Abstract
Influenza A virus (IAV) is an RNA virus that causes respiratory disease in a wide range of mammals including humans and pigs. Cav1.2 is a specific voltage-dependent calcium channel (VDCC) important for the internalization of IAV and VDCC inhibitors can decrease IAV disease severity in mice. In this paper, the distribution pattern of a range of VDCCs by immunohistochemistry and Cav1.2 by in situ hybridization in the porcine respiratory tract is documented for the first time. Furthermore, we showed co-localization of VDCC-positive and IAV-positive cells in experimentally infected pigs. These findings provide new perspectives on the IAV internalization process and pave the way for further research investigating the effect of VDDC inhibitors on the IAV infection dynamics in pigs, which could have relevance to humans too.
Similar content being viewed by others
Introduction
Influenza A virus (IAV) is a negative sense RNA virus that causes respiratory disease in humans and several animal species including pigs. The main IAV receptors are sialic acids (SA) that are linked to glycoproteins in either an alpha 2,3- (α2,3) or alpha 2,6-linkage (α2,6). IAVs isolated from humans and pigs prefer binding to SA-α2,6 while IAVs isolated from birds prefer binding to SA-α2,31,2,3. Most IAVs seem to be species-specific and although humans and pigs have similar receptor distribution in their respiratory tract4,5, it is only some IAV strains that can be transmitted between pigs and humans6. This indicates that other factors are important for effective replication in a new host species. Such factors could be physical factors such as differences in temperatures, pH values, and host proteases in the respiratory tract7,8,9. Sporadic interspecies transmission of swine IAVs to humans most often results in dead-end transmissions, however, the most recent IAV pandemic (H1N1pdm09) occurred due to reassortment events in pigs10,11. The H1N1pdm09 became established in both pigs and humans and it has since then increased the diversity of swine IAVs dramatically, which has complicated the surveillance of IAVs6,12,13. Hence, a complete understanding of the swine and human IAV pathogenesis, to identify possible host factors that allow interspecies transmission, is needed to identify potential zoonotic swine IAVs.
Calcium channels have recently been shown to be important for the internalization of IAV and other human respiratory viruses, such as respiratory syncytial virus, measles, and rhinovirus14. Additionally, the presence of extracellular calcium is important for the internalization and replication of IAV, which has been demonstrated by adding ethylene glycol tetraacetic acid (EGTA, Ca2+ chelator)15,16 and by using specific voltage-dependent calcium channel (VDCC) blockers17,18. VDCCs consist of three families Cav1 (L-type), Cav2 (P/Q, N, and R-types), and Cav3 (T-type). Common to all families of VDCCs is that stimulation by a voltage signal results in an extracellular calcium influx and they consist of an α1 subunit, which is the pore-forming unit of the VDCCs19. One of the L-type VDCC α1 subunits is Cav1.2 and it contains four N-glycosylations sites, which can attach terminal SA20. Furthermore, it has been shown that hemagglutinin, the surface protein important for the attachment of IAV to cells, binds to Cav1.2. Interestingly, this binding was significantly reduced by a sialidase pre-treatment, but not by adding EGTA, indicating that the sialic acids of Cav1.2 are important for viral attachment18. Moreover, Fujioka et al.18 showed that a knockdown of Cav1.2 significantly reduced IAV internalization. Overall, the previous studies strongly suggest that Cav1.2 is an important factor in the internalization of IAV. Additionally, treatment of mice with Diltiazem, a specific Cav1.2 inhibitor, significantly decreased the disease severity of IAV18,21. FDA has approved Diltiazem for the treatment of hypertension and coronary artery spasm22 and, therefore, a phase II clinical trial on Diltiazem as a treatment for severe IAV was already initiated in 201723, however, the results of these trials are still pending.
Cav1.2 has been detected by in situ methods in the lower respiratory tract in fetal lungs of humans and mice24, in adult cardiomyocytes of mice25 and rats26, and in adult murine brain tissues21. Furthermore, the Cav1 and Cav2 families of VDCCs have been recognized by CavPan α1-antibodies in the adult murine lung by immunohistochemistry (IHC)14. In the present report, we document for the first time, the presence of Cav1 and Cav2 families by CavPan α1-antibodies and specifically Cav1.2 by in situ hybridization in the respiratory tract of pigs. These findings provide new perspectives on the swine IAV internalization process and open up for further investigation on the effect of VDCC-blockers as a treatment for IAV in pigs, which potentially could be translatable to humans.
Methods
Ethics declaration
The porcine and murine tissues originated from two experimental studies27,28 approved by the Danish Animal Experimentation Council (protocol no. 2020-15-0201-00502 and no. 2012-15-2934-00256, respectively).
VDCC and IAV IHC
Immunohistochemically, CavPan α1-antibodies stain the α1 genes from the Cav1 and Cav2 families and are therefore considered a pan-reactive VDCC reagent. The amino acid sequence used to produce the CavPan α1-antibody corresponds to amino acids 1506 to 1524 of the rat Cav1.2 and was compared to the corresponding amino acid sequences of Cav1.2 from pig, mouse, human, guinea pig, ferret, chicken, and duck origin.
Heart, nose, trachea, and lung tissues from six to seven-week-old pigs (N = 3, one control pig and two IAV-inoculated pigs) were used for the examination of VDCC immunohistochemically, together with brain tissue from one adult mouse, which was used as a positive control. The formalin-fixed tissues were embedded in paraffin wax and sliced into 2–3 µm sections. The sections were deparaffinized and after each of the following steps, the sections were washed two times for 5 min in tris-buffered saline (TBS). The sections were blocked by endogen peroxidase 0.6% (CAS: 7722-84-1, VWR International, Søborg, Denmark), pre-treated with 0.1% trypsin, and blocked with ultra v-block (TL-125-HLJ, Thermo Scientific, Waltham, Massachusetts, USA). CavPan α1-antibody (ACC-004, Almone Labs, Jerusalem, Israel) diluted 1:2000 was added to the sections overnight at 4 °C. The method of detection was Ultravision ONE HRP polymer reagent (TL-125-HLJ, Thermo Scientific) which was added to the sections for 30 min. A pre-adsorption control was performed as described above, but by mixing the CavPan α1 blocking peptide (BLP-CC004, Almone Labs) and the CavPan α1-antibody. An isotype control (IgG) (X0903, Agilent Technologies, Santa Clara, California, USA), diluted to the same content of protein as the primary reagent in 1% bovine serum albumin (BSA)/TBS, instead of CavPan α1-antibody was also performed. The staining was developed by adding 3,3′-Diaminobenzidine (DAB) for 6 min, and sections were counterstained by Mayer’s hematoxylin (AMPQ00254.5000, VWR International, Radnor, Pennsylvania, USA) and mounted with glycerol-gelatine.
At least two representative images of each of the tissues and lung compartments (bronchi, bronchioles, and alveoli) were acquired. Lamina epithelialis in the nose, trachea, bronchi, bronchioles, and alveoli was manually selected as a region of interest (ROI) in ImageJ29. The percentage of pixels above the threshold in the selected area (% area) was calculated by using the plugin “Color Deconvolution2” and by setting a threshold of 126 for hematoxylin and 120 for DAB30. The % area of CavPan α1 DAB was normalized to hematoxylin (% area DAB/% area hematoxylin) and the % area of isotype DAB normalized to hematoxylin was subtracted.
Parallel sections of two IAV-infected pigs, inoculated with an IAV circulating in pigs (swine-adapted) and an IAV circulating in humans (human-adapted), were stained for IAV in the nose, trachea, and lung tissues. The sections were deparaffinized and blocked for endogenous peroxidase as described above and hereafter pretreated with 0.018 g proteinase (P8038, Sigma-Aldrich, Missouri, USA) diluted in 100 ml TBS for 5 min. Additionally, the sections were blocked for 5 min by Ultra V Block (TA-125-UB, Epredia, Michigan, United States) and anti-IAV (nucleoprotein (NP)) antibodies (HYB 340-05, SSI-antibodies, Copenhagen S, Denmark) diluted 1:50000 in 1% BSA/TBS were added to the sections overnight at 4 °C. A Primary Antibody Enhancer (TL-125-PB, Epredia) was added to the sections for 20 min followed by a Large Volume HRP Polymer (TL-125-PH, Epredia) for 30 min. The staining was developed by adding AEC vector (SK-4200, Vector Laboratories, California, USA) for 10 min and counterstaining by Mayer’s hematoxylin (AMPQ00254.5000, VWR International). The sections were mounted with glycerol-gelatine.
Cav.1.2 Reverse transcriptase-qPCR
The presence of Cav1.2, in frozen lung tissue, obtained from three pigs that had tested negative for IAV, and in HEK 293 T cells was investigated. The HEK 293 T cells were used as a positive control for the reverse transcriptase-qPCR (RT-qPCR) assay since the Cav1.2 channels previously have been documented in these cells18. Briefly, 200 µl of HEK 293 T cells with Eagle’s Minimum Essential Medium (MEM) (Gibco) were collected and treated as previously described for nasal swabs31. RLT-buffer (QIAGEN, Hilden, Germany) which contained 2-mercaptoethanol (Merck, Darmstadt, Germany) was added to 70 mg frozen lung tissues, lysed in TissueLyser LT (QIAGEN) at 30 Hz for 3 min and centrifuged at 9651 × g for 3 min. The RNA extraction was performed as described previously31. RT-qPCR was performed by using Qiagen one-step RT-PCR kit (QIAGEN, Hilden, Germany) with primers for Cav1.2 developed by ref. 18. The PCR cycles were as follows: 50 °C for 40 min, 95 °C for 15 min, and 45 cycles of (94 °C 40 s, 60 °C 40 s, 72 °C 40 s) and 72 °C for 10 min. The amplified Cav1.2 was detected by SYBR green.
Cav1.2 Sanger sequencing
The RT-qPCR product was purified with a High Pure PCR product Purification kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer’s protocol. PCR products of one of the pig lung tissues together with primers (the same as used for the RT-qPCR) were sent for Sanger sequencing at LGC Biosearch Technologies (Berlin, Germany). The resulting sequences were imported into the program CLC Main Workbench version 22.0 (QIAGEN, Hilden, Germany) and trimmed manually. The forward and reverse reads were assembled and the consensus sequence was checked for identity to publically available sequences using the tool BLAST against NCBI Genbank. The sequence was uploaded to NCBI Genbank with the accession number OQ973297.
In situ hybridization
The consensus sequences were used to create probes for the in situ hybridization to investigate the Cav1.2 mRNA expression in porcine lung tissues (N = 1). The probe sequences correspond to nucleotides 3532–3625 (between exon 27-exon 28) on the sus scrofa calcium voltage-gated channel subunit alpha1 C/Cav1.2 (CACNA1C) predicted coding nucleotide sequence Genbank ID XM_021092981.1 and were ordered from by Eurofins Genomics (Germany GmbH). The sequences were as follows; sense, 5′ GCT GGA CAA GAA CCA GCG GCA GTG ‘3 and antisense, 5′ CAC TGC CGC TGG TTC TTG TCC AGC 3′. The probes were labeled with CY3 fluorophore on both ends and HPLC purified. The in situ hybridization was performed using Shandon racks as earlier described with minor modifications32. The sections were pretreated with 10% Proteinase K diluted in TBS (124568, Sigma-Aldrich, Missouri, USA) for 10 min at 37 °C and flushed two times with Mili-Q water after deparaffinization. To investigate the amount of autofluorescent signal in the porcine tissues the method described above was performed but without adding the probes. The images of the porcine lung tissues were obtained with an LSM 900 microscope with an Airyscan2 detector attached and a 40x/1.2 objective. All images were acquired, processed, and analyzed with the ZEN software by using the same settings for the CY3 channel on all images. The images were analyzed in ImageJ in the same manner as described for CavPan α1 IHC but by using the “split channels” function, selecting the surface of lamina epithelialis, and setting a threshold of 181 for DAPI and 6; 158 for the Cav1.2 probes.
Results
VDDCs and Cav1.2 are expressed on the surface of the porcine respiratory epithelium
The HEK 293 T cells and the porcine lung tissues tested positive for Cav1.2 when tested by RT-qPCR. The consensus sequence (OQ973297) of the porcine lung tissue corresponds to nucleotides 3532-3625 on sus scrofa calcium voltage-gated channel subunit alpha1 C/Cav1.2 (CACNA1C) predicted coding nucleotide sequence (Genbank ID XM_021092981.1). The porcine consensus sequence had the highest sequence similarity with the human Cav1.2 (97% identity) followed by the guinea pig and the ferret Cav1.2 sequences (96 % and 95% identity, respectively, Supplementary file S1).
All of the selected species showed 100% homology in the Cav1.2 amino acid sequence targeted by the CavPan α1 antibody (Supplementary file 2). The immunohistochemical staining for CavPan α1 and the negative absorption control is shown in Fig. 1. The absorption control showed no staining in the murine brain and porcine heart, nasal and tracheal tissues, and only weak irregular staining was found in the bronchial epithelium. Furthermore, the application of the isotype IgG-antibody control in murine brain tissue and porcine heart, nose, trachea, and lung tissues is shown in Fig. S1. Although the isotype control was negative in the murine brain tissue, unspecific staining was observed in the porcine heart, nose, bronchi, and bronchioles, but it was significantly lower than the staining intensity of CavPan α1. The CavPan α1 staining of the murine brain showed strong staining in the molecular, granular, and Purkinje cell layers of the cerebellum and in neurons (Fig. 1). Furthermore, there was a moderate to strong positive staining of CavPan α1 throughout the porcine heart tissue with the highest staining intensity of the Purkinje fibers. In the porcine respiratory tissues, the strongest staining was observed in the epithelial layer and the glands in the submucosal layer. Furthermore, there was also a weaker positive staining of the smooth muscle cells surrounding the bronchi and bronchioles. The % CavPan α1 area in lamina epithelialis of the different porcine respiratory tissues and lung compartments is presented in Fig. 2. Briefly, the highest % CavPan α1 area was observed in the nose and bronchi followed by the alveoli and the bronchioles, which also showed a higher variation of staining. The lowest expression of CavPan α1 was observed in the trachea.
The presence of CavPan α1 (brown staining) in murine brain, porcine heart, nasal, tracheal, and lung tissues was visualized immunohistochemically by CavPan α1-antibodies. A negative control was performed by a homologous absorption test on parallel sections of the murine brain together with the porcine heart and respiratory tissues.
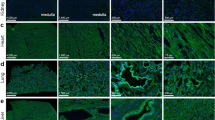
Cav1.2 mRNA visualized by in situ hybridization is presented in Fig. 3. Positive staining on the surface of the terminal bronchioles epithelial cells was observed with the anti-sense probe, whereas the sense probe showed a weaker signal. The anti-sense and sense probes showed no to weak positive staining of the alveoli, and as no difference between the two probes was observed, this signal only was interpreted as a background signal. In general, an autofluorescent signal of the erythrocytes was observed but otherwise, no autofluorescent signal was found (Fig. S2).
A The presence of Cav1.2 (red staining) in porcine lung tissues was visualized by in situ hybridization and a CY3 fluorophore-labeled anti-sense Cav1.2 probe. A negative control was performed using a sense probe. B The percentage of area with pixels above the threshold in the bronchiolar epithelium (% Cav1.2 area) observed with the anti-sense and sense probes. C The percentage of area with pixels above threshold in the alveoli (% Cav1.2 area) observed with the anti-sense and sense probes. Images were obtained by an LSM 900 microscope and an Airyscan2 detector. Blue: nuclear DAPI staining. White scale bar indicates 20 µm.
Cells expressing CavPan α1 co-localize with cells positive for IAV in experimentally infected pigs
Parallel sections of immunohistochemical staining for IAV and CavPan α1 from two IAV-experimentally infected pigs (one with swine-adapted IAV and one with human-adapted IAV) are presented in Fig. 4. Co-localization of IAV-positive and CavPan α1-positive cells was present in the respiratory epithelium throughout the respiratory tract. Furthermore, the leukocytes present in the bronchial and bronchiolar exudate also stained positive for CavPan α1 but were only rarely positive for IAV.
Discussion
The most interesting finding was that α1 subunits of Cav1 and Cav2 families (CavPan α1) are expressed in the respiratory epithelium throughout the porcine respiratory tract and co-localize with IAV-positive cells in experimentally IAV-infected pigs. The expression of Cav1.2 was observed in the porcine terminal bronchioles but not in the alveoli.
The anti-CavPan α1 staining of the adult murine brain correlates to previous findings of non-fetal mice25,33, and the CavPan α1 in the porcine heart is in agreement with findings in cardiomyocytes of adult mice25. Both staining methods showed staining of the porcine lungs that correlate to the findings in fetal human and fetal/adult murine lung tissue18,24. Anti-CavPan α1 binds to both the Cav1 (Cav1.1–1.4) and Cav2 families (Cav2.1– 2.3) and showed staining of the smooth muscle cells surrounding the bronchi and bronchioles of the lungs, whereas the Cav1.2 antisense-probe specifically binds to Cav1.2 and showed no staining of the smooth muscle cells. This difference could be explained by Cav2.2 showing a stronger staining intensity of the smooth muscle cells compared to Cav1.2 in the fetal human lung, which only showed weak staining of the smooth muscle cells24.
The IAV tropism in pigs are epithelial cells throughout the respiratory tract, and occasionally in glands in the submucosal layer and alveolar macrophages5,34,35,36,37,38,39. The lowest expression of α1 subunits of Cav1 and Cav2 families (CavPan α1) was observed in the trachea, which correlates with a previous study that found a lower viral load in the trachea compared to porcine nose and lung tissues40. An explanation for a lower viral load in the trachea remains to be clarified since the pig IAV host receptor (SA-α2,6) is expressed in the trachea (5, 36, 38), however, one explanation could be a lower expression of α1 subunits of Cav1 and Cav2 families. Furthermore, a higher viral load is mainly found in the bronchiolar epithelium compared to the alveoli5,35,36,38, which correlates with the Cav1.2 mRNA expression. Thus, Cav1.2 specifically could be an important factor for IAV internalization in pigs. Further investigations of α1 subunits of Cav1 and Cav2 families and specifically Cav1.2 distribution in the adult human lung tissues are needed to elucidate if pigs and humans have different distribution patterns in the respiratory tissues. Additionally, we observed a high within-tissue variation of the expression of α1 subunits of Cav1 and Cav2 families and if this is explained by variance in the IHC staining or due to other factors such as the presence of neutrophils expressing cav2.3 and/or IAV-induced necrosis of the epithelial cells, remain unknown41.
The results of the present study strongly suggest that the pig is a suitable model to investigate VDCC inhibitors’ effect on the treatment of IAV in porcine cell lines. Additionally, optimized RT-qPCR or situ hybridization methods are needed to quantify the expression of Cav1.2 throughout the porcine respiratory tract and to evaluate if Cav1.2 is up- or down-regulated during an IAV infection. Squamous metaplasia is often induced after IAV-infections in humans and squamous metaplasia has been shown to decrease the expression of Cav1.2 in the nasopharynx of humans, suggesting that Cav1.2 is less expressed in humans with recent IAV infections42,43.
The co-localization of IAV and VDCCs might be translatable to humans since the pathogenesis of IAV infection in pigs resemble human, pigs show clinical signs similar to humans44, and humans also express Cav1.2 in their fetal respiratory tract. Additionally, an L-type VDCC inhibitor prevents the internalization and spread of SARS-Cov-2, thus it could be speculated that Cav1.2 is important for other respiratory viruses as well45.
Data availability
Images and ROIs used for the semi-quantification are uploaded to the Open Science Framework (https://osf.io/8fz6j/, https://doi.org/10.17605/OSF.IO/8FZ6J) otherwise data is presented within the paper or in the supplementary files.
References
Gambaryan, A. S. et al. Receptor-binding properties of swine influenza viruses isolated and propagated in MDCK cells. Virus Res. 114, 15–22 (2005).
Song, H. et al. Avian-to-human receptor-binding adaptation by influenza A virus hemagglutinin H4. Cell Rep. 20, 1201–1214 (2017).
Liu, M. et al. Human-type sialic acid receptors contribute to avian influenza A virus binding and entry by hetero-multivalent interactions. Nat. Commun. 13, 4054 (2022).
Nicholls, J. M., Bourne, A. J., Chen, H., Guan, Y. & Peiris, J. S. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir. Res. 8, 73 (2007).
Trebbien, R., Larsen, L. E. & Viuff, B. M. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol. J. 8, 434 (2011).
Ryt-Hansen, P. et al. Co-circulation of multiple influenza A reassortants in swine harboring genes from seasonal human and swine influenza viruses. Elife 10, e60940 (2021).
Rajao, D. S., Vincent, A. L. & Perez, D. R. Adaptation of human influenza viruses to swine. Front. Vet. Sci. 5, 347 (2019).
Russell, C. J., Hu, M. & Okda, F. A. Influenza hemagglutinin protein stability, activation, and pandemic risk. Trends Microbiol. 26, 841–853 (2018).
Böttcher-Friebertshäuser, E., Klenk, H. D. & Garten, W. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog. Dis. 69, 87–100 (2013).
Mena, I. et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 5, e16777 (2016).
European Centre for Disease Prevention and Control. Zoonotic influenza. In: ECDC. Annual epidemiological report for 2022. Stockholm: ECDC. (2023).
Henritzi, D. et al. Surveillance of European domestic pig populations identifies an emerging reservoir of potentially zoonotic swine influenza A viruses. Cell Host Microbe 28, 614–627 (2020).
Watson, S. J. et al. Molecular epidemiology and evolution of influenza viruses circulating within European swine between 2009 and 2013. J. Virol. 89, 9920–9931 (2015).
Chen, X., Cao, R. & Zhong, W. Host calcium channels and pumps in viral infections. Cells 9, 94 (2019).
Bao, M. N. et al. Influenza A viruses enter host cells via extracellular Ca2+ influx-involved clathrin-mediated endocytosis. ACS Appl. Bio Mater. 4, 2044–2051 (2021).
Fujioka, Y. et al. A Ca(2+)-dependent signalling circuit regulates influenza A virus internalization and infection. Nat. Commun. 4, 2763 (2013).
Huang, Y. et al. Nisoldipine inhibits influenza A virus infection by interfering with virus internalization process. Viruses 14, 2738 (2022).
Fujioka, Y. et al. A sialylated voltage-dependent Ca2+ channel binds hemagglutinin and mediates influenza A virus entry into mammalian cells. Cell Host Microbe 23, 809–818.e5 (2018).
Buraei, Z. & Yang, J. The ß subunit of voltage-gated Ca2+ channels. Physiol. Rev. 90, 1461–1506 (2010).
Lazniewska, J. & Weiss, N. Glycosylation of voltage-gated calcium channels in health and disease. Biochim Biophys. Acta Biomembr. 1859, 662–668 (2017).
Pizzorno, A. et al. Repurposing of drugs as novel influenza inhibitors from clinical gene expression infection signatures. Front. Immunol. 10, 60 (2019).
Bausch Health Companies Inc, CARDIZEM® CD, U.S. Food and Drug Administration website. https://www.accessdata.fda.gov/scripts/cder/daf/.
Efficiency of Antagonist Drugs of the Cellular Transcriptomic Signature of Influenza A Virus Infection. (FLUNEXT). ClinicalTrials.gov identifier: NCT03212716. Updated February 6, 2023. Accessed February 7, 2023. https://clinicaltrials.gov/ct2/show/NCT03212716.
Brennan, S. C. et al. Fetal calcium regulates branching morphogenesis in the developing human and mouse lung: involvement of voltage-gated calcium channels. PLoS One 8, e80294 (2013).
Splawski, I. et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 119, 19–31 (2004).
Eden, M. et al. Myoscape controls cardiac calcium cycling and contractility via regulation of L-type calcium channel surface expression. Nat. Commun. 7, 11317 (2016).
Kristensen, C. et al. Experimental infection of pigs and ferrets with “pre-pandemic,” human-adapted, and swine-adapted variants of the H1N1pdm09 influenza A virus reveals significant differences in viral dynamics and pathological manifestations. PLoS Pathog (in press).
Sørensen, D. B. et al. Time-dependent pathologic and inflammatory consequences of various blood sampling techniques in mice. J. Am. Assoc. Lab. Anim. Sci. 58, 362–372 (2019).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Landini, G., Martinelli, G. & Piccinini, F. Colour deconvolution: stain unmixing in histological imaging. Bioinformatics 37, 1485–1487 (2021).
Ryt-Hansen, P. et al. Longitudinal field studies reveal early infection and persistence of influenza A virus in piglets despite the presence of maternally derived antibodies. Vet. Res. 50, 36 (2019).
Jensen, H. E. et al. Fluorescence in situ hybridization for the tissue detection of bacterial pathogens associated with porcine infections. In: Cunha, M., Inácio, J. (eds) Veterinary Infection Biology: Molecular Diagnostics and High-Throughput Strategies. Methods in Molecular Biology. Humana Press, New York, NY. 1247, https://doi.org/10.1007/978-1-4939-2004-4_17 (2015).
Xiao, H., Chen, X. & Steele, E. C. Jr. Abundant L-type calcium channel Ca(v)1.3 (alpha1D) subunit mRNA is detected in rod photoreceptors of the mouse retina via in situ hybridization. Mol. Vis. 13, 764–771 (2007).
Brookes, S. M. et al. Replication, pathogenesis and transmission of pandemic (H1N1) 2009 virus in non-immune pigs. PLoS One 5, e9068 (2010).
Van Poucke, S. G., Nicholls, J. M., Nauwynck, H. J. & Van Reeth, K. Replication of avian, human and swine influenza viruses in porcine respiratory explants and association with sialic acid distribution. Virol. J. 7, 38 (2010).
Chan, R. W. et al. Infection of swine ex vivo tissues with avian viruses including H7N9 and correlation with glycomic analysis. Influenza Other Respir. Viruses 7, 1269–1282 (2013).
Jung, K., Ha, Y. & Chae, C. Pathogenesis of swine influenza virus subtype H1N2 infection in pigs. J. Comp. Pathol. 132, 179–184 (2005).
Eriksson, P. et al. Characterization of avian influenza virus attachment patterns to human and pig tissues. Sci. Rep. 8, 12215 (2018).
Olaniyi, M. O., Adebiyi, A. A., Ajayi, O. L., Alaka, O. O. & Akpavie, S. O. Localization and immunohistochemical detection of swine influenza A virus subtype H1N1 antigen in formalin-fixed, paraffin-embedded lung tissues from naturally infected pigs. Beni. Suef. Univ. J. Basic Appl. Sci. 9, 16 (2020).
Schwaiger, T. et al. Experimental H1N1pdm09 infection in pigs mimics human seasonal influenza infections. PLoS One 14, e0222943 (2019).
Zhu, B. et al. F0F1 ATP synthase regulates extracellular calcium influx in human neutrophils by interacting with Cav2.3 and modulates neutrophil accumulation in the lipopolysaccharide-challenged lung. Cell Commun. Signal 18, 19 (2020).
Kuiken, T. & Taubenberger, J. K. Pathology of human influenza revisited. Vaccine 26, 59–66 (2008).
Nishi, K. et al. Epipharyngeal abrasive therapy down-regulates the expression of Cav1.2: a key molecule in influenza virus entry. In Vivo 36, 2357–2364 (2022).
Starbæk, S. M. R. et al. Animal models for influenza A virus infection incorporating the involvement of innate host defenses: enhanced translational value of the porcine model. ILAR J. 59, 323–337 (2018).
Straus, M. R. et al. Inhibitors of L-type calcium channels show therapeutic potential for treating SARS-CoV-2 infections by preventing virus entry and spread. ACS Infect. Dis. 7, 2807–2815 (2021).
Acknowledgements
This work was supported by the Novo Nordic Foundation (FluZooMark: NNF19OC0056326). The funder played no role in study design, data collection, analysis and interpretation of data or the writing of the manuscript. We acknowledge the Core Facility for Integrated Microscopy, Faculty of Health and Medical Sciences, University of Copenhagen with the support of the in situ hybridization images.
Author information
Authors and Affiliations
Contributions
C.K. and L.E.L. contributed with the conception of the data. C.K. performed the acquisition and the analysis of the data. All authors contributed to the interpretation of the work. C.K. wrote the main manuscript and H.E.J., R.T., P.R.-T., and L.E.L. have substantively revised it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kristensen, C., Jensen, H.E., Trebbien, R. et al. Co-localization of influenza A virus and voltage-dependent calcium channels provides new perspectives on the internalization process in pigs. npj Viruses 1, 8 (2023). https://doi.org/10.1038/s44298-023-00009-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44298-023-00009-x