Abstract
Gene expression quantification on cultured cells using the reverse transcription quantitative polymerase chain reaction (RT-qPCR) typically involves an RNA purification step that limits sample processing throughput and precludes parallel analysis of large numbers of samples. An approach in which cDNA synthesis is carried out on crude cell lysates instead of on purified RNA samples can offer a fast and straightforward alternative. Here, we evaluate such an approach, benchmarking Ambion's Cells-to-CT kit with the classic workflow of RNA purification and cDNA synthesis and demonstrate its good accuracy and superior sensitivity.
Similar content being viewed by others
Introduction
Gene expression quantification on cultured cells using the reverse transcription quantitative polymerase chain reaction (RT-qPCR) is an important tool in molecular biological and biomedical research. In vitro functional analysis through gene expression measurements can serve as a readout for qualitative or quantitative assessment of the effect of diverse treatments such as RNAi and drug compound administration. Going from cell cultures to gene expression results typically involves harvesting of cells, isolation of RNA, removal of co-purified contaminating DNA through DNase treatment, cDNA synthesis and finally qPCR. Cell harvesting and RNA isolation are the rate limiting steps in this workflow. The limited throughput of harvesting procedures and classical RNA extraction methods -either phenol/chloroform based RNA extraction and ethanol precipitation or column based solid phase extraction- characterized by time consuming and laborious protocols, preclude the inclusion of large numbers of samples in a single cell culture experiment. However, the demand for high-throughput gene expression analyses on cell cultures is dramatically increasing, amongst others by application of genome wide RNAi and compound library screening.
To accommodate this need, an elegant method was introduced for high-throughput synthesis of cDNA to be used for qPCR gene expression analyses of 96- and 384-well cell culture samples, excluding the need for cell harvesting and RNA purification. The method uses crude cell lysates as input for reverse transcription, significantly increasing processing speed and throughput and offering possibilities towards automation. Different kits, such as the Cells-to-CT kit (Ambion), the RealTime ready Cell Lysis kit (Roche) and the CellsDirect kit (Invitrogen), are available on the market, all of them compatible with both DNA binding dye or hydrolysis probe qPCR detection chemistry. Apart from a handful reports on the sensitivity, specificity and compatibility with automation1,2, little is known about the performance in terms of accuracy of this new method. Here, we performed an extensive characterization of the workflow by evaluating the Cells-to-CT kit and benchmarked it to the hitherto gold standard workflow of RNA purification and cDNA synthesis. We demonstrate superior sensitivity, good accuracy and effective DNase treatment when using crude cell lysates. Both low and high-throughput RT-qPCR experiments on cells grown in microtiter plates may substantially benefit from this new workflow.
Results
Accuracy
First, the ability of the Cells-to-CT workflow to accurately quantify relative gene expression levels and resulting fold changes between samples was assessed. To this end, cDNA from duplicate cultures of four neuroblastoma cell lines, two bearing amplification of the MYCN transcription factor gene (NGP and IMR-32) and two without the amplification (SH-EP, SK-N-AS), was prepared using either the classic or the Cells-to-CT workflow. Relative expression levels of 10 genes of interest (DKK3, INHBA, PLAT, RGS4, MYC, MTHFD2, MYCN, TGFBI, PMP22, NTRK2) known to be differentially expressed between cells with and without MYCN-amplification3 were then quantified, yielding a theoretical number of 80 expression level data points for both workflows. The fraction of missing data was low and comparable for both methods and occurred for genes with very low expression, as expected. For the classic and the Cells-to-CT workflow, respectively 78 and 76 relative expression levels could be effectively determined. Fold changes were calculated among all cell lines (28 paired comparisons for 4 cell lines in duplicate). Theoretically 280 fold changes could be calculated and when taking into account the missing data, 266 and 254 fold changes could be effectively calculated for the classic and the Cells-to-CT workflow, respectively. The assessment of four different cell lines and ten genes results in a wide coverage of fold changes, ranging from no change to more than a thousand fold change. Relative expression levels and fold changes were log2 transformed for further data-analysis.
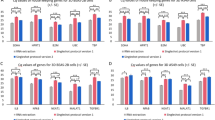
High and significant correlations could be observed between Cq values (Fig. 1b, Pearson r = 98%; Spearman r = 97%), log2 normalized relative quantities (Fig. 1a,b & suppl. Fig. 1, Pearson r = 96%, Spearman r = 98%) and log2 fold changes (Fig. 1b, Pearson r = 99%, Spearman r = 98%) obtained with both methods. A cumulative distribution plot of the difference in log2 fold change determined with both methods (Δlog2 fold change) shows that this difference is smaller than 0.5 for more than 63%, smaller than 1 for more than 83% and smaller than 1.5 for more than 93% of the fold changes observed (Fig. 2a). Similar conclusions can be drawn from a Bland-Altman plot for the log2 fold changes (Fig. 2b), with a 95% confidence interval of the true difference that ranges from 1.57 to −1.76. Bland-Altman plots are often regarded as better measures to assess the agreement of two methods than simple correlation analyses.
Accuracy.
(a) Log2 normalized relative quantities quantified with either the classic workflow (black) or the Cells-to-CT workflow (grey) for 6 genes (DKK3, INHBA, PLAT, RGS4, MYC, MTHFD2). (b) Correlation between Cq values, log2 normalized relative quantities and log2 fold changes quantified with either the classic or the Cells-to-CT workflow.
Accuracy.
(a) Cumulative distribution of Δlog2 fold changes. (b) Bland-Altman plot for log2 fold changes. The difference in log2 fold change measured with both methods is plotted against the average of these log2 fold changes. The 95% confidence interval of the true difference is delineated by the upper (ULA) and lower limit of agreement (ULA). (c) Enrichment analysis. Each data point in the cumulative distribution plot of Δlog2 fold changes results from two log2 fold changes, each calculated from two log2 relative quantities that both in turn result from five Cq values (one gene of interest and four reference genes). Data points in purple are data points for which at least 1 of the 20 Cq values giving rise to this data point is higher than the specified cutoff.
Log2 fold changes that showed suboptimal concordance, here defined as having a delta log2 fold change greater than 1, were significantly enriched in fold changes calculated from high Cq values (p-value Fisher's Exact Test < 0.001; Fig. 2c), in high log2 fold changes (suppl. Table 1) and in log2 fold changes for specific qPCR assays (suppl. Table 2). High Cq value was the only independent predictor variable of suboptimal concordance in this data set.
Sensitivity and linearity
In order to compare the sensitivity of working with cell lysates versus working with purified RNA as input for reverse transcription, a cell dilution series of the neuroblastoma cell line SH-EP (20,000, 10,000, 5,000 and 2,500 cells) with duplicate samples was either subjected to classic RNA isolation immediately after dilution series preparation or seeded in 96-well cell culture plates and lysed with Cells-to-CT reagents after 6 h of adherence. An approach like this, with immediate RNA isolation and quasi immediate Cells-to-CT cell lysis, guarantees minimal differences in cell input to both methods, as adherence time is below the doubling time of SH-EP cells and at the same time meets the requirement to have completely adhered cells in cell culture plates for Cells-to-CT lysate preparation.
As raw Cq values, rather than normalized relative expression levels, are the subject of comparison here whereby no normalization occurs, ideally reverse transcription on both RNA samples and cell lysates is performed in the same run and with the same RT procedure. Therefore, the Cells-to-CT reverse transcription kit, which can cope with both types of input, was used on both purified RNA as well as cell lysates. To normalize the cDNA input for the qPCR reactions to the input cell number used for RNA isolation or Cells-to-CT lysis, equivalent fractions of either total RNA isolate or cell lysate (18% of total volume) were used as input for reverse transcription. qPCR was performed for four genes (SDHA, HPRT1, GAPDH, UBC). To avoid the introduction of inter-run variation, all qPCR reactions were performed in the same run using the same mastermix and PCR conditions (Eurogentec protocol).
For every point in the cell dilution series and for every gene tested the Cq values were on average 4 cycles earlier when using cell lysates (Fig. 3), clearly illustrating the superior sensitivity over working with purified RNA fractions. Furthermore, just as for classic RNA isolation, within a range of cell numbers frequently used for seeding in 96-well cell culture experiments, cDNA yields were linear.
DNase treatment
To evaluate the performance of DNase treatment on crude cell lysates, single replicate Cells-to-CT lysates were prepared both in the presence and the absence of DNase I in the lysis buffer for four neuroblastoma cell lines (SH-EP, SK-N-AS, NGP, IMR-32). The crude cell lysates were directly used as input for qPCR using four DNA specific assays (NEUROD1, XRCC3, PLAT, MTHFD2).
DNase treatment appeared to be efficient. A complete or almost complete eradication of the DNA specific signal could be observed with all assays in all cell lines (Fig. 4).
Discussion
Cell harvesting and RNA purification are the rate limiting steps in going from cell cultures to cDNA and thus eventually to gene expression levels and fold changes using RT-qPCR. An alternative method for cDNA synthesis that eliminates these steps and uses crude cell lysates instead of purified RNA as an input for reverse transcription could offer a fast and straightforward alternative, significantly increasing sample processing throughput. Here, we show for the first time the reliability and accuracy of this approach by extensively evaluating one of the commercially available kits. Other kits on the market should undergo the same level of performance assessment prior to their routine use, following the testing framework presented here.
We clearly observed high concordance between RT-qPCR gene expression results coming from cell lysates or purified RNA as input for the reverse transcription and this over a wide range of expression differences measured for various genes in multiple cell lines. For normalized relative expression levels and fold changes, both the correlation and the agreement of values was high, demonstrating that both methods could be used interchangeably with great confidence. Due to a combination of a difference in sensitivity and in input amounts into qPCR reactions between both workflows for cDNA synthesis, raw Cq values did not show a high agreement. Rather, there was a constant and systematic bias that could be eliminated through normalization. In this data set, only 17% of the fold changes would have been differentially called by more than a factor 2 (i.e. a log2 fold difference of 1) when applying one method or the other. However, as this 17% almost exclusively consists of large fold changes, this does not pose a serious problem, as the exact magnitude of the fold change is often of less importance. In addition, the 17% suboptimally correlating results are also highly enriched in log2 fold changes resulting from high Cq values. Inherent to qPCR, the variation increases with higher Cq values and the resulting fold changes are thus less accurate and precise. Hence, the discrepancies in log2 fold changes observed are most likely not related to the inaccuracy of one method or the other, but due to this phenomenon inherent to qPCR. It should be noted that the level of agreement between both methods observed is in fact an underestimation of their true concordance, as the nature of the techniques makes it impossible to execute both workflows on the same cell culture sample and thus correlated data obtained here originate from cell culture replicates cultured in different plate formats (i.e. 6-well for the classic workflow using purified RNA and 96-well plates for the Cells-to-CT workflow).
Next to a good accuracy, a higher sensitivity for the lysate approach compared to the classic workflow could be demonstrated. This may be explained by the fact that fewer steps and no purification are required when working with cell lysates, meaning there is less chance to introduce bias in a given sample's mRNA content.
Finally, we positively evaluated the performance of DNase treatment on crude cell lysates, with either a complete or an almost complete removal of the contaminating genomic DNA. Incomplete digestion of DNA only poses a problem for lowly expressed genes, as the remaining DNA specific signal may contribute relatively more to the total signal. Longer DNase I incubation times than the ones proposed by the manufacturer may further improve the efficiency of the digestion. Of note, DNase digestion on purified RNA is also often incomplete4, so it is probably not related to the complex environment of a crude cell lysate. Incomplete DNase digestion potentially also partially explains the discrepancies observed for log2 fold changes involving high Cq values and thus lowly expressed genes.
Apart from the improvements in terms of speed, throughput and ease of sample handling, another main advantage of working with cell lysates is its better compatibility with the smaller cell culture plate formats (i.e. 96-well and even 384-well format). Together with the higher sensitivity this enables substantial downscaling of reactions, hereby lowering reagent usage and overall experiment cost. In addition, together with the fact that cell harvesting is not required anymore, it allows total automation of cDNA synthesis from cell cultures1. Classic phenol/chloroform extraction is not amendable to automation. Other conventional methods for RNA isolation have been adapted to high-throughput platforms. Examples are the use of glass fiber filter plates where RNA binds to the filter in the presence of high salt and alcohol concentrations or the use of magnetic microspheric beads to selectively bind nucleic acids. However, these methods still require a great deal of user interaction.
In conclusion, we here provide solid evidence for the accuracy of RT-qPCR gene expression quantification using cell lysates as an input for reverse transcription, along with a confirmation of the superior sensitivity of this approach.
Methods
The procedures in this section describe both a classic workflow (RNA purification) and the Cells-to-CT workflow for gene expression quantification. Specific adaptations made to both workflows to test particular aspects are described where relevant in the Results section.
Cell culture
Neuroblastoma cell lines (SH-EP, SK-N-AS, NGP, IMR-32) were cultured in RPMI-1640 (Invitrogen) supplemented with fetal calf serum (10%), L-Glutamine (1%), penicillin/streptomycin (1%), kanamycine (1%) and HEPES (25 mM). To synthesize cDNA according to the classic workflow, cells were seeded in 6-well culture plates at a density of 250,000 cells/well. Alternatively, to synthesize cDNA using the Cells-to-CT kit, cells were seeded in 96-well culture plates at a density of 10,000 cells/well. cDNA synthesis was carried out 48 h after cell seeding.
cDNA synthesis
The classic workflow for cDNA synthesis consists of cell harvesting by scraping, phenol/guanidine thiocyanate-chloroform based lysis of cells in combination with silica membrane based RNA extraction using the miRNeasy Mini Kit (Qiagen), on-column DNase I digestion using the RNase-Free DNase Set (Qiagen), followed by oligo(dT)/random primer mediated reverse transcription using the iScript cDNA Synthesis Kit (Bio-Rad) with 500 ng of RNA as input. Alternatively, the Cells-to-CT kit (Ambion) was used to produce DNase I digested cell lysates and perform cDNA synthesis. All kits were used according to manufacturer's instructions.
RT-qPCR
RT-qPCR gene expression quantifications were performed and reported according MIQE guidelines5. All reactions were performed in 384-well plates using the LightCycler 480 instrument (Roche). Reactions with classic cDNA as input were performed in a total volume of 8 µl, comprising 4 µl 2x SYBR Green I Master Mix (Eurogentec), 2.5 ng cDNA (total RNA equivalents) and 235 nM of each primer (final concentration). For gene expression analysis of the Cells-to-CT cDNA, a downscaled version of the qPCR protocol supplied with the Cells-to-CT kit was used, with reactions performed in 10 µl, comprising 5 µl Power SYBR Green Master Mix supplied with the kit, 1/50th of the cDNA sample and 300 nM of each primer (final concentration).
All qPCR reactions were performed in duplicate and Cq values were averaged. Liquid handling was done using a pipetting robot (TECAN Evo 100). All qPCR assays (suppl. Table 3 & 4) were extensively validated in silico using the RTPrimerDB primer evaluation pipeline6 and empirically validated, checking both primer efficiency and specificity. Calculation of normalized relative expression levels was done using the qbasePLUS software version 1.57 (Biogazelle). Normalization was performed using four stably expressed neuroblastoma reference genes (AluSq, HPRT1, TBP and YWHAZ) validated using the genormPLUS 8module in qbasePLUS.
Statistics
All statistical analyses were performed using the R statistical programming environment (version 2.10.1).
References
Abruzzese, R., Fekete, R. & Kephart, D. Cells-to-CT Kits: Next Generation Gene Expression Analysis Workflows that Eliminate Sample Purification. Journal of the Association for Laboratory Automation 15, 362–368 (2010).
Tavares, R. L. C., Xu, K., Zhang, C. & Freitas, V. A specific and quick gene expression study in mouse ES cells. J Assist Reprod Genet 24, 366–372 (2007).
Vermeulen, J. et al. RNA pre-amplification enables large-scale RT-qPCR gene-expression studies on limiting sample amounts. BMC Res Notes 2, 235 (2009).
Tavares, L., Alves, P. M., Ferreira, R. B. & Santos, C. N. Comparison of different methods for DNA-free RNA isolation from SK-N-MC neuroblastoma. BMC Res Notes 1, 140 (2011).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55, 611–622 (2009).
Lefever, S., Vandesompele, J., Speleman, F. & Pattyn, F. RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res 37, D942–D945 (2009).
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F. & Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8, R19 (2007).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, RESEARCH0034 (2002).
Author information
Authors and Affiliations
Contributions
G.V.P. executed the experiments, performed the analyses and drafted the manuscript. P.M. and J.V. supervised the study, revised the manuscript and approved the final version of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Van Peer, G., Mestdagh, P. & Vandesompele, J. Accurate RT-qPCR gene expression analysis on cell culture lysates. Sci Rep 2, 222 (2012). https://doi.org/10.1038/srep00222
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00222
This article is cited by
-
Direct lysis of 3D cell cultures for RT-qPCR gene expression quantification
Scientific Reports (2023)
-
Genes expressed at low levels raise false discovery rates in RNA samples contaminated with genomic DNA
BMC Genomics (2022)
-
Methanandamide diminish the Porphyromonas gingivalis lipopolysaccharide induced response in human periodontal ligament cells
BMC Oral Health (2020)
-
Integrated morphologic and molecular analysis of Trichomonas vaginalis, Mycoplasma hominis, and human papillomavirus using cytologic smear preparations
Parasitology Research (2018)
-
Response of human periodontal ligament stem cells to IFN-γ and TLR-agonists
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.