Abstract
Meningococcal factor H binding protein (fHbp) is an important vaccine antigen for prevention of disease caused by capsular group B strains. The protein has been sub-classified into three variant groups. Most anti-fHbp antibodies are variant group-specific and recognize epitopes on the C-terminal domain. We report a murine IgG1 mAb, JAR 41, which broadly cross-reacted with fHbp sequence variants from all variant groups. The mAb bound to the surface of live meningococci with fHbp from each of the three variant groups. In combination with second non-bactericidal anti-fHbp mAbs, JAR 41 elicited complement-mediated bactericidal activity in vitro and augmented passive protection against meningococcal bacteremia in human fH transgenic rats. The epitope was located on a conserved region of the N-terminal portion of the fHbp molecule opposite that of fH contact residues. The data underscore the importance of broadly cross-reactive, surface-exposed epitopes on the N-terminal domain in the design of protective fHbp vaccines.
Similar content being viewed by others
Introduction
Factor H binding protein (fHbp) is an important antigen for vaccines being developed for prevention of meningococcal disease caused by capsular group B strains, for which currently there is no broadly protective vaccine (reviewed in1,2). In mice and humans, vaccines containing recombinant fHbp elicited serum bactericidal antibody activity3,4,5,6,7, which is a serologic surrogate of protection against developing meningococcal disease8,9,10. Based on amino acid sequence homology and antigenic cross-reactivity, fHbp has been sub-classified into two sub-families11 or three variant groups12. fHbp also can be classified into modular groups based on different combinations of five variable segments, each of which is encoded by genes from one of two lineages13. In general, bactericidal activity of anti-fHbp antibodies was largely confined to meningococcal strains with fHbp amino acid sequence variants from the same variant group or sub-family as that of the vaccine antigen7,12,14.
In previous studies, our laboratory has characterized panels of anti-fHbp mAbs prepared in mice immunized with fHbp from variant groups 1, 2 or 315,16. While an occasional mAb reacted with fHbp sequence variants from more than one variant group, to date, none of the mAbs reacted with sequence variants from all three variant groups. In this study, we describe a new anti-fHbp mAb, designated JAR 41, which was isolated from a human factor H (fH) transgenic mouse that had been immunized with a recombinant fHbp from variant group 117. The new mAb recognized a surface-exposed, broadly cross-reactive epitope present in all fHbp sequence variants tested from variant groups 1, 2 or 3. Thus, JAR 41 may be useful for developing assays to measure strain expression of fHbp, which is an important determinant of susceptibility to anti-fHbp bactericidal activity18,19,20,21. Information on the JAR 41 epitope also may be important for the design of improved broadly protective fHbp vaccines14,22.
Results
Anti-fHbp mAb JAR 41 is broadly cross-reactive
By ELISA, JAR 41 had similar concentration-dependent binding to fHbp ID 1 (variant group 1) as that of a control anti-fHbp, JAR 5, which is specific for variant group 1 (Figure 1a). Two other anti-fHbp mAbs, JAR 11 and JAR 35, which bind fHbp from variant groups 2 and 3, but not from group 1, served as negative IgG controls. JAR 41 also showed similar concentration-dependent binding to fHbp ID 77 (variant group 2) and fHbp ID 28 (variant group 3) as that of the respective positive controls, variant group-specific mAbs (Figures 1b and 1c, respectively). There was no reactivity of these two proteins with JAR 5, which was specific for fHbp variant group 1.
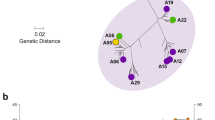
As of December 2011, more than 500 unique fHbp amino acid sequence variants had been deposited in the public database, http://pubmlst.org/neisseria/fHbp/. To select sequence variants representative of fHbp diversity for measuring the breadth of JAR 41 reactivity, we performed a phylogenic analysis of 531 sequences in the public database in the form of a network diagram produced using the program SplitsTree 423 (Figure 2a). Each unique sequence is represented by a “twig” (node), which are distributed on the network into two sub-families, A and B, as described by Fletcher et al7, or three variant groups, 1, 2 or 3, as described by Masignani et al12. Sub-family A contains the sequences in variant groups 2 and 3 and sub-family B contains the sequences in variant group 1. The proteins also can be classified into ten modular groups based on the respective lineages of five variable segments13,18.
Relatedness of unique fHbp amino acid sequences.
Network analysis illustrating the relatedness of unique fHbp amino acid sequences, as generated using the program SplitsTree23. Panel a, the relatedness of 531 unique fHbp amino acid sequences available from the public database www.pubmlst.org. Each “twig” (node) represents a unique sequence. Sub-families, as described by Murphy et al11; variant groups as described by Masignani et al12 and modular groups (denoted using roman numbers as described by Beernink and Granoff13. Modular Group X is a new group reported in this work. Panel b, the relatedness of the 21 fHbp sequence variants, which were selected for production of recombinant proteins (See Table 1). Each filled circle represents a unique sequence variant designated by fHbp ID number from the public database http://pubmlst.org/neisseria/fHbp.
We selected 21 amino acid sequence variants representative of the known fHbp amino acid sequence diversity for expression of recombinant proteins for testing JAR 41 reactivity (Table 1). This panel included variants from both sub-families A and B, all three variant groups12 and all ten modular groups13,18 (Figure 2b). Note that fHbp ID 283, which is a natural hybrid of variant groups 1 and 3, has A, B and C segments from lineage 2 and D and E segments from lineage 1. This variant was assigned to a new modular group (X).
JAR 41 showed similar respective concentration-dependent binding with all of the amino acid sequence variants in variant groups 1, 2 and 3 (Figure 3a–c) and with two natural hybrids, ID 207 and ID 283 (Figure 3d). Binding was specific for fHbp in that JAR 41 did not bind to two unrelated recombinant meningococcal surface proteins, NadA and GNA1030 (Figure 3e). Also, with each of the sequence variants tested by ELISA, we included negative control IgG anti-fHbp mAbs that were specific for fHbps from heterologous variant groups, which showed negligible binding to the sequence variant tested for binding with JAR 41 (See for example, Figure 1).
Binding of JAR 41 to 21 fHbp sequence variants as measured by ELISA.
Panel a, recombinant fHbps from variant group 1. Panel b, recombinant fHbps from variant group 2. Panel c, recombinant fHbps from variant group 3. Panel d, natural hybrids between variant groups 1 and 2, or 1 and 3. Panel e, solid lines, Data from JAR 41 binding to all 21 recombinant sequence variants (solid lines); or with with negative control recombinant proteins NadA and GNA1030 (dashed lines).
The JAR 41 epitope is exposed on the surface of live meningococci
By flow cytometry, JAR 41 bound to the surface of live bacteria from N. meningitidis strains. Figure 4a compares binding of JAR 41 (25 µg/ml) to wild-type group B strain H44/76 (fHbp ID 1, variant group 1) and an isogenic mutant of strain H44/76 with lower fHbp expression (H44/76-LE). There is ∼4-fold lower binding with the mutant strain. There was no detectable binding to either strain with a control IgG mAb, JAR 11 (specific for fHbps in variant groups 2 or 3). Binding of an anti-PorA mAb was similar with both the wild-type and mutant (Figure 4b), which is evidence that similar numbers of wild-type and mutant cells were tested for JAR 41 binding.
Binding of mAbs to live N. meningitidis strains as measured by flow cytometry.
Panel a, Comparison of JAR 41 binding (25 µg/ml) to wild-type strain H44/76 (H44/76-WT, solid black line) with fHbp ID 1 in variant group 1, or an isogenic mutant with lower expression of fHbp ID 1 (H44/76-LE, blue dashed line). Solid green line, H44/76-WT incubated with 25 µg/ml of a negative control IgG mAb (JAR 11, specific for fHbp in variant groups 2 and 3); green-shaded area, H44/76-LE incubated with JAR 11. Panel b, binding of a control anti-PorA mAb. Orange line, H44/76-WT; orange-shaded area, H44/76-LE. Panels c-f, Concentration-dependent binding of JAR 41 with strain H44/76-WT (panel c), mutant strain H44/76-LE (panel d), strain 8047 with fHbp in variant group 2 (panel e), or M1239 with fHbp in variant group 3 (panel f). Panels c, d and e, thick black line, 25 µg/ml of JAR 41; dark grey-shaded area, 5 µg/ml; dashed black line, 1 µg/ml; thin black line, 0.2 µg/ml; light grey-shaded area, 25 µg/ml JAR 41 with a negative control H44/76 fHbp knock-out mutant; solid blue line, 25 µg/ml of JAR 5, which is specific for fHbp in variant group 1 and a negative control mAb for variants 2 and 3. Panel f, thick black line, 50 µg/ml of JAR 41 with wild-type strain M1239 with fHbp in variant group 3; dark shaded area, 5 µg/ml of JAR 41 (lower concentrations of JAR 41 were not tested); blue line, 50 µg/ml of JAR 5 (specific for fHbp variant group 1); Light gray shade area, 50 µg/ml of JAR 41 incubated with M1239 fHbp knock-out mutant.
We next tested binding of JAR 41 to strains H44/76 and its isogenic mutant using a range of mAb concentrations (Figures 4c and 4d, respectively). As little as 0.2 µg/ml of JAR 41 showed significant binding to the H44/76-WT or -LE mutants over that of background binding, which was determined by testing 25 µg/ml of JAR 41 with a H44/76 fHbp knockout mutant (light gray shaded area). JAR 41 also bound to the surface of group B strains 8047 (fHbp ID 77, variant group 2) and M1239 (fHbp ID 28, variant group 3) (Figures 4e and 4f, respectively). For each of these strains the respective JAR 41 binding was similar when tested at the two highest concentrations (5 µg/ml or 25 µg/ml for 8047, or 5 and 50 µg/ml for M1239 [grey-shaded area and solid lines, respectively]). Strain 8047 also was tested with lower concentrations of the mAb. Binding above that of the IgG negative control (an IgG anti-fHbp mAb specific for variant group 1) was detected with as little as 0.2 µg/ml of JAR 41.
JAR 41 elicits human complement-mediated bactericidal activity with second anti-fHbp mAbs
JAR 41 was not bactericidal when tested individually at concentrations of up to 100 µg/ml against any of the four test strains (H44/76-WT [Figure 5a], the H44/76 LE mutant [Figure 5b], 8047 [Figure 5c] or M1239 [data not shown]). Activation of the classical complement pathway by IgG requires binding of two IgG mAbs to appropriately spaced epitopes such that the respective IgG Fc regions can engage C1q24. When an individual mAb binds to a sparse antigen, there may be insufficient immune complex formed to activate bacteriolysis, whereas combinations of more than one mAb may be sufficient24. When tested in combination with other non-bactericidal anti-fHbp mAbs specific for fHbp in variant group 1, JAR 41 elicited cooperative bactericidal activity against strains H44/76 (Figure 5a) or the H44/76 LE mutant with lower fHbp expression (Figure 5b). In contrast, JAR 41 was not bactericidal in combination with anti-fHbp mAbs specific for variant groups 2 or 3 when tested against strains 8047 (fHbp variant group 2, Figure 5c) or M1239 (fHbp variant group 3, data not shown). Both of these strains were susceptible to bactericidal activity elicited by anti-fHbp mAb JAR 4 when tested with the anti-fHbp mAbs specific for variant groups 2 or 3 (for example, JAR 4 + JAR 11 or JAR 13 against strain 8047, Figure 4c).
Cooperative bactericidal activity elicited by JAR 41, individually or in combination with second anti-fHbp mAbs.
Panel a, strain H44/76 (fHbp ID 1, variant group 1). Panel b, mutant of strain H44/76 engineered to have lower expression of fHbp ID 1 as that of the wild-type strain (See Figure 4a). Panel c, strain 8047 (fHbp ID 77, variant group 2). BC50%, concentration of mAb (µg/ml) resulting in 50% decrease in CFU/ml after 1 hr incubation with human complement, compared to time 0. The combinations contained 1∶1 mixtures (i.e, BC50% of 1 µg/ml contained 0.5 µg/ml of each of the mAbs in the combination). The anti-capsular mAb (SEAM 12)41 or anti-PorA mAbs (P1.7, panels a and b; or P1.2, panel c) served as positive controls. Error bars represent range of results from two to three independent assays.
JAR 41 augments passive protective activity of anti-fHbp mAb JAR 5 against experimental meningococcal bacteremia
Binding of human fH to the surface of meningococci down-regulates complement activation, which enables the organism to evade host defenses25,26,27. Binding is specific for human fH27. We recently developed a human fH transgenic infant rat model to investigate the effect of human fH on meningococcal bacteremia28. After challenge of wild-type rats, strain H44/76 was cleared rapidly from the blood stream whereas the presence of human fH in the transgenic animals enhanced bacteremia. In the present study we used the human fH transgenic rat model to investigate the ability of JAR 41 given individually or in combination with other non-bactericidal anti-fHbp mAbs to confer passive protection against bacteremia caused by challenge with wild-type group B strain H44/76.
Despite lack of bactericidal activity in vitro, pretreatment of human fH transgenic infant rats with 25 µg of JAR 5 conferred partial protection against bacteremia, as evidenced by a lower geometric mean CFU/ml in blood obtained 6 hrs after IP challenge, compared to that of control rats that received PBS (p<0.05, Experiments 1 and 2, Figure 6). In contrast, 25 µg/rat of JAR 41 did not confer significant protection in either experiment (p>0.05). To determine whether JAR 41 could augment JAR 5 protective activity, both antibodies (12.5 µg/rat of each mAb) were given in combination to groups of human fH transgenic infant rats. In both experiments, none of these animals had bacteremia 6 hrs after the IP challenge. In Experiment 2, in which we tested larger numbers of animals, the augmentation of protection by the combination of JAR 41 and JAR 5 was statistically significant (6 of 9 with bacteremia in the JAR 5 or JAR 41 alone groups, vs. 0 of 8 in the JAR 5+JAR 41 combination group, p = 0.009).
JAR 41 augments passive protective activity of anti-fHbp mAb JAR 5 against bacteremia caused by group B strain H44/76 in human fH transgenic infant rats.
Panel a, Experiment 1, 8- to 9-day old rats challenged IP with 4900 CFU/rat; Panel b, Experiment 2, 6- to 7-day old rats challenged IP with 760 CFU/rat. In both experiments, rats were given a total dose of 25 µg of mAb or phosphate buffered saline alone (PBS, negative control) 1 hr before the bacterial challenges. The combination contained 12.5 µg of each mAb. Blood cultures were obtained 6 hrs after challenge. Compared to PBS, rats given JAR 5 alone had a lower geometric mean CFU/ml (Experiment 1, p = 0.04 and Experiment 2, p<0.03)) but there was no protection by JAR 41 alone (p>0.05). In Experiment 2, the combination of JAR 41 and JAR 5 (0/8 with bacteremia) had greater protective activity than JAR 5 alone (6 out of 9 with bacteremia, p = 0.009, Fisher's exact test). ND, not done.
JAR 41 recognizes an epitope that overlaps that of anti-fHbp mAb JAR 4
While the combination of JAR 4 with JAR 5, or JAR 4 with mAb502, was bactericidal against group B strain H44/76, JAR 41 was not bactericidal in combination with JAR 4 (Figure 5a). One possible explanation was that JAR 4 and JAR 41 recognized overlapping epitopes. To investigate this question, we measured the ability of JAR 4 to inhibit binding of alkaline phosphatase (AP)-conjugated JAR 41 to fHbp (Supplemental Figure S1a). The positive control, unconjugated JAR 41, gave >95% inhibition of binding while the negative control, unconjugated JAR 5, gave <5% inhibition. At the highest concentration of mAb tested (100 µg/ml), JAR 4 gave ∼70% inhibition of binding of AP-JAR 41 to fHbp. We also measured the ability of unconjugated JAR 41 (IgG1) to inhibit binding of unconjugated JAR 4 (IgG2a) using an AP-conjugated secondary antibody that was specific for IgG2a (Supplemental Figure S1b). JAR 41 gave >95% inhibition of binding of JAR 4 to fHbp. The ability of JAR 41 and JAR 4 to inhibit binding of each other to fHbp implied that the respective epitopes overlapped.
The fHbp epitope recognized by JAR 41 is eliminated by substitution of Ala for Asp at residue 25 in fHbp
We used a mutant fHbp ID 1 library displayed on the surface of yeast to identify amino acid residues that potentially affect JAR 41 binding. The yeast library was sorted to select yeast clones that had lost their ability to bind JAR 41, but which retained their ability to bind JAR 3 (which verified surface-expression of the construct). Individual JAR 41-negative/JAR 3-positive yeast colonies were expanded and the fHbp gene inserts were sequenced. After eliminating sequences containing large deletions or multiple amino acid substitutions, we aligned the predicted amino acid sequences of 13 full-length mutant fHbp genes (Supplemental Figure S2). The mutations that abolished JAR 41 binding in the yeast library included three substitutions for Asp at position 25 (D25G, D25H, D25A), one substitution for Lys at position 27 (K27N) and one substitution for His at position 26 (H26N, which was present in combination with D25A). Substitutions at residue K27 and, to a lesser extent, D25A, corresponded to the region of the N-Terminal domain of fHbp previously shown to be important for binding of JAR 4 to fHbp29.
To confirm the importance of the individual residues on binding of JAR 41, we used site-directed mutagenesis to prepare mutant recombinant proteins with individual alanine substitutions at these positions. While the D25A mutation abolished binding of JAR 41 (Figure 7a, circles with dashed line), there was no significant effect of the K27A or H26A substitutions on JAR 41 binding to the recombinant mutant proteins. It is possible that the lack of an effect of K27A is explained by the different substituted residue (K27N in yeast, vs. K27A in the recombinant protein). The lack of an effect of H26A can be explained by the absence of the D25A substitution, since in yeast the mutant H26A fHbp that did not bind JAR 41 also contained D25A, which individually was sufficient to eliminate JAR 41 binding.
Effect of amino acid substitutions on binding of mAbs to recombinant fHbp.
 (Xs with solid line), wild-type fHbp ID 1;
(Xs with solid line), wild-type fHbp ID 1;  (Open squares with dotted line), H26A;
(Open squares with dotted line), H26A;  (Open triangles with dashed line), K27A;
(Open triangles with dashed line), K27A;  (Open circles with solid line), D25A. Panel a, JAR 41. Panel b, JAR 4. Panel c, JAR 1. Panel d, JAR 5.
(Open circles with solid line), D25A. Panel a, JAR 41. Panel b, JAR 4. Panel c, JAR 1. Panel d, JAR 5.
Disruption of JAR 41 binding with the D25A mutant was not a result of disruption of overall protein folding since the recombinant D25A mutant protein retained epitopes needed for binding with two control anti-fHbp mAbs, JAR 1 and JAR 5 (Figures 7c and 7d, respectively). JAR 1 recently was shown to recognize an epitope involving residue 20430 and JAR 5 recognized an epitope involving residues 121 and 12216 (Figure 7d). Interestingly, the D25A substitution also had minimal effect on binding of JAR 4 (Figure 7b), which has an epitope on the N-terminal domain29 that overlaps that of JAR 41.
Discussion
Binding of most previously described anti-fHbp mAbs was limited to sequence variants belonging to the same variant group or sub-family as that of the vaccine antigen15,16,31. To our knowledge only one anti-fHbp mAb (MN86-994-11) was reported to cross-react with fHbp variants from all three variant groups32. Insufficient details, however, were provided on this mAb to determine the actual breadth of reactivity, functional activity, or amino acid residues affecting the epitope.
In the present study, the ability of JAR 41 to bind equally well by ELISA to all 21 recombinant fHbp sequence variants tested and to bind by flow cytometry to the surface of live bacteria with fHbp from variant groups 1, 2, or 3 indicated that the epitope was nearly universal and was surface-exposed and accessible to antibody.
The fHbp molecule consists of two domains of antiparallel β-strands connected by a linker33,34. The epitope recognized by JAR 41 appears to be located on the N-terminal domain since a single amino acid substitution, Ala for Asp at residue 25 of fHbp ID 1 (D25A), eliminated JAR 41 binding. Among the 531 known amino acid sequence variants in the public fHbp peptide database (http://pubmlst.org/neisseria/fHbp/, as of December 2011), Asp at residue 25 was invariant. Further, the adjacent residues in this region of the N-terminal domain also are highly conserved.
We determined that the JAR 41 epitope overlapped that of a previously reported anti-fHbp mAb, JAR 4, which was specific for fHbp in variant groups 1 and 229. The molecular architecture of fHbp is modular with five variable segments, designated A, B, C, D and E, each of which is derived from one of two lineages13. Among the 21 fHbp sequence variants tested (Supplemental Figure S3), JAR 4 bound only to variants containing “A” segments derived from lineage 1 (previously referred to as alpha lineage13). In contrast, JAR 41 bound to all variants tested with A segments from lineage 1 or lineage 2 (previously referred to as beta lineage).
The residues of fHbp in contact with fH have been identified in a crystal structure of fHbp in a complex with a fragment of fH35. The fH contact residues were located on a surface opposite that containing Asp25, which affected the JAR 41 epitope and Asp25 and Lys27, which affected the JAR 4 epitope (See cartoon, Supplemental Figure S4). The respective locations were consistent with ELISA data showing that neither JAR 416,29 nor JAR 41 mAb inhibited binding of fH to fHbp (data not shown).
JAR 41 individually did not elicit human complement-mediated bactericidal activity, which is typical of most reported murine anti-fHbp mAbs15,16,24,29. fHbp is relatively sparsely distributed15 and it is possible that the distance between most of the fHbp molecules on the bacterial surface exceeds that required for optimal engagement of C1q by binding of individual mAbs. In contrast, binding of two IgG antibodies that recognize appropriately spaced non-overlapping epitopes may permit more efficient engagement of C1q and activation of classical complement pathway bacteriolysis15,16,24,29. Since binding of fH to N. meningitidis increases resistance of the organism to complement-mediated killing25,26, the ability of at least one of the anti-fHbp mAbs in the pair to inhibit fH can increase mAb bactericidal activity36.
While JAR 41 bound to the surface of strains H44/76, 8047 and M1239 with fHbp in variant groups 1, 2 or 3, respectively (Figure 4), the mAb elicited cooperative complement-mediated bactericidal activity only against strain H44/76 with fHbp in variant group 1 and an isogenic mutant, H44/76-LE with lower fHbp ID 1 expression (Figure 5). The amount of expression of fHbp variant group 1 by the mutant appeared to be similar to that of the two strains with fHbp in variant groups 2 or 3 that were resistant to anti-fHbp JAR 41 cooperative bactericidal activity. Most likely the resistance of these two test strains reflected testing bactericidal activity with different second anti-fHbp mAbs, which were specific for fHbp in variant groups 2 or 3, than those used to test the strains with fHbp variant group 1. Conceivably, different proximity and/or spatial orientations of JAR 41 bound in combination with JAR 5 or mAb502 in strain H44/76, as compared with JAR 11 or 13 in strain 8047, may have permitted more efficient engagement of C1q and activation of the classical complement pathway with strain H44/76.
In the present study, we used a recently described human fH transgenic infant rat model28 to investigate the ability of JAR 41, individually or in combination with JAR 5, to confer passive protection against meningococcal bacteremia caused by group B strain H44/76. We chose to use the transgenic infant rat model because binding of fH to N. meningitidis is specific for human fH27,28. Thus, measuring the ability of anti-fHbp mAbs to confer passive protection in the transgenic model likely provided information relevant to protection in humans where bound human fH can down-regulate complement activation on N. meningitidis.
Passive administration of JAR 5 conferred partial protection against meningococcal bacteremia in the transgenic rats (Figure 6). This protective activity was of interest since JAR 5 individually did not elicit human complement-mediated bactericidal activity (Figure 5) but in previous studies was shown to block binding of human fH to fHbp16. In contrast, JAR 41, which was not bactericidal and which did not block fH binding, was not protective against bacteremia in the transgenic rat model. JAR 41, however, augmented protective activity of JAR 5 (sterile blood cultures in all the rats given the combination). This result was consistent with the ability of JAR 41 to elicit cooperative complement-mediated bactericidal activity when tested with JAR 5.
In summary, JAR 41 is a new murine IgG1 mAb that recognizes a broadly cross-reactive, surface-exposed epitope on the N-terminal domain of fHbp from variant groups 1, 2 and 3. The mAb elicits cooperative human complement-mediated bactericidal activity in combination with other non-bactericidal anti-fHbp mAbs and augments passive protective activity against meningococcal bacteremia. The JAR 41 epitope overlapped that of JAR 4, which also had cooperative bactericidal activity but was specific for fHbp from variant groups 1 and 2. Collectively, the data demonstrate the importance of including cross-reactive epitopes in N-terminal domain of fHbp in the design of optimally protective fHbp vaccines.
Methods
Production of anti-fHbp mAb JAR 41
A human fH transgenic BALB/c mouse was immunized with three injections of a recombinant fHbp vaccine as part of a previous study17. The fHbp sequence variant was from variant group 1 and assigned ID 1 as designated in the public fHbp peptide database at http://pubmlst.org/neisseria/fHbp/. The mouse received three injections of the vaccine, IP, along with aluminum hydroxide adjuvant followed by a fourth injection without adjuvant. Splenocytes were isolated three days later for fusion to the mouse myeloma cell line P3X63-Ag8.653 using established methods15. Cells were plated into 96-well plates. After two weeks of incubation, cell culture supernatants were tested for reactivity to recombinant fHbp ID 1 by ELISA (see below). Cells from positive wells were expanded, cloned by limiting dilution and the supernatants confirmed for reactivity to fHbp. The cloning was repeated 2 times to ensure monoclonality.
Control anti-fHbp mAbs
The following murine anti-fHbp mAbs were used as controls in different binding and functional assays: JAR 1 (IgG3), JAR 3 (IgG3), JAR 4 (IgG2a), JAR 5 (IgG2b)15,16 and mAb502 (IgG2a)31,37 were from mice immunized with fHbp ID 1 in variant group 1; JAR 11 (IgG2a) and JAR 13 (IgG2a) were from a mouse immunized with fHbp ID 16 in variant group 216 and JAR 31 (IgG2b) (Unpublished data of DMG) and JAR 3516 were from a mouse immunized with fHbp ID 28 in variant group 3.
Purification of recombinant fHbp
We prepared 21 different recombinant fHbp amino acid sequence variants from variant groups 1, 2 or 3 (Table 1) as previously described30. The proteins were expressed from the T7 promotor using the E. coli plasmid pET21b (Novagen, Madison, WI) as described previously16. The recombinant proteins were purified by immobilized metal ion chromatography using Ni-NTA agarose (Qiagen, Valencia, CA) as described previously16. Purified fHbps were dialyzed against PBS, sterilized by filtration (Millex 0.22 µm; Millipore, Billerica, MA) and stored frozen at −30°C. The protein concentrations were determined by UV absorbance (Nanodrop 1000, Wilmington, DE) based on the extinction coefficient calculated from the amino acid sequence38.
Anti-fHbp ELISA
Binding of anti-fHbp mAbs to recombinant fHbp (rfHbp) was measured by ELISA, which was performed as described previously36 with minor modifications. In brief, 2 µg/ml of purified recombinant fHbp diluted in PBS was used to sensitize the microtiter plates. After blocking, serial dilutions of the mAbs were added and incubated at 4°C overnight. The wells were washed and bound antibody was detected with goat anti-mouse IgG conjugated with alkaline phosphatase. The ability of JAR 41 to inhibit binding of fH to fHbp was also measured by ELISA as previously described36.
Epitope mapping using yeast display
Randomly mutated sequences of fHbp allele 1 (http://pubmlst.org/neisseria/fHbp/), which encodes fHbp amino acid sequence variant ID 1, were generated by error-prone PCR. In brief, MnCl2 concentrations were titrated and conditions selected such that the average number of nucleotide substitutions per fHbp gene copy resulted in 1 to 3 amino acid substitutions. The fragments were ligated into pUC18, expanded in E. coli, excised, inserted into the yeast expression vector pYD1 (Invitrogen, Carlsbad, CA) and transfected into the yeast EBY100 strain (Invitrogen) to form the mutated fHbp expression library. The mutated fHbp peptides were displayed on the surface of yeast as Aga2 fusion proteins39,40. The library was expanded overnight at 30°C, transferred into galactose-containing YNB medium (Yeast Nitrogen Base w/o amino acids [Difco]) to induce recombinant protein expression and incubated for 48 hrs at 20°C. Bulk yeast cultures were simultaneously stained with anti-fHbp mAb, JAR 41, which was detected with FITC-conjugated antibody to mouse IgG and a chimeric human IgG1 mouse anti-fHbp mAb, JAR 336, which was detected with DyLight 649-conjugated antibody to human kappa light chain. JAR 3 and JAR 41 recognized non-overlapping epitopes as evident by lack of inhibition of binding by ELISA of either mAb to fHbp ID 1 by the other mAb (data not shown). The yeast cultures were sorted by flow cytometry (FACSaria, BD) to select yeast clones that had lost their ability to bind JAR 41, but which retained the ability to bind JAR 3 (which verified surface-expression of the construct). The sorted yeast were plated and incubated at 30°C to isolate single colonies. The individual clones were re-grown, their binding profiles verified by flow cytometry and the sequence of the respective fHbp insert determined to identify amino acid substitutions that had resulted in the loss of the JAR 41 epitope. Approximately 20 JAR 41-negative/JAR 3-positive yeast clones were analyzed and the sequences of their inserts aligned to identify altered residues that led to loss of JAR 41 binding.
Construction of site-specific mutants
Recombinant fHbp mutants with single amino acid substitutions were constructed using the QuikChange II kit (Stratagene, La Jolla, CA) using the manufacturer's protocols as previously described16. The mutagenesis reactions were transformed into chemically competent E. coli DH5α (Invitrogen, Carlsbad, CA) and independent mutant clones were verified by DNA sequencing (Davis Sequencing, Davis, CA).
Flow cytometry
Binding of the murine mAbs to the surface of live encapsulated bacteria was measured by flow cytometry, which was performed as described previously36. The group B test strains were H44/76 (B:15:P1.7,16; ST-32) with fHbp ID 1; a previously described isogenic mutant, which had been engineered to have decreased expression of fHbp ID 119; 8047 (B:2b:P1.5-1, 2-2; ST-8), which expressed fHbp ID 77 in variant group 2; and M1239 (B:14:P1.23,14; ST-41/44), which expressed fHbp ID 28 in variant group 3. The bacterial cells were grown in Mueller Hinton broth culture, harvested by centrifugation, washed and resuspended in buffer as described elsewhere36.
Serum bactericidal assay
Human complement-mediated bactericidal activity was measured as previously described17 using group B strains H44/76, 8047 and M1239 described above. The complement source was serum from a healthy adult who participated in a protocol that was approved by the Children's Hospital Oakland Institutional Review Board (IRB). Written informed consent was obtained from the subject. The serum had normal total hemolytic complement activity and no detectable serum bactericidal antibodies against the test strains. The serum was depleted of IgG using a protein G column (HiTrap Protein G, GE Life Sciences, Piscataway, NJ) as previously described17 to remove non-bactericidal antibodies that might augment anti-fHbp mAb activity4.
The bactericidal activity (BC50%) was defined by the mAb concentration that resulted in a 50% decrease in CFU/ml after 60-min incubation in the reaction mixture compared with CFU/ml in negative control wells at time zero.
Passive protective activity against bacteremia
Details of the human fH transgenic rat meningococcal bacteremia model have been recently described28. For measurement of passive protective activity, at time 0 human fH transgenic rats, ages 8 to 9 days (experiment 1) or 6 to 7 days (experiment 2), were administered 25 µg IP of anti-fHbp mAb JAR 5 or JAR 41, or a combination of JAR 5 + JAR 41 (12.5 µg of each). Control rats received buffer alone (phosphate buffered saline, PBS) or 12.5 µg of an anti-PorA mAb (P1.7, experiment 1 only). Total volume of mAb or buffer for each animal was 100 µl. At 1.5 hrs, animals were challenged IP with 4900 CFU (experiment 1) or 760 CFU (experiment 2) of group B strain H44/76. At 7.5 hrs, blood samples were obtained by cardiac puncture into syringes containing 25 U heparin. 1, 10 and 200 µl aliquots of each blood sample were spread onto chocolate agar plates, which were then incubated overnight at 37°C in 5% CO2. Colony counts were obtained the following day to determine CFU/ml. The lower limit of detection was 5 CFU/ml.
Statistical analyses
Statistical analyses were performed using Prism for Mac version 5.0d (GraphPad Software, La Jolla, CA). Comparisons of the proportions of rats that developed bacteremia after pre-treatment with different mAbs or PBS were performed using the Fisher exact test. For the purpose of calculating the geometric mean CFU/ml of blood, sterile blood cultures were assigned a value of half of the lower limit of detection (i.e., <5 CFU/ml was assigned a value of 2.5 CFU/ml). Comparisons of the geometric mean CFU of bacteria per ml of blood between any two groups of rats were performed on log-transformed values using the Student t-test. All probability values reported are two-tailed.
Live vertebrate experiments
All experiments in mice and infant rats were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, U.S.A. The protocols were approved by the Children's Hospital & Research Center at Oakland Institutional Animal Care and Use Committee. Blood collection was performed under anesthesia and all efforts were made to minimize suffering.
Change history
20 March 2013
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has not been fixed in the paper.
References
Granoff, D. M. Review of meningococcal group B vaccines. Clin. Infect. Dis. 50 Suppl 2, S54–65 (2010).
Sadarangani, M. & Pollard, A. J. Serogroup B meningococcal vaccines-an unfinished story. Lancet Infect. Dis. 10, 112–124 (2010).
Giuliani, M. M. et al. A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. U. S. A. 103, 10834–10839 (2006).
Vu, D. M., Wong, T. T. & Granoff, D. M. Cooperative serum bactericidal activity between human antibodies to meningococcal factor H binding protein and Neisserial heparin binding antigen. Vaccine. 29, 1968–1973 (2011).
Snape, M. D. et al. Immunogenicity of two investigational serogroup B meningococcal vaccines in the first year of life: a randomized comparative trial. Pediatr. Infect. Dis. J. 29, e71–79 (2010).
Findlow, J. et al. Multicenter, open-label, randomized phase II controlled trial of an investigational recombinant meningococcal serogroup B vaccine with and without outer membrane vesicles, administered in infancy. Clin. Infect. Dis. 51, 1127–1137 (2010).
Fletcher, L. D. et al. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect. Immun. 72, 2088–2100 (2004).
Frasch, C. E., Borrow, R. & Donnelly, J. Bactericidal antibody is the immunologic surrogate of protection against meningococcal disease. Vaccine. 27 Suppl 2, B112–116 (2009).
Borrow, R., Balmer, P. & Miller, E. Meningococcal surrogates of protection--serum bactericidal antibody activity. Vaccine. 23, 2222–2227 (2005).
Granoff, D. M. Relative importance of complement-mediated bactericidal and opsonic activity for protection against meningococcal disease. Vaccine. 27 Suppl 2, B117–125 (2009).
Murphy, E. et al. Sequence diversity of the factor H binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J. Infect. Dis. 200, 379–389 (2009).
Masignani, V. et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J. Exp. Med. 197, 789–799 (2003).
Beernink, P. T. & Granoff, D. M. The modular architecture of meningococcal factor H-binding protein. Microbiology. 155, 2873–2883 (2009).
Beernink, P. T. & Granoff, D. M. Bactericidal antibody responses induced by meningococcal recombinant chimeric factor H-binding protein vaccines. Infect. Immun. 76, 2568–2575 (2008).
Welsch, J. A., Rossi, R., Comanducci, M. & Granoff, D. M. Protective activity of monoclonal antibodies to genome-derived neisserial antigen 1870, a Neisseria meningitidis candidate vaccine. J. Immunol. 172, 5606–5615 (2004).
Beernink, P. T. et al. Fine antigenic specificity and cooperative bactericidal activity of monoclonal antibodies directed at the meningococcal vaccine candidate, factor H-binding protein. Infect. Immun. 76, 4232–4240 (2008).
Beernink, P. T. et al. A meningococcal factor H binding protein mutant that eliminates factor H binding enhances protective antibody responses to vaccination. J. Immunol. 186, 3606–3614 (2011).
Pajon, R., Beernink, P. T., Harrison, L. H. & Granoff, D. M. Frequency of factor H-binding protein modular groups and susceptibility to cross-reactive bactericidal activity in invasive meningococcal isolates. Vaccine. 28, 2122–2129 (2010).
Pajon, R., Fergus, A. M., Koeberling, O., Caugant, D. A. & Granoff, D. M. Meningococcal factor H binding proteins in epidemic strains from Africa: Implications for vaccine development. PLoS Negl. Trop. Dis. 5, e1302 (2011).
Donnelly, J. et al. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc. Natl. Acad. Sci. U. S. A. 107, 19490–19495 (2010).
Jiang, H. Q. et al. Broad vaccine coverage predicted for a bivalent recombinant factor H binding protein based vaccine to prevent serogroup B meningococcal disease. Vaccine. 28, 6086–6093 (2010).
Scarselli, M. et al. Rational design of a meningococcal antigen inducing broad protective immunity. Sci. Transl. Med. 3, 91ra62 (2011).
Huson, D. H. & Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006).
Welsch, J. A., Ram, S., Koeberling, O. & Granoff, D. M. Complement-dependent synergistic bactericidal activity of antibodies against factor H-binding protein, a sparsely distributed meningococcal vaccine antigen. J. Infect. Dis. 197, 1053–1061 (2008).
Madico, G. et al. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J. Immunol. 177, 501–510 (2006).
Schneider, M. C. et al. Functional significance of factor H binding to Neisseria meningitidis. J. Immunol. 176, 7566–7575 (2006).
Granoff, D. M., Welsch, J. A. & Ram, S. Binding of complement factor H (fH) to Neisseria meningitidis is specific for human fH and inhibits complement activation by rat and rabbit sera. Infect. Immun. 77, 764–769 (2009).
Vu, D. M. et al. Enhanced bacteremia in human factor H transgenic rats infected by Neisseria meningitidis. Infect. Immun. 80, 643–650 (2012).
Beernink, P. T., Lopasso, C., Angiolillo, A., Felici, F. & Granoff, D. A region of the N-terminal domain of meningococcal factor H-binding protein that elicits bactericidal antibody across antigenic variant groups. Mol. Immunol. 46, 1647–1653 (2009).
Giuntini, S., Beernink, P. T., Reason, D. C. & Granoff, D. M. Monoclonal antibodies to meningococcal factor H binding protein with overlapping epitopes and discordant functional activity. PloS One, In press (2012).
Giuliani, M. M. et al. The region comprising amino acids 100 to 255 of Neisseria meningitidis lipoprotein GNA 1870 elicits bactericidal antibodies. Infect. Immun. 73, 1151–1160 (2005).
McNeil, L. K. et al. Detection of LP2086 on the cell surface of Neisseria meningitidis and its accessibility in the presence of serogroup B capsular polysaccharide. Vaccine. 27, 3417–3421 (2009).
Cantini, F. et al. Solution structure of the factor H-binding protein, a survival factor and protective antigen of Neisseria meningitidis. J. Biol. Chem. 284, 9022–9026 (2009).
Mascioni, A. et al. Structural basis for the immunogenic properties of the meningococcal vaccine candidate LP2086. J. Biol. Chem. 284, 8738–8746 (2009).
Schneider, M. C. et al. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. .Nature 458, 890–893 (2009).
Giuntini, S., Reason, D. C. & Granoff, D. M. Complement-mediated bactericidal activity of anti-factor H binding protein monoclonal antibodies against the meningococcus relies upon blocking factor H binding. Infect. Immun. 79, 3751–3759 (2011).
Scarselli, M. et al. Epitope mapping of a bactericidal monoclonal antibody against the factor H binding protein of Neisseria meningitidis. J. Mol. Biol. 386, 97–108 (2009).
Gasteiger, J. Chemoinformatics: a new field with a long tradition. Anal. Bioanal. Chem. 384, 57–64 (2006).
Chao, G., Cochran, J. R. & Wittrup, K. D. Fine epitope mapping of anti-epidermal growth factor receptor antibodies through random mutagenesis and yeast surface display. J. Mol. Biol. 342, 539–550 (2004).
Boder, E. T. & Wittrup, K. D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997).
Granoff, D. M. et al. Bactericidal monoclonal antibodies that define unique meningococcal B polysaccharide epitopes that do not cross-react with human polysialic acid. J. Immunol. 160, 5028–5036 (1998).
Beernink, P. T. et al. Prevalence of factor H-binding protein variants and NadA among meningococcal group B isolates from the United States: implications for the development of a multicomponent group B vaccine. J. Infect. Dis. 195, 1472–1479 (2007).
Acknowledgements
This work was supported in part by Public Health Service grants R01 AI 046464 and AI 082263 from the National Institute of Allergy and Infectious Diseases, NIH. D.M.G. also is supported in part by an endowment established by the Clorox Company. The laboratory work at Children's Hospital Oakland Research Institute was performed in a facility funded by Research Facilities Improvement Program grant C06 RR 016226 from the National Center for Research Resources, NIH. We are grateful to Denise Playdle Green for expert technical assistance.
Author information
Authors and Affiliations
Contributions
R.P. performed the phylogenic analyses of fHbp sequence variants and prepared Figure 2 and Supplementary Figure 4. D.C.R. sorted the fHbp ID 1 yeast display mutants and assisted with mapping the JAR 41 epitope. D.M.V. and D.M.G. designed the study, wrote the main manuscript text and prepared the remaining figures and table 1. All authors reviewed and contributed to writing of the manuscript.
Ethics declarations
Competing interests
D.M.G. holds a paid consultancy with Novartis Vaccines and Diagnostics. D.M.G. R.P. and D.M.V. are inventors on patents or patent applications in the area of meningococcal B vaccines. D.C.R. report no conflicts of interest.
Electronic supplementary material
Supplementary Information
Supplementary Figures S1 to S4
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Vu, D., Pajon, R., Reason, D. et al. A Broadly Cross-Reactive Monoclonal Antibody Against an Epitope on the N-terminus of Meningococcal fHbp. Sci Rep 2, 341 (2012). https://doi.org/10.1038/srep00341
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00341
This article is cited by
-
A homopolymeric adenosine tract in the promoter region of nspA influences factor H-mediated serum resistance in Neisseria meningitidis
Scientific Reports (2019)
-
Structural basis for cooperativity of human monoclonal antibodies to meningococcal factor H-binding protein
Communications Biology (2019)
-
Crystal structure reveals vaccine elicited bactericidal human antibody targeting a conserved epitope on meningococcal fHbp
Nature Communications (2018)
-
Human protective response induced by meningococcus B vaccine is mediated by the synergy of multiple bactericidal epitopes
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.