Abstract
Organoids bridge the gap between 2D cell lines and in vivo studies. With their 3D organization and cellular heterogeneity, adult stem cell-derived organoids closely resemble their tissue of origin. The development of CRISPR-mediated genome engineering and the recent additions of base and prime editing to the CRISPR toolbox have greatly simplified the generation of exact, isogenic models for Mendelian diseases. Here, we review recent developments in CRISPR-mediated genome engineering and its application in human adult-stem-cell-derived organoids in the construction of isogenic disease models. These models allow accurate qualification of the impact of allelic disease variants observed in patients. Furthermore, we discuss the use of organoids as models for safety and efficacy of CRISPR for gene repair. Although transplantation of repaired tissue remains challenging, benchmarking CRISPR tools in organoids can bring genome engineering one step closer to patients.
Key points
-
CRISPR–Cas9-mediated genome engineering acts by introducing double-stranded DNA breaks into the genome. The damage repair process can be used for gene knockout or precise targeted introduction of exogenous DNA.
-
Next-generation CRISPR tools, including base and prime editing, allow for induction of precise base changes and small insertions and deletions, bypassing potentially deleterious double-stranded DNA breaks.
-
Owing to their 3D organization, adult-stem-cell-derived organoids closely resemble the tissue of origin and are therefore a good model system to study human health and disease.
-
CRISPR–Cas9-mediated genome engineering can be used to create isogenic models to investigate the onset, cause and treatment of human diseases.
-
CRISPR tools can be benchmarked for efficiency and safety by studying gene repair ex vivo in adult-stem-cell-derived organoids, facilitating CRISPR–Cas9 clinical translation.
-
Ex vivo repaired adult-stem-cell-derived organoids can potentially be transplanted into patients to relieve disease phenotypes.
Similar content being viewed by others
Introduction
Variant sequences in the genome have a fundamental role in the onset, course and outcome of hundreds of human diseases. A combination of the increasing number of genome-wide association studies and the decreasing price of whole-exome and whole-genome sequencing has been driving the identification of genetic disease variants1,2. Although the number of detected genetic variants increases, it remains difficult to accurately qualify their impact on disease progression. To obtain a deeper understanding of the molecular mechanisms that underlie genetic variations and to develop novel therapeutic strategies, isogenic human disease models hold great promise. These models consist of human cells that are engineered to accurately model the genetic variant and are matched with wild-type controls with the same genetic background. The development of clustered regularly interspaced short palindromic repeats (CRISPR) as an effective and versatile tool for genome engineering has greatly advanced the generation of isogenic in vitro models of human disease3,4,5.
Classical 2D tissue culture techniques have been used extensively to better understand the homeostasis and pathophysiology of the human body6. 2D cell lines are easy to maintain and are amenable to CRISPR–Cas9-mediated genome engineering. However, such cell lines — typically derived from malignancies — do not reflect the cellular complexity of the organ they are derived from. To overcome these issues, the development of more complex in vitro tissue culture techniques has gained traction. These efforts have ultimately led to the development of organoids. These ‘mini-organs’ exhibit faithful micro-anatomy and are grown in a matrix that allows for 3D expansion of stem cells, which give rise to cell types present in the native tissue7. Current organoid culturing technology exploits either induced pluripotent stem cells or adult stem cells (ASCs). Organoids derived from induced pluripotent stem cells are taken through an extensive fate-specialization procedure mimicking the embryonic developmental trajectory of the organ of interest8. ASC-derived organoids, the subject of this Review, model the adult homeostatic state of organs. They can be derived from most wild-type human and murine epithelial tissues, including large and small intestine9 (Fig. 1a), stomach10, kidney11, pancreas12, breast13, endometrium14 and cervix15, liver16,17 (Fig. 1b), upper airway and lung18,19, taste bud20, lacrimal gland21, prostate and bladder22,23 and thyroid24,25 (Fig. 1c,d). ASC-derived organoids do not require an extensive maturation process, are genetically stable and can be passaged indefinitely. Moreover, organoids can be clonally expanded from a single adult stem cell, aiding the generation of CRISPR-mediated isogenic 3D cultures that closely resemble the tissue of origin26. Besides variant impact qualification, these isogenic organoid models can be used for drug efficacy screening and de novo drug discovery (Fig. 1d).
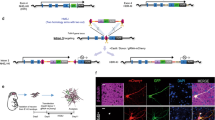
Confocal images of adult-stem-cell-derived organoids. a, Small intestine organoids. Nuclei are stained by DAPI (turquoise) and actin by phalloidin (red). b, Fetal hepatocyte organoids. Nuclei are stained by DAPI (orange) and actin by phalloidin (blue). c, Murine thyroid organoids. Nuclei are stained by DAPI (purple), actin is stained by phalloidin (green) and the hormone carrier protein thyroglobulin (Tg) is stained in red. Scale bars in panels a to c are 50 µm. d, Organoids can be derived from most epithelial tissues of murine and human origin. By using CRISPR engineering, putative disease variants can be introduced into the genome. By pairing up with wild-type controls, an isogenic system is created that can be used for drug screening, variant impact qualification and drug discovery. Part a, image courtesy of Joep Beumer. Part b, image courtesy of Shashank Gandhi. Part c, image courtesy of Jelte van der Vaart.
In this Review we provide the rationale behind the generation of isogenic disease models in human ASC-derived organoids. First, we review the recent advances in CRISPR-mediated genome engineering that enable efficient induction of mutations and genetic variants in the genome. Then, we discuss the efforts made to use these technologies in organoids for modelling and repair of genetic variants that cause disease. Next, we provide technical considerations to generate genetically altered organoids and create complex isogenic disease models. Finally, we provide an outlook on the combination of CRISPR–Cas9-mediated genome engineering and ASC-derived organoids.
CRISPR–Cas-mediated genome engineering
CRISPR is superior to previously developed strategies aimed at altering the genome (Box 1) and has quickly been adopted by laboratories all over the world. To date, six classes of Cas genes have been described, of which the class II CRISPR system (which includes Cas9) is the most studied27. In conventional CRISPR–Cas9-mediated genome engineering, the effector protein Cas9 is guided towards a genomic target site by an RNA sequence called the guide RNA (Fig. 2). This guide RNA consists of a CRISPR RNA (crRNA) sequence, complementary to the target site, and a trans-activating CRISPR RNA (tracRNA) that is needed for crRNA maturation and binding to Cas9. The guide RNA was simplified by creating a chimeric crRNA–tracRNA fusion, yielding a single-guide RNA (sgRNA) for target recognition3. The target site in the genome consists of two elements, the protospacer and a short essential sequence directly downstream of the target site, called the protospacer-adjacent motif (PAM). The PAM motif of the most frequently used Cas9 (derived from the bacterium Streptococcus pyogenes, SpCas9) is NGG3. Upon binding of the sgRNA, the Cas9–sgRNA complex screens the genome for PAMs28. After a suitable PAM is found, sgRNA complementarity to the protospacer is tested by opening the DNA around the target site in an R-loop into two single DNA strands (ssDNA). These ssDNA strands are individually cleaved by the RuvC and HNH domain of Cas9, resulting in a double-stranded DNA break (DSB). The cell recognizes this DSB and has two main pathways to repair the damage. In most cases, non-homologous end joining (NHEJ) is initiated. In this error-prone process, the two DNA ends are quickly ligated together, which often results in a small deletion or insertion (indel) at the cut site29. The CRISPR–Cas9-induced indel, if out-of-frame, results in early termination of the targeted protein. Alternatively, the homology-directed repair (HDR) pathway is initiated by supplementing the reaction with an exogenous DNA repair template that contains homology to the DNA adjacent to the DSB and the edit of interest. Therefore, by hijacking the endogenous DNA repair pathways, CRISPR can be used for genome engineering in a sgRNA-mediated manner3,4,5 (Fig. 2).
Upon binding of the Cas9–single-guide RNA (sgRNA) complex to the genomic target site, which consists of a protospacer and a protospacer-adjacent motif (PAM), the genome is opened in an R-loop, resulting in two single-stranded DNA (ssDNA) strands. These ssDNA strands are individually cleaved by the RuvC and HNH domain of Cas9, resulting in a double-stranded DNA break (DSB). The cell has two endogenous repair mechanisms to resolve DSBs. DSB repair by non-homologous end joining (NHEJ) results in the induction of indels at the target site, which can be used to knock out genes of interest. Homology-directed repair (HDR) can be used to introduce exogenous DNA, containing the genetic alteration of interest at the target site.
There are key downsides to using conventional CRISPR–Cas9 for the construction of isogenic disease models. First, because the human genome consists of three billion base pairs, the chances are great that the sgRNA will initiate DSBs at off-target sites30. To overcome this issue, high-fidelity variants of Cas9 such as SpCas9-HF31 and hifi-Cas932 have been developed. Alternatively, off-target free sgRNAs can be selected using profiling strategies such as Guide-seq or Circle-seq prior to use in experiments33,34. Next, even if an off-target free sgRNA is used, on-target DSB repair can result in undesired editing outcomes, such as the induction of large deletions, insertions and translocations35. In extreme cases, CRISPR–Cas9 could lead to chromothripsis, a process of chromosome shattering and massive structural variation downstream of the sgRNA target site36. Finally, HDR upon CRISPR-mediated DSB induction is required for modelling specific genetic variants. However, HDR can only occur during the G2 and S phase of the cell cycle when sister chromatids are present37, which thereby risks NHEJ repair pathway domination. Cell cycle synchronization and addition of NHEJ inhibitors are two strategies to push the cell towards HDR-mediated DSB repair instead38,39. Nevertheless, because both alleles are often cleaved, the end result of an HDR experiments is often correct variant introduction on one allele whereas the second allele is knocked out owing to the cell’s bias towards using the NHEJ for DNA repair29. Because only about 2.4% of disease-causing variants are indels, alternative strategies of genome engineering that do not require DSB induction have been pursued40.
DSB-free genome editing
A single histidine residue at position 840 of the HNH domain of SpCas9 cleaves the PAM strand, whereas the aspartate at position 10 in the RuvC domain cleaves the opposite strand3. Mutating both amino acids to alanines (D10A and H840A) results in nuclease-inactive or ‘dead’ Cas9 (dCas9). dCas9 still recognizes its target site and opens up the DNA in an R-loop but does not induce DSBs. The binding of dCas9 to its target site alone can function as a repressor of transcription and is dubbed CRISPR interference (CRISPRi)41. Alternatively, dCas9 can be used as a vehicle to localize DNA effector proteins to the genome. Examples of this strategy are CRISPR activators (CRISPRa)42 and CRISPR–DNMT3 fusion proteins for targeted methylation43. To induce genetic variants, DNA-alteration enzymes are fused to dCas9 to overcome the limitations associated with DSB induction in genome engineering. These ongoing strategies could facilitate CRISPR-based genome-engineering clinical translation (Box 2).
Base editing
The first base editor fuses dCas9 to the rat cytidine deaminase apolipoprotein B mRNA editing catalytic polypeptide-like (rAPOBEC1), which catalyses the conversion from cytidine to uracil. The cell repairs this uracil into thymidine, resulting in a construct (BE1) that replaces a C•G by a T•A base pair, called a cytosine base editor (CBE)44. First-generation CBEs were inhibited by uracil glycosylation. Therefore, second-generation base editors (BE2) were developed by fusing an uracil glycosylase inhibitor (UGI) to the dCas9–rAPOBEC1 fusion45. To increase editing efficiency, dCas9 can be converted into a nickase SpCas9-D10A. In this optimized base editor architecture (BE3), the strand that is not modified by rAPOBEC1 is cleaved. The cell detects the nick and initiates DNA repair to resolve the damage. The strand containing the base change is then used as a template for repairing the nick, yielding stable integration of the edit with an efficiency between 15% and 75% depending on the sgRNA44. The BE3 architecture was further improved by fusing an additional UGI in combination with linker optimization, resulting in the fourth-generation cytosine base editor (BE4). BE4s have improved editing efficiency (by around 50%), with two-fold reduction of unintended byproducts such as indels and point mutations46. Subsequent codon optimization47 and ancestral reconstitution48 led to a CBE architecture that currently enables the most robust base editing in 2D cell lines, organoids and in vivo by improving expression and nuclear localization of the proteins49 (Fig. 3a). A similar base editor, that enables C-to-T base changes, was developed by fusing cytidine deaminase 1 (CDA1) to SpCas9D10A in a system called Target-AID50. This base editor has a shifted activity window (from positions 4–8 in the sgRNA for CBE to positions 1–5 in target-AID)50. Moreover, C-to-G and C-to-A changes are frequently observed in Target-Aid. These unwanted byproducts also occur in first-generation BE3, but have been resolved in newer iterations of CBE such as BE4 (ref. 46).
a, A cytosine base editor consists of a nickase Cas9-D10A fused to a cytidine deaminase and a tandem repeat of uracil glycosylase inhibitors (UGI). After binding of the Cas9–single-guide RNA (sgRNA) complex, the DNA opens up in an R-loop, which enables cytidine deamination and conversion into uracil. After nicking of the non-edited opposite strand, the U•G base pair is repaired into a T•A base pair, effectively resulting in C>T base editing. b, An adenine base editor consists of a nickase Cas9-D10A fused to a heterodimer of TadA, an adenine deaminase. The asterisk indicates the evolved TadA variant and further exemplifies the heterodimer state of the fusion protein. After R-loop generation by binding of the Cas9–sgRNA complex to the target site, adenine is deaminated, effectively turning it into inosine. The inosine residue is converted into guanine by nicking of the non-edited strand, after which DNA repair is guided towards the correct A>G edit. c, The cytidine and adenine deaminases function only on single-stranded DNA. Therefore, base editor activity is limited to a small editing window within the R-loop that spans from roughly the 4th to the 8th base from the start of the sgRNA.
The opposite base change can be performed with the use of adenine base editors. For example, tRNA-specific adenosine deaminase, TadA, is a protein that enables editing of adenine residues in the DNA51. Because wild-type TadA does not act on DNA, the protein was evolved using a process called phage-assisted continuous evolution (PACE)52. Fusion of the seventh generation of evolved TadA in a heterodimer with a wild-type TadA to SpCas9-D10A results in an adenine base editor (ABE) with an A-to-G editing efficiency of up to 50% depending on the target site, which is comparable or higher than that of third-generation CBEs44,51. As opposed to CBEs, the TadA heterodimer in ABEs deaminates adenine residues, which are then converted to inosine. Upon cleavage of the non-edited strand and resolving of the DNA mismatch, the inosine residue is converted into guanine, effectively resulting in a A•T to G•C edit (Fig. 3b). The applicability of ABEs was further improved by codon optimization and additional PACE-mediated directed evolution, resulting in optimized eighth-generation base editors with a 1.5–3.2-fold improvement in editing efficiency for ABE8 (ref. 53), and a 9.4–24-fold increase for ABE8e54, depending on the sgRNA and nucleotide location in the editing window49.
The deaminases fused to Cas9 in base editors function only on ssDNA. Therefore, base editors act only on a few bases of the single-stranded R-loop that is generated upon Cas9 target recognition3,4,5. This so-called ‘editing window’ roughly spans four nucleotides, between positions 4 and 8 from the 5′ end of the protospacer (Fig. 3c). This ssDNA dependence greatly reduces the sgRNA-mediated off-target effects of base editors but requires very specific localization of Cas9 to induce the desired edits. Relaxing the PAM requirements and increasing the target space of Cas9 has resulted in a series of evolved SpCas9 variants. For example, PACE and structural guided evolution resulted in xCas9 and SpCas9-NG, which recognize an NGN PAM55,56. Further structural modification led to nearly PAMless SpCas9 variants that target NRN (where R = A or G) and NYN (where Y = C or T)57. An alternative strategy to increase the target space of base editors relies on Cas9 homologues such as Streptococcus aureus (PAM = NNGRRT). Other approaches resulting in evolved SpCas9 variants and SpCas9 homologues with alternative PAM requirements and editing windows have also been developed40,58.
However, base editors have limitations. Although ABEs essentially yield zero off-target effects, genome-wide profiling of CBEs has shown genome-wide C>T mutations owing to the overexpression of APOBEC in the cell59,60. Evolved APOBEC domains can be used in CBE to decrease these side effects61. Moreover, CBE-induced uracil residues sometimes yield adenine and guanine residues instead of the desired thymidine owing to unwanted cellular uracil DNA glycosylation during the base excision DNA repair pathway44,47. Despite these editing outcomes being undesired, this observation has led to the development of new classes of base editor. For example, removal of UGI from the CBE architecture pushes DNA repair towards guanine instead of thymidine and has allowed the development of C>G base editors62,63. Furthermore, not all desired point mutations can currently be generated by base editors. Finally, base editors cannot introduce indels or larger genetic variants. Prime editors have been developed to overcome these limitations and allow for more versatile DSB-free genome engineering.
Prime editing
The rationale behind prime editing is to bring exogenous DNA with the edit of interest close to the Cas9 binding site. In the first generation of prime editors (PE1), a reverse transcription (RT) domain derived from the Moloney murine leukaemia virus was fused to nickase SpCas9-H840A64. The RT domain converts RNA into DNA and finds its template in the 3′ extension of the specially designed sgRNA, called the prime-editing guide RNA (pegRNA), that guides the Cas9 in PE1 to the target site. Upon target recognition, the PAM-containing strand is nicked by the active HNH domain of Cas9-H840A. Then, the pegRNA extension binds to the nicked strand at the primer-binding site (PBS), after which the RT domain of PE1 uses the remaining pegRNA (RT template) to synthesize a 3′-DNA flap containing the edit of interest. This DNA-flap is resolved by cellular DNA repair processes integrating the edit of interest. Efficiencies of prime editing can be further enhanced by using a rationally evolved variant of the RT domain (PE2) and by inducing a proximal second nick in the opposing DNA strand, guided by a second (PE3) guide RNA64 (Fig. 4). However, the use of a PE3-guide in prime editing comes with a cost as indel numbers are substantially higher compared to PE2 (6.8% average indels for the sgRNA with the highest editing efficiency)64. This issue can be resolved by using a PE3b-guide that matches the edited strand, resulting in a second nick once the edit is made65. In addition to all transition and transversion mutations, the first description of prime editing reported the induction of deletions of up to 80 and insertions of up to 44 base pairs64. For efficient use of prime editing, extensive optimization of the pegRNA and PE3 guide is required. The length of both the PBS and the RT, as well as the distance between the pegRNA and PE3-guide nick influence the editing efficiencies of prime editing. Optimization can be easily performed in the HEK293T cell line, but is more difficult in organoids or in vivo. However, once fully optimized, prime editing is the most versatile DSB-free genome-engineering technology to date.
The prime-editing guide RNA (pegRNA) complexes with the nickase SpCas9-H840A–RT prime-editing fusion protein and binds to the target DNA. Upon protospacer-adjacent motif strand cleavage by SpCas9-H840A, the primer-binding site of the pegRNA extension binds the single-stranded DNA, upon which the reverse transcriptase (RT) synthesizes a 3′-DNA flap containing the edit of interest. This 3′-flap is resolved by cellular DNA processes, which can be further enhanced by introducing a proximal second nick in the opposing DNA strand, guided by a second (PE3) guide RNA. The red scissors indicate the nick site of the SpCas9-H840A. PAM, protospacer-adjacent motif; PBS, primer-binding site.
Prime editing holds great promise owing to its versatility in potential edits; however, the need for optimizing pegRNA and PE3-guides limits its application in organoids. To overcome this issue, three key modifications have been made to the prime-editing system. First, the use of two pegRNAs in trans with overlapping RT domains increases prime-editing efficiencies in human cells as well as plants66,67,68. Second, engineered pegRNAs can have evopreq or tmpknot domains fused to the 3′ end. These domains increase the stability of the pegRNA, which can increase prime-editing efficiency69. Finally, including the R221K and N394K amino acid changes increases the nuclease function of SpCas9, resulting in a more efficient PE2Max70.
Isogenic organoid disease models
Because ASC-derived organoids more closely resemble their tissue of origin compared to 2D cell lines, they are more suitable for the study of human physiology. The rapid developments of CRISPR-mediated genome engineering now allow for rapid generation of isogenic organoid models that harbour specific mutations that have a role in the onset and course of human diseases.
Tumorigenesis and cancer
The majority of CRISPR-generated isogenic ASC-derived organoid models currently focus on tumorigenesis and carcinogenesis. Two similar studies in human intestinal organoids recreated the Vogelstein model of sequential driver mutation accumulation in colorectal tumorigenesis71,72,73. By removing selected growth factors or adding small molecule inhibitors, organoids with mutations in APC (removal of Wnt), TP53 (addition of Nutlin), KRAS (removal of epidermal growth factor, EGF), SMAD4 (removal of Noggin) and PIK3CA (addition of MEK-inhibitors) can be generated. Subcutaneous transplantation of these growth-factor-independent organoids in mice results in metastasizing carcinomas72,73. Inspired by these first two studies, multiplexed genome engineering was applied in ASC-derived organoids with subsequent transplantation into mice to elucidate the minimal requirements for tumorigenesis in other tissues. For example, subcutaneous transplantation of CRISPR-mediated knockout of PTEN, TP53, RB1 and NF1 in breast organoids results in tumour formation resembling oestrogen- and progesterone-receptor-positive and human epidermal growth factor receptor 2 (HER2)-negative luminal B breast cancers in mice74. Furthermore, CRISPR–Cas9-mediated knockouts of TP53, SMAD4, PTEN, NF1 and BAP1 were generated in cholangiocyte organoids to elucidate the role of the tumour suppressor BAP1 in cholangiocarcinoma75. Loss of BAP1 results in impaired chromatin accessibility and thus gene expression, crucial for epithelial integrity in the organoids and in mice75. Two studies generated isogenic models for pancreatic ductal adenocarcinoma (PDAC) in human ASC-derived ductal pancreas organoids. When combined with oncogenic KRASG12V, CRISPR-based multiplexed mutation of TP53, CDKN2A and SMAD4 results in organoids with PDAC phenotypes76, whereby overexpression of KRAS leads to organoids mimicking PDAC precursor states77. Two independent studies created CRISPR–Cas9-mediated knockout models of DNA repair genes. Mutational signature analysis of human colonic organoids with loss-of-function mutations in MLH1 revealed the predominant occurrence of COSMIC signature 20 associated with errors made during normal DNA replication65,78. Moreover, knockout of NTHL1 in colonic organoids results in an increase in C>T transitions, which resembles COSMIC signature 30, whereas XPC knockout generates organoids deficient in nucleotide excision repair, yielding COSMIC signature 8 (ref. 79). In a more sophisticated approach, the common fusion genes DLG1–BRAF, PTPRK–RSPO3 and EIF3E–RSPO2 were modelled into human colon organoids80. Co-transfecting two sgRNAs that target both loci of interest results in complex genomic rearrangements only in organoids that lacked TP53 expression80. CRISPR–Cas9 strategies can be similarly used to create single- or double-mutant isogenic knockout models of TP53 in human hepatocyte organoids81, ARID1A in human gastric organoids82, RB1 in human intestinal organoids to model neuroendocrine neoplasms83 and RNF43 to model early-onset colorectal cancers84.
Multiple genes can also be CRISPR-screened in a single experiment. For example, performing a small targeted CRISPR screen in human intestinal organoids enables mapping of RASGAP dependencies in colorectal cancer progression. Only loss of NF1 results in enhanced RAS-ERK signal amplification85. To increase the throughput of CRISPR, genome-wide CRISPR screening platforms have been developed86,87,88, allowing for positive and negative survivability screens while assessing loss-of-function mutations across all genes in the genome. Furthermore, protocols for genome-wide CRISPR screening have been developed for use in ASC-derived organoids. For example, a positive selection genome-wide CRISPR screen performed in WT, APC-KO and APC-KO; KRASG12D mutant intestinal organoids, identified genes involved in a previously undescribed link between TGFβ and WNT signalling, revealing PBRM189 and ARID1A and SMARCA490 as novel hits driving TGFβ resistance. These studies emphasize the possibility of genome-wide CRISPR screening in 3D models.
To model mutations observed in cancer patients accurately, simple CRISPR–Cas9-mediated knockouts by indel formation is not sufficient. Different mutations in the same cancer gene can have drastically different effects, as shown for TP5391 and KRAS92, highlighting the need for specific mutations instead of ‘blunt’ CRISPR–Cas9-mediated knockouts. For example, CRISPR-based prime editing for cancer modelling can be performed by introducing common TP53 mutations in human colon and hepatocyte organoids. Besides prime-editing-mediated induction of TP53R175H and TP53R249S, it is possible to directly compare ABE to prime editing by introducing the same mutation using both techniques. Targeting of TP53Y220C results in organoids that are able to grow on medium containing the mouse double minute 2 homologue (MDM2) inhibitor nutlin-3, which kills wild-type organoids by stabilizing TP53 (ref. 93). ABE substantially outperforms prime editing with 1.5–2-fold increased editing efficiencies but induces undesired additional base changes93. Similarly, introducing common in-frame deletions in CTNNB1 exon 3 in human cholangiocyte organoids generates mutant organoids, which can grow without exogenous Wnt94. These results highlight the efficacy of DSB-free genome engineering in organoids for cancer modelling.
Isogenic disease models beyond cancer
An example of combining CRISPR with ASC-derived organoids beyond cancer applications is the creation of isogenic models of DGAT1 loss in intestinal organoids as a model for congenital diarrhoeal disorder95, resulting in mutant organoids being more susceptible to lipid-induced cell death compared to their controls95. Another example comprises a genome-wide positive CRISPR screen to study confounding factors contributing to ulcerative colitis in mice. Despite wild-type organoids dying when treated with interleukin-17A (IL17A), organoids that harbour mutations in IL17RA and NFKBIZ upon treatment are enriched96. Furthermore, organoids are a great model to study the 2019 pandemic causing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)97,98,99. To elucidate essential host factors for SARS-CoV-2 entry, a targeted CRISPR screen generated knockouts of 19 previously implicated genes100. Interestingly, SARS-CoV-2 is not able to infect intestinal organoids harboring mutations in the host genes ACE2 and TMPRRS2, whereas none of the other 17 target genes shows a substantial decrease in infection potential of the virus100.
Assessing CRISPR-mediated gene repair
Because ASC-derived organoids more closely resemble their tissue of origin than do 2D cell lines, they hold the promise of mapping the efficacy and safety of therapeutic genome engineering in vitro prior to in vivo application. However, genome-engineering tools are benchmarked for efficiency and safety in conventional 2D cell lines such as HEK293T and U2OS4,5,44. Despite the ease of handling of these in vitro cultures, on- and off-target efficiencies can differ vastly from the cell types that are targeted in patients. For example, Cas9 binding can be influenced by methylation of CpG islands and chromatin accessibility, which differs greatly between cell types101,102. Therefore, testing CRISPR efficiency and safety in the target cell type in ASC-derived organoids could provide more accurate prediction of CRISPR-based genome engineering in patients. Furthermore, transplantation of ASC-derived organoids might complement whole-organ transplantation17,103. In vitro CRISPR-repaired autologous organoids could similarly be transplanted back to patients after rigorous off-target determination by whole-genome sequencing for safety purposes (Box 2).
Cystic fibrosis
The first hereditary disease to be repaired in human stem cells with the use of CRISPR–Cas9-mediated genome engineering was cystic fibrosis104. Cystic fibrosis is caused by various mutations in the cystic fibrosis transmembrane conductance regulator gene (CFTR), with the deletion of phenylalanine-508 being the most common (F508del)105. ASC-derived intestinal organoids model the function of the CFTR channel, in this case, through the forskolin-induced swelling assay, which correlates with clinical disease severity106,107,108. Sequencing-based screening of co-transfected Cas9 with a targeting sgRNA towards exon 11 and a donor plasmid containing a repaired F508 CFTR sequence and an intronic insertion of puromycin reveals correction of the F508del mutation in 17 out of 89 sequenced organoid clones (19.1%). These repaired organoids restore the forskolin-induced swelling response to wild-type levels104. Another strategy relies on repairing deleterious splice site mutations that disrupt CFTR function: 3272–26 kb A>G and 3849+10 kb C>T. Instead of directly repairing the point mutations, an allele-specific disruption of the mutation can be chosen by using Cas12a, a type 5 Cas protein109,110. Lentiviral transduction of the Cas9 and sgRNA into intestinal organoids derived from patients with cystic fibrosis results in 40% allele-specific indel induction depending on the corrected splicing as measured by forskolin-induced swelling110.
An intestinal organoid biobank was subsequently established containing 664 organoid lines that represent 154 distinct CFTR mutations111. From this biobank, organoids that could be repaired by SpCas9-ABE were selected. Intestinal organoids harbouring the R785* mutation were transfected with base editing reagents, after which forskolin-induced swelling revealed functional repair in about 9% of the transfected organoids. The target space of base-editor-mediated CFTR repair was increased by using xCas9-ABEs to repair R553*, R1162* and W1282* mutations in intestinal and upper airway organoids111. Similarly, prime editing in intestinal organoids allows DSB-free repair of the F508del mutation93. The lack of genome-wide off targets as measured by whole-genome sequencing of intestinal organoids after CFTR repair with base editing and prime editing underlies the safety of DSB-free genome engineering for therapeutic purposes93,111. To increase the in vitro editing efficiency of prime editing, a fluorescent prime editing and enrichment reporter called fluoPEER was developed112. In FluoPEER, mCherry is expressed if active prime editing occurs within the cell. Fluorescence-activated cell sorting (FACS)-based selection of mCherry-positive cells facilitates the generation of isogenic prime-edited organoids. Using FluoPEER, the repair efficiency of CFTRF508del is increased to 80%, enabling reparation of the elusive CFTRG542* mutation112.
Diseases beyond cystic fibrosis
Applying prime editing in patient-derived isogenic intestinal organoids allows restoration of the most common mutation in DGAT1, c.629_631delCTT,p.s210del, which causes a defect in fatty acid storage in lipid droplets, as measured by survivability after fatty acid addition to the culture medium94. Similar results were observed in an experiment repairing ATP7B mutations in liver organoids of patients with Wilson disease94. Subsequently, using the FluoPEER system effectively corrected mutations in ABCB4 and ATP8B1 responsible for intrahepatic cholestasis112. CRISPR-engineered isogenic organoids can also be used to study primary cilia dyskinesia disease. Using an organoid differentiation protocol allows visualization of cilia defects in airway organoids from patients with primary cilia dyskinesia113. From a mini-biobank of patient-derived organoids, organoids harbouring a splicing mutation in the cilia gene DNAH11 can be repaired using prime editing with efficiencies of up to 85%. Owing to limitations in clonal outgrowth of human airway organoids, no morphological analysis of repaired organoids could be performed113.
Technical considerations
Because conventional and next-generation CRISPR tools have primarily been developed for 2D cell lines, translation into 3D cell cultures is not straightforward. Therefore, key considerations need to be addressed before using ASC-derived organoids to create isogenic disease models.
The right genome-engineering tool
To circumvent undesired on-target and off-target effects of conventional CRISPR–Cas9, the use of next-generation CRISPR tools, in this case, base editing and prime editing, is advisable. CBE can be effectively used for the introduction of stop codons in the genes114, mediating C•G to T•A base changes turning arginine (CGA to TGA), glutamine (CAA/CAG to TAA/TAG) and tryptophan (TGG to TAG/TGA/TAA) into stop codons. According to the CRISPR-STOP method, because CBE does not require DSBs, lower levels of apoptosis are observed, resulting in cells that are less stressed upon transfection of genome-editing components. If no suitable sgRNAs are available for the CBE-mediated introduction of stop codons, ABE can be used to disrupt either the start codon115 or splice sites116 to effectively create a gene knockout. Importantly, using CRISPR-STOP eliminates the need for Sanger deconvolution or sub-cloning to determine individual indel outcomes of both alleles, as is required for conventional CRISPR–Cas9-NHEJ, resulting in a more efficient genotyping process117.
We recommend first designing the CBE-mediated stop-codon insertion before designing ABE-mediated methods because the cell could still express the protein of interest owing to alternative splicing or use of an alternative start site. Moreover, it is important to realize that the DNA-altering fusion proteins in base editors function in a specific sequence context. For example, a machine-learning protocol called BE-Hive reports that guanine residues in front of the cytosine substantially decrease the levels of base editing when using BE4118. Alternatively, modifications of CBE and ABE, such as evoA-BE4 and ABE-CP, can be used to perform base edits in alternative sequence contexts119. Moreover, base editors function most effectively within the editing window that roughly spans from the 4th to the 8th nucleotide from the start of the sgRNA (Fig. 3c). If a specific edit is required, but editing of additional nucleotides within the editing window would result in unwanted amino acid changes, prime editing is the preferred strategy because the RT-template can be designed to exclusively incorporate the edit of interest.
Delivery and selection
The delivery of genome-editing agents into organoids is considerably more difficult than in popular 2D cell lines. Simple lipofection of plasmid DNA can reach up to 95% transfection efficiency in HEK293T, whereas efficiencies in 3D organoids differ greatly per line and can be as low as 1.5%111. Lentiviral transduction can be a more efficient strategy. However, even if titrated properly, the viral genome is prone to integrate multiple times, which might influence organoid fitness90. Electroporation of ribonucleoprotein complexes into organoids substantially increases transfection efficiencies120, but it decreases flexibility, because for each subsequent next-generation CRISPR tool, a new protein has to be produced. The low transfection efficiencies in organoids do not limit successful isogenic model generation as long as strategies to select for either the transfected or functionally edited organoids is taken into account during the experimental setup. For functional selection, CRISPR-engineered organoids are selected for based on the introduced genetic variant, which can be based either on survivability or phenotypic changes upon genome engineering. Examples of functional selection based on survivability are the removal of Wnt and Rspondin-1 for selecting WNT pathway mutants such as APC and removal of EGF in combination with the addition of the EGFR inhibitor gefitinib or MEK inhibitors for the selection of oncogenic KRAS or PIK3CA mutations in intestinal organoids, respectively72,73. TP53 mutations can be selected by the addition of MDM2 inhibitor Nutlin-3, which enables straightforward mutagenesis of multiple cancer genes by CRISPR multiplexing with TP53 (ref. 75). An example of morphology-based functional selection is the swelling response of intestinal organoids that carry either a wild-type (swelling) or mutant (no swelling) CFTR gene93,104,111. If no functional selection is available for the desired edit, transfection selection is the preferred strategy. The choice can be made either by FACS sorting based on fluorescence101 or by antibiotic resistance that is acquired upon integration into the coding sequence of a gene to create a knockout77 or by co-transfection of hygromycin resistance piggyBac119 (Fig. 5).
Patient-derived organoids are dissociated into single cells, upon which genome-engineering agents are delivered by electroporation, lipofection or lentiviral transduction. Selection for edited cells can be performed based on transfection selection or functional selection. After Sanger validation, clonal isogenic organoid pairs can be used for variant impact qualification, drug discovery and screening and assessment of the safety of genome-engineering approaches.
Outlook
Conventional CRISPR engineering through active Cas9 nucleases that create DSBs in the genome is highly efficient; however, it can induce DNA damage and be detrimental to the cell. To overcome these issues, next-generation CRISPR tools have been developed that no longer require the induction of a DSB to induce genetic alterations. The first class, base editors, allow for the introduction of either C>T or A>G base changes and have already proved their potential in vitro and in vivo. The need for more versatile genome-editing tools has led to the development of prime editors that can induce all transition and transversion point mutations, as well as introduce DNA insertions and deletions. These developments have allowed modelling or repair of over 90% of all genetic variants described in human disease, simply by selecting the most optimal genome-engineering tool for the desired genetic alteration. The application of CRISPR tools can be extended to organoids that can be derived from ASCs from both healthy and diseased donors. Complex ASC-derived isogenic disease models have been developed to reveal the mechanisms of disease progression.
Although creating knockouts of interest in ASC-derived organoids is not difficult, modelling single-nucleotide variants and larger genomic alterations remains a challenge. CBE and ABE have proved to be efficient for genome engineering and disease modelling of organoids; however, simple and robust application of prime editing could substantially increase the scope of single-nucleotide variants modelling because it can mediate all base changes. The suggested improvements to prime-editing strategies enhance the editing efficiency in HEK293T cells, but it remains to be seen whether they also prove to be effective in ASC-derived organoids, where editing efficiencies appear to be more difficult to predict.
Even if further development of the current genome-engineering toolbox results in more robust genome engineering, we can still engineer ‘only’ 90% of the genetic variants observed in patients using the current toolset. The remaining 10% of disease-causing mutations involve larger chromosomal alterations such as larger inversion, deletions and insertions, up to the loss or duplication of a complete chromosome. Efforts are ongoing to increase the target scope of genome engineering. Two new iterations of prime editing could be part of the solution. Insertions of up to 5 kb and inversion of DNA pieces of up to 40 kb can be achieved by pairing prime editing with site-specific recombinases66. Combining prime editing with integrases increases the potential of genomic integration up to 36 kb without the need for DSBs121.
Genome-wide CRISPR screens have already been used in organoids to obtain biological insight in tumour development and colitis. However, to perform a high-quality CRISPR screen, a library saturation of up to 500-fold is needed, requiring tens of millions of transfected or transduced adult stem cells90. Scalability in ASC-derived organoids is expensive owing to the cost of 3D matrices and growth factors. To resolve this issue, a 3D matrix consisting of only 5% basement membrane extract (compared to the conventional 50–100%) has been benchmarked for CRISPR screens in organoids, resulting in easy and, most importantly, cheap expansion of cancer organoid cells122. This protocol adaptation could further simplify genome-wide CRISPR screens in organoids derived from different tissues. Moreover, most genome-wide CRISPR libraries still use NHEJ to create genetic knockouts. Genome-wide base editor screens for DSB-free screening of disease variants have been developed123, which could be expanded to ASC-derived organoids to enable high-throughput and accurate qualification of genetic variants in the future.
The combination of CRISPR and ASC-derived organoids could also benefit the clinical translation of genome engineering. Despite its high efficiency, safety remains the biggest concern for CRISPR–Cas9-mediated in vivo genome engineering. ASC-derived organoids can address safety concerns because gene repair can be performed ex vivo followed by rigorous off-target analysis. Moreover, organoids can be rapidly expanded, and the safely corrected clone can then be expanded and transplanted back into the patient to repopulate the affected organ. However, tissue-specific transplantation protocols do not yet exist for most tissues. Transplantation of ASC-derived organoids have originally focused on the first established organoid system, the mouse intestinal organoids4,124,125, whose success laid the foundations of choloangiocyte, thyroid and salivary gland organoids transplantation25,126,127.
Despite transplantation into humans not being common practice at present, ASC-derived organoids could already improve the safety of genome-engineering technologies. Clinical trials applying CRISPR as a therapeutic strategy have already started, with one standing out in particular. Systemic injection of lipid nanoparticles containing SpCas9 mRNA and a sgRNA targeting TTR, whose mutation is associated with transthyretin amyloidosis, substantially decreased the baseline serum TTR in all subjects128. Prior to injecting human subjects with CRISPR reagents, the efficacy of the strategy was assessed in cynomolgus monkeys after careful selection of the sgRNA based on on-target efficiency and specificity. However, because this treatment aims to target the entire human liver, billions of cells must undergo CRISPR engineering with nuclease-active Cas9s. It is almost impossible to control off- and on-target adverse effects in such a vast number of cells, because uncontrolled cell growth can be induced by a single chromosomal rearrangement, thereby compromising safety. Moreover, despite validating specificity in cynomolgus monkeys, no safety experiment has been performed in the cells of patients. ASC-derived organoids could fill this gap.
With the development of DSB-free genome-engineering strategies, the majority of safety concerns can be addressed. Although the first generations of CBEs exhibited extensive off-target effects owing to overexpression of APOBEC, ABEs have not shown any genome-wide off-target effects59. Upgraded iterations of CBEs have also reported reduced sgRNA-independent off-target effects129. Furthermore, despite the undesirable on-target edits observed in prime editing, edits at off-target sites are mostly absent130. One likely reason is the Cas9-nickase architecture, which requires two nicks close to each other to induce a DSB131. Therefore, we envision that these DSB-free ‘next-generation CRISPR tools’ will take over the therapeutic space.
CRISPR-based genome engineering in organoids holds great promise for disease modelling and for patient care in the future. The rapid development of new genome-engineering technologies that allow the scalable induction of increasingly complex DNA mutations further highlights the applicability of CRISPR in ASC-derived organoids.
Citation diversity statement
We acknowledge that papers authored by scholars from historically excluded groups are systematically under-cited. Here, we have made every attempt to reference relevant papers in a manner that is equitable in terms of racial, ethnic, gender and geographical representation.
References
Visscher, P. M. et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017).
Xuan, J., Yu, Y., Qing, T., Guo, L. & Shi, L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 340, 284–295 (2013).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). This article contains the first description of the CRISPR–Cas9 system as a potential tool for RNA-programmable genome engineering.
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Kapałczyńska, M. et al. 2D and 3D cell cultures — a comparison of different types of cancer cell cultures. Arch. Med. Sci. 14, 910–919 (2018).
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Kim, J., Koo, B. K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011).
Barker, N. et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010).
Schutgens, F. et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol. 37, 303–313 (2019).
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013).
Linnemann, J. R. et al. Quantification of regenerative potential in primary human mammary epithelial cells. Development 142, 3239–3251 (2015).
Boretto, M. et al. Development of organoids from mouse and human endometrium showing endometrial epithelium physiology and long-term expandability. Development 144, 1775–1786 (2017).
Lõhmussaar, K. et al. Patient-derived organoids model cervical tissue dynamics and viral oncogenesis in cervical cancer. Cell Stem Cell 28, 1380–1396.e6 (2021).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Hu, H. et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606.e19 (2018).
Sachs, N. et al. Long‐term expanding human airway organoids for disease modeling. EMBO J. 38, e100300 (2019).
Nikolić, M. Z. et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. eLife 6, e26575 (2017).
Ren, W. et al. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl Acad. Sci. USA 111, 16401–16406 (2014).
Bannier-Hélaouët, M. et al. Exploring the human lacrimal gland using organoids and single-cell sequencing. Cell Stem Cell 28, 1221–1232.e7 (2021).
Mullenders, J. et al. Mouse and human urothelial cancer organoids: a tool for bladder cancer research. Proc. Natl Acad. Sci. USA 116, 4567–4574 (2019).
Karthaus, W. R. et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175 (2014).
van der Vaart, J. et al. Adult mouse and human organoids derived from thyroid follicular cells and modeling of Graves’ hyperthyroidism. Proc. Natl Acad. Sci. USA 118, e2117017118 (2021).
Ogundipe, V. M. L. et al. Generation and differentiation of adult tissue-derived human thyroid organoids. Stem Cell Rep. 16, 913–925 (2021).
Sato, T. et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). This article describes the first adult-stem-cell-derived organoid cultures derived from the mouse intestine.
Wright, A. V., Nuñez, J. K. & Doudna, J. A. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164, 29–44 (2016).
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014).
Ghezraoui, H. et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell 55, 829–842 (2014).
Zhang, X. H., Tee, L. Y., Wang, X. G., Huang, Q. S. & Yang, S. H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4, e264 (2015).
Kleinstiver, B. P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Vakulskas, C. A. et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 24, 1216–1224 (2018).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Tsai, S. Q. et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Leibowitz, M. L. et al. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet. 53, 895–905 (2021).
Branzei, D. & Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9, 297–308 (2008).
Maruyama, T. et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 33, 538–542 (2015).
Lin, S., Staahl, B. T., Alla, R. K. & Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Chavez, A. et al. Highly-efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015).
Vojta, A. et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 44, 5615–5628 (2016).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). This article reports the first base-editing system by fusing cytidine deaminase APOBEC to nickase- and nuclease-inactive Cas9 allowing for C-to-T base editing.
Cascalho, M. Advantages and disadvantages of cytidine deamination. J. Immunol. 172, 6513–6518 (2004).
Komor, A. C. et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774 (2017).
Zafra, M. P. et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 36, 888–896 (2018).
Koblan, L. W. et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–848 (2018).
Levy, J. M. et al. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 4, 97–110 (2020).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). This article describes the first adenine base editor that allows for A-to-G base editing without the need for DSBs.
Esvelt, K. M., Carlson, J. C. & Liu, D. R. A system for the continuous directed evolution of biomolecules. Nature 472, 499–503 (2011).
Gaudelli, N. M. et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900 (2020).
Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020).
Hu, J. H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
Nishimasu, H. et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 9, 1259–1262 (2018).
Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR–Cas9 variants. Science. 368, 290–296 (2020).
Yu, S.-Y. et al. Increasing the targeting scope of CRISPR base editing system beyond NGG. CRISPR J. 5, 187–202 (2022).
Pavlov, Y. I. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 8, 647–656 (2019).
Zuo, E. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 292, eaav9973 (2019).
Yu, Y. et al. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat. Commun. 11, 2052 (2020).
Kurt, I. C. et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 39, 41–46 (2021).
Koblan, L. W. et al. Efficient C•G-to-G•C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat. Biotechnol. 39, 1414–1425 (2021).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). This article presents prime editing as a tool that can potentially repair 89% of all disease-causing mutations observed in humans without the need for DSBs.
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Anzalone, A. V. et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 40, 731–740 (2021).
Lin, Q. et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 39, 923–927 (2021).
Choi, J. et al. Precise genomic deletions using paired prime editing. Nat. Biotechnol. 40, 218–226 (2022).
Nelson, J. W. et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-01039-7 (2021).
Chen, P. J. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e29 (2021).
Fearon, E. F. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Drost, J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015).
Matano, M. et al. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262 (2015).
Dekkers, J. F. et al. Modeling breast cancer using CRISPR-Cas9-mediated engineering of human breast organoids. J. Natl. Cancer Inst. 112, 540–544 (2020).
Artegiani, B. et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell 24, 927–943.e6 (2019).
Seino, T. et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 22, 454–467.e6 (2018).
Lee, J. et al. Reconstituting development of pancreatic intraepithelial neoplasia from primary human pancreas duct cells. Nat. Commun. 8, 14686 (2017).
Drost, J. et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 358, 234–238 (2017).
Jager, M. et al. Deficiency of nucleotide excision repair is associated with mutational signature observed in cancer. Genome Res. 29, 1067–1077 (2019).
Kawasaki, K. et al. Chromosome engineering of human colon-derived organoids to develop a model of traditional serrated adenoma. Gastroenterology 158, 638–651.e8 (2020).
Artegiani, B. et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat. Cell Biol. 22, 321–331 (2020).
Lo, Y. H. et al. A CRISPR/Cas9-engineered ARID1A-deficient human gastric cancer organoid model reveals essential and nonessential modes of oncogenic transformation. Cancer Discov. 11, 1562–1581 (2021).
Kawasaki, K. et al. An organoid biobank of neuroendocrine neoplasms enables genotype–phenotype mapping. Cell 183, 1420–1435.e21 (2020).
Yan, H. H. N. et al. Organoid cultures of early-onset colorectal cancers reveal distinct and rare genetic profiles. Gut 69, 2165–2179 (2020).
Post, J. B. et al. CRISPR-induced RASGAP deficiencies in colorectal cancer organoids reveal that only loss of NF1 promotes resistance to EGFR inhibition. Oncotarget 10, 1440–1457 (2019).
Bock, C. et al. High-content CRISPR screening. Nat. Rev. Methods Prim. 2, 8 (2022).
Wang, T. et al. Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015).
Shalem, O. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–88 (2014).
Michels, B. E. et al. Pooled in vitro and in vivo CRISPR–Cas9 screening identifies tumor suppressors in human colon organoids. Cell Stem Cell 26, 782–792.e7 (2020).
Ringel, T. et al. Genome-scale CRISPR screening in human intestinal organoids identifies drivers of TGF-β resistance. Cell Stem Cell 26, 431–440.e8 (2020).
Boettcher, S. et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 365, 599–604 (2019).
Stolze, B., Reinhart, S., Bulllinger, L., Fröhling, S. & Scholl, C. Comparative analysis of KRAS codon 12, 13, 18, 61, and 117 mutations using human MCF10A isogenic cell lines. Sci. Rep. 5, 8535 (2014).
Geurts, M. H. et al. Evaluating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids. Life Sci. Alliance 4, 1–12 (2021).
Schene, I. F. et al. Prime editing for functional repair in patient-derived disease models. Nat. Commun. 11, 5352 (2020).
van Rijn, J. M. et al. Intestinal failure and aberrant lipid metabolism in patients with DGAT1 deficiency. Gastroenterology 155, 130–143.e15 (2018).
Nanki, K. et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature 577, 254–259 (2020).
Lamers, M. M. et al. SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 (2020).
Zhou, J. et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 26, 1077–1083 (2020).
Geurts, M. H., van der Vaart, J., Beumer, J. & Clevers, H. The organoid platform: promises and challenges as tools in the fight against COVID-19. Stem Cell Rep. 16, 412–418 (2021).
Beumer, J. et al. A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat. Commun. 12, 5498 (2021).
Veres, A. et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15, 27–30 (2014).
Wu, X. et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32, 670–676 (2014).
Lombaert, I. M. A. et al. Rescue of salivary gland function after stem cell transplantation in irradiated glands. PLoS One 3, e2063 (2008).
Schwank, G. et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658 (2013). This article reports the first proof of the potential clinical application of CRISPR by repairing the most common mutation that causes cystic fibrosis in patient-derived intestinal organoids.
Sosnay, P. R. et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat. Genet. 45, 1160–1167 (2013).
Dekkers, J. F. et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 19, 939–945 (2013).
Dekkers, J. F. et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 8, 344ra84–344ra84 (2016).
Berkers, G. et al. Rectal organoids enable personalized treatment of cystic fibrosis. Cell Rep. 26, 1701–1708.e3 (2019).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Maule, G. et al. Allele specific repair of splicing mutations in cystic fibrosis through AsCas12a genome editing. Nat. Commun. 10, 3556 (2019).
Geurts, M. H. et al. CRISPR-based adenine editors correct nonsense mutations in a cystic fibrosis organoid biobank. Cell Stem Cell 26, 503–510.e7 (2020). This article reports the first proof of DSB-free gene repair in adult-stem-cell-derived organoids by repairing mutations that cause cystic fibrosis in patient-derived organoids without genome-wide off-target effects.
Schene, I. F. et al. Mutation-specific reporter for optimization and enrichment of prime editing. Nat. Commun. 13, 1028 (2022).
van der Vaart, J. et al. Modelling of primary ciliary dyskinesia using patient‐derived airway organoids. EMBO Rep. 22, e52058 (2021).
Kuscu, C. et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat. Methods 14, 710–712 (2017).
Wang, X. et al. Efficient gene silencing by adenine base editor-mediated start codon mutation. Mol. Ther. 28, 431–440 (2020).
Kluesner, M. G. et al. CRISPR–Cas9 cytidine and adenosine base editing of splice-sites mediates highly-efficient disruption of proteins in primary and immortalized cells. Nat. Commun. 12, 2437 (2021).
Conant, D. et al. Inference of CRISPR edits from sanger trace data. CRISPR J. 5, 123–130 (2022).
Arbab, M. et al. Determinants of base editing outcomes from target library analysis and machine learning. Cell 182, 463–480.e30 (2020).
Andersson-Rolf, A. et al. One-step generation of conditional and reversible gene knockouts. Nat. Methods 14, 287–289 (2017).
Sun, D. et al. A functional genetic toolbox for human tissue-derived organoids. eLife 10, e67886 (2021).
Yarnall, M. T. N. et al. Drag-and-drop genome insertion of large sequences without DNA cleavage using CRISPR-directed integrases. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01527-4 (2022).
Price, S. et al. A suspension technique for efficient large-scale cancer organoid culturing and perturbation screens. Sci. Rep. 12, 5571 (2022).
Hanna, R. E. et al. Massively parallel assessment of human variants with base editor screens. Cell 184, 1064–1080.e20 (2021).
Drost, J. & Clevers, H. Translational applications of adult stem cell-derived organoids. Development 144, 968–975 (2017).
Yui, S. et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 18, 618–623 (2012).
Pringle, S. et al. Human salivary gland stem cells functionally restore radiation damaged salivary glands. Stem Cell 34, 640–652 (2016).
Sampaziotis, F. et al. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science 371, 839–846 (2021).
Gillmore, J. D. et al. CRISPR–Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021). This article describes a landmark clinical trial in which patients are injected with nuclease-active Cas9 and a sgRNA targeting the transthyretin gene that causes amyloid plaques in the liver.
Doman, J. L., Raguram, A., Newby, G. A. & Liu, D. R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38, 620–628 (2020).
Aida, T. et al. Prime editing primarily induces undesired outcomes in mice. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2020.08.06.239723v1 (2020).
Shen, B. et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399–402 (2014).
Muller, H. J. Artificial transmutation of the gene. Science 66, 84–87 (1927).
Brenner, S. The genetics of Ceanorhabditis elegans. Genetics 77, 71–94 (1974).
Nüsslein-volhard, C. & Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 (1980).
Doudna, J. A. & Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096–1258096 (2014).
Scherer, S. & Davis, R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl Acad. Sci. USA 76, 4951–4955 (1979).
Smithies, O., Gregg, R. G., Boggst, S. S., Koralewski, M. A. & Kucherlapati, R. S. Insertion of DNA sequences into the human chromosomal β-globin locus by homologous recombination. Nature 317, 230–236 (1985).
Rudin, N., Sugarman, E. & Haber, J. E. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics 122, 519–534 (1989).
Rouet, P., Smih, F. & Jasin, M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14, 8096–8106 (1994).
Epinat, J. C. et al. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 31, 2952–2962 (2003).
Wood, A. J. et al. Targeted genome editing across species using ZFNs and TALENs. Science 333, 307 (2011).
Hu, J. H., Davis, K. M. & Liu, D. R. Chemical biology approaches to genome editing: understanding, controlling, and delivering programmable nucleases. Cell Chem. Biol. 23, 57–73 (2016).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Brouns, S. J. J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–965 (2008).
Akcakaya, P. et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561, 416–419 (2018).
Shirley, J. L., de Jong, Y. P., Terhorst, C. & Herzog, R. W. Immune responses to viral gene therapy vectors. Mol. Ther. 28, 709–722 (2020).
Wu, Z., Asokan, A. & Samulski, R. J. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol. Ther. 14, 316–327 (2006).
Nieuwenhuis, B. et al. Optimization of adeno-associated viral vector-mediated transduction of the corticospinal tract: comparison of four promoters. Gene Ther. 28, 56–74 (2021).
Burger, C. et al. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol. Ther. 10, 302–317 (2004).
Naso, M. F., Tomkowicz, B., Perry, W. L. & Strohl, W. R. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 31, 317–334 (2017).
Liu, P. et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat. Commun. 12, 2121 (2021).
Böck, D. et al. In vivo prime editing of a metabolic liver disease in mice. Sci. Transl. Med. 14, eabl9238 (2022).
Segel M. et al. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 185, 882–889 (2021).
June, C. H., O’Connor, R. S., Kawalekar, O. U., Ghassemi, S. & Milone, M. C. CAR T cell immunotherapy for human cancer. Science. 359, 1361–1365 (2018).
Frangoul, H. et al. CRISPR–Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 384, 252–260 (2021).
Watanabe, S. et al. Transplantation of intestinal organoids into a mouse model of colitis. Nat. Protoc. 17, 649–671 (2022).
Sugimoto, S. et al. An organoid-based organ-repurposing approach to treat short bowel syndrome. Nature 592, 99–104 (2021).
Acknowledgements
The authors thank J. Beumer for providing confocal images of human intestinal organoids, S. Gandhi for providing confocal images of human fetal hepatocyte organoids and J. van der Vaart for providing confocal images of murine thyroid organoids.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
H.C. is inventor on several patents related to organoid technology; his full disclosure is given at https://www.uu.nl/staff/JCClevers/. H.C. is currently head of pharma Research Early Development (pRED) at Roche. H.C. holds several patents on organoid technology. Their application numbers, followed by their publication numbers (if applicable), are as follows: PCT/NL2008/050543, WO2009/022907; PCT/NL2010/000017, WO2010/090513; PCT/IB2011/002167, WO2012/014076; PCT/IB2012/052950, WO2012/168930; PCT/EP2015/060815, WO2015/173425; PCT/EP2015/077990, WO2016/083613; PCT/EP2015/077988, WO2016/083612; PCT/EP2017/054797, WO2017/149025; PCT/EP2017/065101, WO2017/220586; PCT/EP2018/086716, n/a; and GB1819224.5, n/a. M.H.G. is currently a scientist at Xilis BV.
Peer review
Peer review information
Nature Reviews Bioengineering thanks Nicholas Zachos and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Geurts, M.H., Clevers, H. CRISPR engineering in organoids for gene repair and disease modelling. Nat Rev Bioeng 1, 32–45 (2023). https://doi.org/10.1038/s44222-022-00013-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44222-022-00013-5
This article is cited by
-
Inner Ear Organoids: Strengths and Limitations
Journal of the Association for Research in Otolaryngology (2024)
-
One-step generation of tumor models by base editor multiplexing in adult stem cell-derived organoids
Nature Communications (2023)