Abstract
Antibodies specific for histone post-translational modifications (PTMs) have been central to our understanding of chromatin biology. Here, we describe an unexpected and novel property of histone H4 site-specific acetyl antibodies in that they prefer poly-acetylated histone substrates. By all current criteria, these antibodies have passed specificity standards. However, we find these site-specific histone antibodies preferentially recognize chromatin signatures containing two or more adjacent acetylated lysines. Significantly, we find that the poly-acetylated epitopes these antibodies prefer are evolutionarily conserved and are present at levels that compete for these antibodies over the intended individual acetylation sites. This alarming property of acetyl-specific antibodies has far-reaching implications for data interpretation and may present a challenge for the future study of acetylated histone and non-histone proteins.
Similar content being viewed by others
Introduction
The identification and biological characterization of histone post-translational modifications (PTMs) has been the subject of intense recent investigation1,2,3. One of the most studied histone PTMs is lysine acetylation, which typically occurs on the N-terminal “tails” and globular domains of histones and can influence chromatin-based events including transcription, DNA replication, DNA repair and dosage compensation1,4. One mechanism by which lysine acetylation influences chromatin function is by removing positive charges from lysine side chains, thus making local chromatin structure more permissive to specific protein machineries5. Lysine acetylation can also function by serving as a docking site for bromodomain-containing proteins, often found as subunits of histone acetyltransferases (HATs), ATP-dependent chromatin remodelers and transcriptional coactivators6,7. Significantly, recent studies show that bromodomain-containing proteins preferentially recognize poly-acetylated chromatin signatures7,8,9. These studies lend further support to the ‘histone code’ hypothesis, which suggests that histone PTMs function in a combinatorial fashion to regulate chromatin architecture and DNA-templated cellular processes10,11.
Direct investigations of biological functions associated with specific histone PTMs have been facilitated by genetic and biochemical methods and often depend on antibodies to monitor these PTMs. Furthermore, large scale epigenomics efforts, like the ENCODE and modENCODE projects, rely on these antibodies to map the genomic distribution of chromatin signatures12,13,14. Therefore, antibody specificity is of utmost importance for accurate data interpretation. The standard criteria for characterizing antibody specificity typically involves primary reactivity with a single species from cell extracts by immunoblotting that is diminished in the absence or mutation of epitope and that can be competed with recombinant or synthetic antigen9,15,16. Extended criteria often involve characterizing the ability of antibodies to perform in biological assays, like chromatin immunoprecipitation (ChIP), immunohistochemistry, enzyme-linked immunosorbent assay (ELISA) and immunoblots.
Recent studies from our lab and others demonstrate that neighboring PTMs often enhance or perturb the ability of histone antibodies to recognize their intended target9,15,16. Furthermore, these studies have found that histone antibodies often have specific difficulties in recognizing their appropriate epitopes, either due to the inability to distinguish methyl-lysine states (mono-, di- and tri-methylation) or to recognize off-target PTMs. In addition, studies from the modENCODE consortium have found that > 25% of commercial histone antibodies fail basic quality control measures17. Here, we uncover a novel property of histone H4 antibody-antigen recognition (preferential detection of poly-acetylated chromatin signatures) that presents a significant concern with the use of these reagents. Our findings caution interpreting results to date that employ these site-specific acetyl antibodies and suggest more thorough validation of antibodies is needed before they can be labeled as specific.
Results
Site-specific H4 acetyl antibodies prefer poly-acetylated substrates
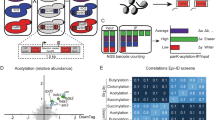
To interrogate the interactions of chromatin-associated proteins and antibodies with combinatorial histone PTMs, we recently developed a peptide microarray platform where > 250 unique biotinylated histone peptides, containing 0–8 possible PTMs, were immobilized on streptavidin-coated glass slides (Supplemental Table 1)9,16. These peptide arrays were probed with a number of commonly used commercial histone acetyl-specific antibodies (Supplemental Table 2) to discern their specificities. We found that acetyl-specific antibodies directed against H3 lysines 9 and 14 (H3K9ac and H3K14ac) generally performed as expected, in that they showed no discernable interaction with unmodified histones and detected their intended PTM alone and in the context of adjacent acetylation events with similar signal intensity (Fig. 1a and Supplemental Fig. 1). Of note, H3S10 phosphorylation (H3S10p) perturbed the recognition of H3K9ac (see peptides 37, 41, 144 and 148 in Supplemental Fig. 1), but had little effect on H3K14ac recognition. H3S10p, enriched on mitotic chromatin18, has been shown to exist on the same histone tail as H3K9ac in cells19,20,21. Our array analysis therefore suggests this H3S10 phosphorylated population of H3K9 acetylated histone tails may be underrepresented in biological assays using this antibody. We also detected weak cross-reactivity of these antibodies with H4 and H2A acetylated peptides (Supplemental Fig. 1).
(a-c) Heat maps summarizing peptide array results for H3 and H4 acetyl antibodies.For each array, the most intense series of peptide spots (12 individual spots per peptide) is assigned a value of 1 (blue) and all values are normalized to this peptide. Values ≥ 0.1 are colored red in panel C to enable interpretation of low signal intensities. Each interaction is presented as an averaged normalized intensity from at least two independent arrays (r2 > 0.9). See Rothbart et al16 for details of the array methodology. Antibody information can be found in Supplemental Table 2. While one antibody for each histone PTM is shown in the figure for representation, other antibodies tested to the same PTMs showed similar findings (Supplemental Table 2).(d) Normalized array signal intensities for H4K12ac antibody binding to the indicated peptides. Values are presented as an average of 24 individual spots (2 arrays) ± s.e.m.
In sharp contrast to above, antibodies designed to recognize H4K5ac, H4K8ac and H4K12ac preferentially bound H4 peptides harboring two or more adjacent acetylation events (Fig. 1b and Supplemental Table 2). Importantly, acetyl recognition was dependent on the single acetylation event that was intended to be recognized by the antibody. For example, the H4K8ac antibody bound to H4K8ac/K12ac and H4K8ac/K16ac, but not to the H4K12ac/K16ac peptide (Fig. 1b). Similar observations were observed for the H4K5ac and H4K12ac antibodies. While poly-acetylated substrates ranked as being the preferred substrates on our arrays for the H4 acetyl antibodies tested (Fig. 1b and Supplemental Table 2), closer examination of the individually acetylated peptides revealed low-intensity (i.e. weaker) interactions that demonstrated these antibodies do indeed recognize their intended PTM preferentially to other single acetylation events on the H4 tail (Fig. 1c). By these criteria, these antibodies would likely be labeled as specific. However, strong preference for poly-acetylated peptides (4- to 20-fold; Table 1 and Supplemental Table 2) suggests an added layer of complexity to the specificity criteria and implies these antibodies may have difficulty distinguishing singly modified epitopes in vivo. Importantly, the antibody we tested that recognizes H416ac was not dramatically influenced by poly-acetylated H4. However, unlike the other site-specific H4 antibodies examined, the H4K16ac antibody cross-reacted with acetylated H3 and H2A peptides (Supplemental Fig. 2).
As acetylation masks the positive charge on lysine ε-amines, we next wondered if poly-acetyl recognition might be a consequence of “charge masking.” To test this idea, we synthesized an H4K12ac peptide in which lysines 5, 8 and 16 were mutated to glutamine, a commonly used acetyl mimic. Importantly, H4K12ac antibody recognition of this peptide did not mimic that seen with a tetra-acetylated peptide (Fig. 1d). These results strongly suggest poly-acetyl antibody recognition is not solely a property of charge neutralization.
Poly-acetylated chromatin signatures are evolutionarily conserved
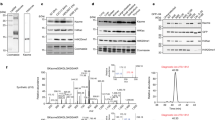
Our array results suggest that poly-acetylated H4 chromatin signatures could be problematic for antibody-based detection of single H4 acetylation events in cells. However, the extent to which poly-acetylated H4 chromatin signatures exist in relation single acetylation events in cells is unknown. We therefore sought to determine and compare the abundance of histone H4 acetylation signatures by mass spectrometry in budding yeast, mouse embryonic stem cells (mESCs), mouse embryonic fibroblasts (MEFs) and the HeLa human cervical carcinoma cell line (Fig. 2a and Supplemental Fig. 3). In all four cell types, 50–60% of the H4 tail (residues 4–17) is unmodified, while H4K16ac marks 20–40%. H4K12ac is the next most abundant single mark (5–15%) in all cells analyzed, followed by H4K5ac (<5%) and the H4K8ac (<2%). Importantly, the presence of poly-acetylation events (i.e. two or more) on the H4 tail is detectable in all cells examined and this signature is often present at levels comparable to both H4K5ac and H4K8ac as single acetylation events (Fig 2a).
(a) Quantitative mass spectrometry to determine the distribution of single- and poly-acetylation of the indicated H4 peptide across species.Poly-ac is represented as a summation of 2 or more acetylations in the context of the single mark. A complete analysis is shown in Supplemental Fig. 3. (b-c) Western blots of HeLa chromatin extracts following antibody incubation with the indicated concentrations of competing peptide.
Site-specific H4 acetyl antibodies preferentially recognize poly-acetylation signatures in bulk chromatin
The biological identification of appreciable poly-acetylated H4 finally led us to determine whether site-specific H4 acetyl antibodies preferentially recognize this poly-acetylated chromatin signature in cells. Using peptide competition assays, we first examined the H4K12ac antibody, which showed a strong (20-fold) preference for poly-acetylated peptides over the single mark by array (Table 1), but conversely was under-represented as a poly-acetylated chromatin signature in vivo in comparison to the single mark by mass spectrometry (Fig 2a). Importantly, a tetra-acetylated H4 peptide was able to compete the H4K12ac antibody at a concentration 10-fold lower than an H4K12ac peptide (Fig 2b). Similar results were seen with an H4K5ac antibody (Fig 2c). Collectively, these results demonstrate that poly-acetylation signatures are prevalent in cells and are the preferred epitope for these site-specific acetyl antibodies.
Discussion
Selectivity issues related to antibodies, especially those targeting histone PTMs, is by no means a new problem. Common concerns include cross-reactivity with other PTMs or states of methyl-lysine and recent studies have begun characterizing how neighboring PTMs influence antibody recognition in a similar manner to effector protein binding9,15,16,22. With so many biological methods relying on antibodies for detection and enrichment, thorough characterization of these reagents is paramount. Our results demonstrate a previously uncharacterized property of histone H4 site-specific acetyl antibodies, an inherent preference for poly-acetylated chromatin signatures. By standard criteria, these antibodies would be judged selective for their intended PTM. They all recognize a single protein band by immunoblot, can be competed with a peptide antigen, do not cross-react with other singly acetylated epitopes and even have been shown to lose reactivity when probing for histones mutated at the target lysine (Figs. 1–2 and data not shown). Using our peptide arrays as a new criterion for antibody characterization, our findings suggest H4K5ac, H4K8ac and H4K12ac antibodies may not recognize their intended target, but instead, all recognize the same poly-acetylated histone signature.
Our unexpected finding regarding the property of H4 acetyl-specific antibodies raises new questions with the interpretation of genome-wide mapping studies of H4 acetylation events across species and cell lines. In general, genomic studies find a high correlation for the positioning of H4K5ac, K8ac and K12ac across genomes23,24,25. However, it is entirely possible that these findings represent the preference of these antibodies for H4 poly-acetylation – a chromatin signature that is present in vivo and at levels that these antibodies would compete for over their individually acetylated counterparts. Given the problematic nature of these antibodies, the true genomic locations and relative distributions of the individual H4K45ac, H4K8ac and H4K12ac marks are likely still unknown. Since our mass spectrometry results show high species conservation and significant abundance of singly acetylated H4 tails, we suggest that the individual H4K5, H4K8 and H4K12 acetylation events might have non-overlapping genomic distributions and functions. Future studies involving improved site-specific H4 acetyl antibodies will be needed to test this hypothesis.
A counter argument to the concern of recognizing poly-acetylation signatures are classic genetic studies in budding yeast showing that mutation of H4 lysines 5, 8 and 12 individually have indistinguishable changes in gene expression profiles or growth defects, while only mutation of lysine 16 has a unique gene expression signature23,26. Combing multiple H4 lysine mutations results in a cumulative effect on gene expression and defective growth - suggesting H4K5, H4K8 and H4K12 acetylation events are functionally redundant and cumulative. However, parallel studies have not yet been performed in more complex organisms where individual acetylation events might play a more significant role. With a new Drosophila melanogaster histone replacement model now available, important and interesting questions such as these can be tested27.
One question remaining is why might these antibodies strongly detect poly-acetylated histones in the first place? The fact that H3 acetyl antibodies do not have strong poly-acetyl preference suggests this problem is specific towards histone H4. A potential explanation may lie in the repetitive sequence surrounding the K5, K8 and K12 acetylation sites on the H4 tail. The GKG motif that surrounds these lysines is repeated on the histone H4 tail. Lysine 16, however, differs, from this pattern (AKR) and the H4K16ac antibody coincidently has the least enhanced preference for poly-acetylated H4. Regardless of the reasons, one would ideally want the H4 acetyl antibodies to behave more similarly to the H3 acetyl antibodies tested, recognizing singly modified acetyl-lysines similarly to poly-acetylated epitopes.
Beyond histones, we note that high-resolution mass spectrometry studies have recently identified thousands of acetylation events across multiple species28,29,30,31. Site-specific acetyl antibodies will undoubtedly be developed for studying biological functions associated with these non-histone acetylation events. Importantly, the same GKG consensus motif that may pose a problem for H4 acetyl antibodies surrounds a number of identified acetylated lysines on non-histone proteins32,33. It remains to be determined whether similar antibody-based detection issues will apply for these non-histone PTMs.
In conclusion, we describe a new and concerning property of site-specific acetyl antibodies that has been previously missed in all other forms of characterization. This re-defined property has implications for past data interpretation and represents a formidable challenge for future studies. This paper therefore serves to encourage more thorough validation of the next generation of acetyl antibodies for the biological community at large.
Methods
Materials
A full list of antibodies used in this study is available in Supplemental Table 2. HeLa cells were cultured in suspension between 5−10 × 105 cells/ml in minimum essential Joklik modified media supplemented with 10% new-born calf serum, 2 mM L-glutamine, 100 units/mL penicillin and 0.1 mg/mL streptomycin34. mESCs were cultured in DMEM high glucose medium supplemented with 15% fetal bovine serum, 2 mM L-glutamine, 0.1 mM non-essential amino acids, 0.1 mM 2-mercaptoethanol, 100 units/mL penicillin, 0.1 mg/mL streptomycin/mL and 1000 units/mL LIF/ESGRO35. MEFs were derived from 14.5 d mouse embryos and cultured in DMEM high glucose medium supplemented with 10% fetal bovine serum, 2 mM L-Glutamine, 100 units/mL penicillin and 0.1 mg/mL streptomycin35.
Peptide arrays
Peptide synthesis and validation, array fabrication and antibody analysis were performed essentially as described9,16.
Mass spectrometry
Sample collection, derivation and mass spectrometry were performed as previously described34 with the following modifications: 60 to 120 µg bulk histones were used for each chemical derivation and analysis; the m/z of double, triple and quadruple acetylations on the H4 4–17 peptide were targeted in the MS runs.
Peptide competitions
Bulk chromatin was isolated from asynchronously growing HeLa cells as described36 with the following modifications: Pelleted cells were lysed in cold buffer containing 10 mM PIPES, pH 7.0, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1x EDTA-free protease inhibitor (Roche), 1x phosphatase inhibitor cocktail (Sigma) and 0.1% Triton X-100. Chromatin fractions were treated with benzonase to solubilize histones prior to SDS-PAGE.
References
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Gardner, K. E., Allis, C. D. & Strahl, B. D. Operating on chromatin, a colorful language where context matters. J Mol Biol 409, 36–46 (2011).
Henikoff, S. & Shilatifard, A. Histone modification: cause or cog? Trends Genet 27, 389–396 (2011).
Shahbazian, M. D. & Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76, 75–100 (2007).
Shogren-Knaak, M. et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Zeng, L. & Zhou, M. M. Bromodomain: an acetyl-lysine binding domain. FEBS Lett 513, 124–128 (2002).
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012).
Ruthenburg, A. J. et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 (2011).
Fuchs, S. M., Krajewski, K., Baker, R. W., Miller, V. L. & Strahl, B. D. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol 21, 53–58 (2011).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Kharchenko, P. V. et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471, 480–485 (2011).
Gerstein, M. B. et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787 (2010).
Bock, I. et al. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics 6, 256–263 (2011).
Rothbart, S. B., Krajewski, K., Strahl, B. D. & Fuchs, S. M. Peptide microarrays to interrogate the "histone code.". Methods in Enzymology 512, In Press (2012).
Egelhofer, T. A. et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol 18, 91–93 (2011).
Hendzel, M. J. et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106, 348–360 (1997).
Zippo, A. et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138, 1122–1136 (2009).
Zippo, A., De Robertis, A., Serafini, R. & Oliviero, S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol 9, 932–944 (2007).
Karam, C. S., Kellner, W. A., Takenaka, N., Clemmons, A. W. & Corces, V. G. 14-3-3 mediates histone cross-talk during transcription elongation in Drosophila. PLoS Genet 6, e1000975 (2010).
Fuchs, S. M. & Strahl, B. D. Antibody recognition of histone post-translational modifications: emerging issues and future prospects. Epigenomics 3, 247–249 (2011).
Kurdistani, S. K., Tavazoie, S. & Grunstein, M. Mapping global histone acetylation patterns to gene expression. Cell 117, 721–733 (2004).
Liu, C. L. et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3, e328 (2005).
Wang, Z. et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40, 897–903 (2008).
Dion, M. F., Altschuler, S. J., Wu, L. F. & Rando, O. J. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A 102, 5501–5506 (2005).
Gunesdogan, U., Jackle, H. & Herzig, A. A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep 11, 772–776 (2010).
Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009).
Kim, S. C. et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23, 607–618 (2006).
Weinert, B. T. et al. Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci Signal 4, ra48 (2011).
Zhang, J. et al. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics 8, 215–225 (2009).
Basu, A. et al. Proteome-wide prediction of acetylation substrates. Proc Natl Acad Sci U S A 106, 13785–13790 (2009).
Smith, K. T. & Workman, J. L. Introducing the acetylome. Nat Biotechnol 27, 917–919 (2009).
Zee, B. M., Levin, R. S., Dimaggio, P. A. & Garcia, B. A. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin 3, 22 (2010).
Sridharan, R. et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377 (2009).
Mendez, J. & Stillman, B. Chromatin association of human origin recognition complex, cdc6 and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20, 8602–8612 (2000).
Acknowledgements
This work was supported in part by grants from the NIH (GM085394) and the North Carolina Biotechnology Center (NCBC) to B.D.S. S.B.R. was supported by postdoctoral grant T32CA09156. Support from a National Science Foundation (NSF) Early Faculty CAREER award and an NIH Innovator award (DP2OD007447) from the Office of the Director (NIH) to B.A.G. is gratefully acknowledged. S.L. was supported by postdoctoral training grant (NIGMS P50GM071508) from the Center for Quantitative Biology at Princeton University grant to the Lewis-Sigler Institute, PI D. Botstein.
Author information
Authors and Affiliations
Contributions
S.B.R. performed array and peptide competition experiments, analyzed the data and prepared figures. S.L., L.M.B and B.A.G. performed and analyzed MS data. K.K. synthesized and validated all peptides for arrays and peptide competitions. M.C.K. provided key reagents and discussed results. S.B.R. and B.D.S. designed the study and wrote the manuscript. All authors participated manuscript review.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Rothbart, S., Lin, S., Britton, LM. et al. Poly-acetylated chromatin signatures are preferred epitopes for site-specific histone H4 acetyl antibodies. Sci Rep 2, 489 (2012). https://doi.org/10.1038/srep00489
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00489
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.