).
Jiao, Y. et al. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (2011).
Yang, Z. & Nielsen, R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17, 32–43 (2000).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40, e49 (2012).
Qian, S., Wang, Y., Ma, H. & Zhang, L. Expansion and Functional Divergence of Jumonji C-Containing Histone Demethylases: Significance of Duplications in Ancestral Angiosperms and Vertebrates. Plant Physiol 168, 1321–37 (2015).
Kang, C. et al. Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca . Plant Cell 25, 1960–78 (2013).
Hollender, C. A. et al. Floral Transcriptomes in Woodland Strawberry Uncover Developing Receptacle and Anther Gene Networks. Plant Physiol 165, 1062–1075 (2014).
Lei, L., Zhou, S. L., Ma, H. & Zhang, L. S. Expansion and diversification of the SET domain gene family following whole-genome duplications in Populus trichocarpa . BMC Evol Biol 12, 51 (2012).
Qian, Y., Xi, Y., Cheng, B., Zhu, S. & Kan, X. Identification and characterization of the SET domain gene family in maize. Mol Biol Rep 41, 1341–54 (2014).
Aquea, F., Vega, A., Timmermann, T., Poupin, M. J. & Arce-Johnson, P. Genome-wide analysis of the SET DOMAIN GROUP family in grapevine. Plant Cell Rep 30, 1087–97 (2011).
Huang, Y., Liu, C., Shen, W. H. & Ruan, Y. Phylogenetic analysis and classification of the Brassica rapa SET-domain protein family. BMC Plant Biol 11, 175 (2011).
Zhu, X., Ma, H. & Chen, Z. Phylogenetics and evolution of Su(var)3-9 SET genes in land plants: rapid diversification in structure and function. BMC Evol Biol 11, 63 (2011).
Schnable, P. S. et al. The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–5 (2009).
Zhang, L. & Ma, H. Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol 195, 248–63 (2012).
Borg, M. & Berger, F. Chromatin remodelling during male gametophyte development. Plant Journal 83, 177–188 (2015).
Zhong, S. et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31, 154–9 (2013).
Xu, J. D., Xu, H. D., Xu, Q. & Deng, X. X. Characterization of DNA Methylation Variations During Fruit Development and Ripening of Sweet Orange. Plant Molecular Biology Reporter 33, 1–11 (2015).
Ndamukong, I., Jones, D. R., Lapko, H., Divecha, N. & Avramova, Z. Phosphatidylinositol 5-phosphate links dehydration stress to the activity of ARABIDOPSIS TRITHORAX-LIKE factor ATX1. PLoS One 5, e13396 (2010).
Slotkin, R. K. & Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8, 272–85 (2007).
Finn, R. D. et al. The Pfam protein families database. Nucleic Acids Res 38, D211–22 (2010).
Marchler-Bauer, A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res 43, D222–6 (2015).
Letunic, I., Doerks, T. & Bork, P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43, D257–60 (2015).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–7 (2004).
Stamatakis, A. Using RAxML to Infer Phylogenies. Curr Protoc Bioinformatics 51, 1–6 (2015).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–4 (2003).
Bailey, T. L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2, 28–36 (1994).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408 (2001).
Amil-Ruiz, F. et al. Identification and validation of reference genes for transcript normalization in strawberry (Fragaria x ananassa) defense responses. PLoS One 8, e70603 (2013).
Acknowledgements
We thank Dr. Jing Ding and Xiaojing Wang for technical support for the bioinformatic analysis. This work was supported by the National Natural Science Foundation of China (31400269) and the Fundamental Research Funds for the Central Universities (KJQN201537) to Tingting Gu.
Author information
Authors and Affiliations
Contributions
T.G., Y.L., R.J.M. and Y.H. wrote the main text and prepared the figures and tables. T.G. and Y.L. designed the project. Y.H., T.G. and R.H. did the experiments and analysis. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Gu, T., Han, Y., Huang, R. et al. Identification and characterization of histone lysine methylation modifiers in Fragaria vesca. Sci Rep 6, 23581 (2016). https://doi.org/10.1038/srep23581
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep23581
This article is cited by
-
Identification of watermelon H3K4 and H3K27 genes and their expression profiles during watermelon fruit development
Molecular Biology Reports (2023)
-
High Temperature Triggers Differential Expression of JUMONJI C (JmjC) Domain-Containing Histone Demethylase Genes in Leaf and Stolon Tissues of Potato (Solanum tuberosum L.) Genotypes
Journal of Plant Growth Regulation (2023)
-
Genome-wide identification and expression analysis of JmjC domain–containing genes in grape under MTA treatment
Functional & Integrative Genomics (2022)
-
A native chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in strawberry fruits
Plant Methods (2020)
-
Bioinformatics and expression analysis of histone modification genes in grapevine predict their involvement in seed development, powdery mildew resistance, and hormonal signaling
BMC Plant Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.