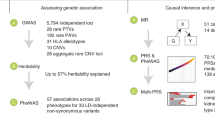
Abstract
Background
The identification of suitable biomarkers is of crucial clinical importance for the early diagnosis of treatment-resistant schizophrenia (TRS). This study aims to comprehensively analyze the association between TRS and blood and urine biomarkers.
Methods
Candidate TRS-related single nucleotide polymorphisms (SNPs) were obtained from a recent genome-wide association study. The UK Biobank cohort, comprising 376,807 subjects with blood and urine biomarker testing data, was used to calculate the polygenic risk score (PRS) for TRS. Pearson correlation analyses were performed to evaluate the correlation between TRS PRS and each of the biomarkers, using calculated TRS PRS as the instrumental variables. Bidirectional two-sample Mendelian randomization (MR) was used to assess potential causal associations between candidate biomarkers with TRS.
Results
Here we identify a significant association between TRS PRS and phosphate (r = 0.007, P = 1.96 × 10−4). Sex subgroup analyses identify seven and three candidate biomarkers associated with TRS PRS in male and female participants, respectively. For example, total protein and phosphate for males, creatinine and phosphate for females. Bidirectional two-sample MR analyses indicate that TRS is negatively associated with cholesterol (estimate = −0.363, P = 0.008). Conversely, TRS is positively associated with total protein (estimate = 0.137, P = 0.027), mean corpuscular volume (estimate = 0.032, P = 2.25 × 10−5), and mean corpuscular hemoglobin (estimate = 0.018, P = 0.007).
Conclusions
Our findings provide insights into the roles of blood and urine biomarkers in the early detection and treatment of TRS.
Plain language summary
People with schizophrenia experience periods of time during which they misperceive reality. Some people with schizophrenia do not respond well to the usual drugs that are used to relieve their symptoms. This type of schizophrenia is known as treatment-resistant schizophrenia (TRS). We looked at differences in the genes (inherited characteristics), blood and urine of a group of people in the UK with schizophrenia to see if people with TRS have particular characteristics that would enable them to be distinguished from patients with schizophrenia who tend to respond to usual treatment. We found several differences in the blood that could be used to predict which people might get TRS, including some that were specific to men or women. These discoveries are important because they can help doctors identify people who are more likely to develop TRS earlier, enabling them to avoid using treatments that might not work well for them.
Similar content being viewed by others
Introduction
Schizophrenia is a complex cognitive and behavioral syndrome that seriously affects the quality of life. It is characterized by emotional, cognitive, perceptual, and thought disorders1,2. Some individuals with schizophrenia experience treatment-resistant symptoms characterized by severe dysfunction in which symptoms do not completely respond to at least two first-line antipsychotic drugs3. Treatment-resistant schizophrenia (TRS) is a complex clinical condition that affects approximately 30% of people living with schizophrenia4. Studies suggest that TRS may have a higher heritability compared to schizophrenia, indicating that TRS may be a more familial phenotype and distinguishable from non-TRS cases based on its genetic underpinnings5. Patients with TRS have poorer prognosis and worse functional outcomes compared to patients with other severe psychiatric disorders6. By elucidating the genetic predisposition to specific biomarkers associated with TRS, it may be possible to develop more accurate predictive models for identifying individuals at risk of developing TRS. This could ultimately lead to earlier interventions and improved outcomes.
Peripheral blood and urine biomarkers are frequently measured to diagnose and evaluate chronic disease conditions7. Many biochemical indicators in peripheral blood and urine have been found to be abnormal in patients with mental diseases, such as schizophrenia8, major depressive disorder9,10, autism spectrum disorder11, and anxiety12. A recent large observational study demonstrated that higher concentrations of tau protein in peripheral blood were associated with cognitive degeneration in patients with Alzheimer’s disease (AD)13. It is noteworthy that peripheral blood biomarkers hold promise as a substitute for the central nervous system in characterizing psychiatric disorders, although their role has yet to be widely applied in clinical practice. Urine is also a convenient and appropriate substance for use in diagnostic or predictive tests for disease, as subtle changes in urine are accumulated in the blood and are unaffected by homeostasis mechanisms14.
Early detection and diagnosis of schizophrenia and TRS are crucial for their prevention and treatment. Screening for suitable biomarkers is essential for early diagnosis. In the past decade, considerable efforts have been made to identify reliable biomarkers for the early detection of schizophrenia. Blood biomarkers are considered as a viable option because the dysregulation of epigenetic patterns, gene expression, metabolic and inflammatory molecules in peripheral blood have unique patterns in individuals with schizophrenia15. Abnormal metabolic, immune, and hormonal alterations have been found in the blood of patients with schizophrenia16, such as inflammatory biomarkers (C-reactive protein and interleukins)17 and neurotrophic biomarkers (BDNF protein)18. Additionally, measurements of urinary bufotenine levels in patients with schizophrenia and healthy controls without psychotic symptoms found that increased urine levels of the endogeneous psychogenic molecule bufotenine may play a vital role in schizophrenia19. Studies have also demonstrated the genetic basis of serum and urine biomarkers and their causal effects on psychiatric disorders7. Although the information contained in blood and urine is not entirely comprehensive, the observed changes may still be used for diagnostic and monitoring purposes.
The polygenic risk score (PRS) is a powerful tool for predicting an individual’s genetic inclination and the severity of mental disorders20. This is achieved by weighting and calculating the effect size of SNPs20. The PRS has been widely used in exploring the correlations between the genetic susceptibility of multiple diseases in phenome-wide association studies21,22.
In this study, we comprehensively analyze the correlation between blood and urine biomarkers with TRS. The TRS PRS are calculated using genotype data from the UK Biobank (UKB) cohort. Pearson correlation analyses are performed to investigate the correlation between each blood or urine biomarker and TRS. Bidirectional two-sample Mendelian randomization analyses are performed to validate the candidate correlations between biomarkers and TRS. In the analysis of TRS PRS-associated blood and urine biomarkers across the total UKB sample, a significant association is found with phosphate at the Bonferroni correction threshold. Six biomarkers exhibit candidate association signals with TRS PRS. Bidirectional Mendelian randomization analyses reveal significant associations between TRS and various biomarkers. TRS is positively associated with mean corpuscular volume but negatively associated with total protein according to IVW MR analyses. MR-Egger analyses suggest an opposite causal direction between TRS and total protein and reveal a causal relationship between TRS and cholesterol. Weighted median MR analyses confirm the associations observed in IVW analyses and reveal additional associations with lymphocyte count and mean corpuscular hemoglobin. Our study provides insight into the application of polygenic risk scores and highlights the importance of blood and urine biomarkers in the early diagnosis of TRS.
Methods
Biomarker phenotypes in serum and urine in UKB cohort
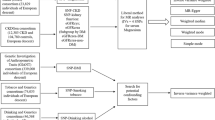
The phenotypic and genotypic data used in this study were obtained from the UKB, which conducted a large prospective cohort study from 2006 to 201023. The UKB performed laboratory testing of commonly measured biomarkers in serum (Category 100080) and urine (Category 100083) on a cohort with extensive phenotype and genome-wide genotype data, including the unrelated individuals in this study23. Health-related records of each participant, including age and sex, were collected through either a screenshot questionnaire or verbal interview within the assessment center. The urine assays category contains information on the assays that have been performed on the UKB urine samples (https://biobank.ndph.ox.ac.uk/showcase/label.cgi?id=100083), while the blood assays category contains information on the assays that have been performed on the UKB blood samples and their results (https://biobank.ndph.ox.ac.uk/showcase/label.cgi?id=100080), including blood count (Category 100081) and blood biochemistry (Category 17518). Blood count contains results of hematological assays that were performed on whole blood before further processing, including data on acquisition time and number of freeze-thaw cycles, such as basophils, eosinophils, monocytes, and neutrophils. Blood biochemistry contains a range of key biochemistry markers that were measured in blood samples collected at recruitment and at repeat assessment. The ethical approval of UKB was granted by the National Health Service National Research Ethics Service (reference 11/NW/0382). All participants gave informed consent for participation in the UKB. Permission to access and analyze UKB data was approved under UKB project 46478. The need to seek additional ethical approval for our study from our University was waived because the study involved the secondary use of data.
UK Biobank genotyping, imputation and quality control
Genome-wide genotyping was performed in UKB individuals using either the Affymetrix UKB Axiom array or Affymetrix UK BiLEVE Axiom, which included 812,428 SNPs. Imputation was conducted using IMPUTE2 with the reference panel of the UK10K projects, Haplotype Reference Consortium, and 1000 Genomes23. For quality control on the genotype data, the UKB excluded the SNPs with INFO < 0.9, Hardy–Weinberg equilibrium (HWE) testing p values < 0.0001, minor allele frequencies (MAF) < 0.01 and genotyping call rate <95%. A total of 488,377 individuals and 805,426 SNPs were kept after applying quality control (QC). After removing individuals who reported inconsistencies between self-reported sex and genetic sex, as well as those with missing covariate information, 376,807 individuals of white British ancestry subset (UK Biobank field ID: 21000)23 were retained for further analysis23.
Polygenic risk score datasets of treatment resistance in schizophrenia
The GWAS summary statistics of TRS were obtained from Pardiñas et al.4. The study analyzed the GWAS of TRS based on a sample size of 10,501 individuals with TRS and 20,325 individuals without TRS. All individuals with TRS were administered clozapine in the UK, in accordance with the National Institute for Health and Care Excellence guidelines for TRS, after the failure of at least two trials of antipsychotics24. Due to the involvement of different datasets25 and genotyping arrays26 in this analysis, the processing of TRS and non-TRS GWAS samples was conducted separately on data generated from the original studies. Imputations were performed using the SHAPEIT/IMPUTE2 workflow27,28. The combination results of non-TRS GWAS by the Psychiatric Genomics Consortium were analyzed using the fixed-effects program in METAL29. The combined samples with SNPs called in <20,000 and any strand ambiguous markers with minor allele frequency (MAF) ≥ 40% were removed. Detailed information on genotyping, imputation, quality control, and statistical analysis is available in the eMethods by Pardiñas et al.4. To generate association statistics that accurately reflect differences between TRS and non-TRS groups, Pardinas et al. utilized the test for interaction proposed by Altman and Bland30. This test is similar to a fixed-effect test for moderators in the meta-analytic setting4.
PRS calculation of treatment resistance in schizophrenia in UK Biobank cohort
The clumping algorithm in PRSice-2 (version 2.3.5) was configured to identify any SNPs within 250 kb in linkage disequilibrium (LD) with an r2 threshold greater than 0.1 and ‘clump’ them together to be represented by the most significantly associated SNP within each LD block, known as the index SNP31. PRSice-2 was employed to generate PRS for TRS, utilizing recent large GWAS summary statistics of TRS4. Significance values and effect sizes from the UKB cohort were used to generate the best PRS model. In brief, the PRS for each individual was calculated by summing the effect sizes from all the SNPs included in the best model. To generate the best-fit PRS of TRS, clozapine use (UKB Data-Field 20003) was used as a proxy measure of TRS, with the top 10 principle components (PC) of population structure, age, and sex included as covariates. The PRS analysis was conducted, comparing 45 participants using clozapine to a larger group of 376,762 participants not using clozapine, irrespective of their schizophrenia diagnosis status. The best model was derived by testing the inclusion of SNPs (401–89,618 SNPs) from a range of P value thresholds from 5 × 10−8 to 1 with an incremental interval of 0.0005 (--interval 0.0005 --lower 5e−08), to determine which threshold gave the largest Nagelkerke’s R2 value. These SNPs were then used to generate PRS for each individual in UKB cohorts. The performance of the best model for the selection of SNP markers was evaluated by the area under the receiver operating characteristic (ROC) curve (AUC) using the pROC R package32.
Statistics and reproducibility
The PRS of TRS were initially adjusted for the PC1–PC10 using linear regression models. The rstandard function was then utilized to calculate the standardized residual of PRS, which is obtained by dividing the residual by its standard deviation (SD). The measurements of 59 blood and 4 urine biomarkers were adjusted for potential confounding variables, including sex (excluded in sex stratification analysis) and age, through linear regression models. The aim of this model was to extract the residual biomarker measurements and eliminate the effects of these covariates from subsequent Pearson correlation analysis. The resulting residuals were then utilized as the phenotypic values of biomarkers for the subsequent analyses. Pearson correlation analyses were conducted to investigate the correlation between each biomarker and TRS phenotype, with standardized PRS calculated as instrumental variables. Males and females were divided into separate groups to facilitate a better analysis of differences between sex stratification groups. The significant P value thresholds of Pearson correlation should be 7.94 × 10−4 (0.05/63 independent biomarkers) after strict Bonferroni correction. All statistical analyses, including AUC, linear regression, and Pearson correlation analyses, were performed using R software (version R 4.3.0).
Bidirectional two-sample Mendelian randomization analyses
To validate potential causal associations between blood and urine biomarkers with TRS, we performed bidirectional two-sample MR analyses for significantly correlated blood and urine biomarkers. The present study utilizes candidate blood and urine biomarkers derived from GWAS summary statistics, which are publicly available at https://gwas.mrcieu.ac.uk/. The GWAS summary statistics dataset of TRS was obtained from the GWAS data by Pardiñas et al.4. Initially, we extracted SNPs that showed genome-wide statistical significance in association with exposure phenotypes. Subsequently, two-sample MR analyses were carried out using R (version 4.3.0) and the TwoSampleMR package33. Exposure and outcome GWAS summary statistics were harmonized by aligning summary statistics to infer positive strand alleles using allele frequencies for palindromes to ensure that the effect of an SNP on the exposure, and the effect of that same SNP on the outcome, corresponds to the same allele.
We performed fixed-effects analysis of genetic instruments using inverse variance-weighted (IVW) MR34. The MR-Egger regression analysis and weighted median MR approach were utilized to evaluate the robustness of our findings35,36. To assess horizontal pleiotropy, we tested for the presence of statistically significant (P < 0.05) heterogeneities in MR-Egger analyses using the Cochran Q statistic37. In assessing consistency and robustness, we sought estimates that substantially agreed in direction and magnitude (overlapping confidence intervals) across complementary MR methods.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Descriptive characteristics of study participants
This cross-sectional study included a total of 376,807 participants aged between 39 and 73 years, with a mean age of 56.99 years and a standard deviation of 7.93 years. The study population consisted of 202,434 male and 174,373 female subjects. Biomarker test results from blood and urine assays were obtained from all 376,807 study participants and were utilized for further analysis. The detailed statistical information on serum and urine biomarkers in this study is presented in Supplementary Data 1.
PRS Nagelkerke R 2 and AUC
The PRS models suggest that a stricter cutoff may result in the missing of informative SNPs, while a looser cutoff may introduce noise by including SNPs with spurious TRS association (Supplementary Data 2). The best-fit PRS at a P value threshold = 0.0015 (R2 = 0.007) was utilized for downstream analysis (Supplementary Fig. 1A). A screening test is considered better than pure chance if the AUC value is >0.538. Using the PRS of the best model as the predictor and the TRS participant group as the outcome, the AUC was 0.61 (95% CI: 0.48–0.74). With the threshold of PRS cutoff 0.252, the sensitivity (true positive rate, TPR) for TRS prediction is 52.17%, and the specificity (true negative rate, TFR) for TRS prediction is 78.26% (Supplementary Fig. 1B).
TRS PRS-associated blood and urine biomarkers in total, male and female population
Upon analyzing the UKB total sample, we identified a significant association between TRS PRS and phosphate (r = 0.007, P = 1.96 × 10−4) at the Bonferroni correction threshold (Fig. 1). Furthermore, we identified six biomarkers that have candidate association signals with TRS PRS, such as gamma-glutamyltransferase (r = −0.005, P = 0.002), reticulocyte percentage (r = 0.004, P = 0.008), and total protein (r = −0.004, P = 0.021). Detailed information on Pearson correlation results for the total population is presented in Supplementary Data 3. Upon testing for association with TRS PRS in male and female participants, no blood or urine biomarker exhibited statistical significance after strict Bonferroni correction (Fig. 1). However, in the male participants of the UKB cohort, we identified seven biomarkers that were potentially associated with TRS PRS at the general P threshold (Supplementary Data 4), such as total protein (r = −0.008, P = 0.003), gamma-glutamyltransferase (r = −0.006, P = 0.012), and phosphate (r = 0.006, P = 0.018). In the female participants, we observed three candidate biomarkers for TRS PRS (Supplementary Data 5), including creatinine (r = −0.007, P = 0.001), phosphate (r = 0.007, P = 0.004), and reticulocyte percentage (r = 0.005, P = 0.029).
Bidirectional Mendelian randomization analyses
Table 1 presents fundamental information on the GWAS summary data used in bidirectional two-sample MR. The sample size, number of significant SNPs, and heritability are provided for each trait analyzed. The traits investigated in this study include TRS, phosphate, gamma glutamyltransferase, reticulocyte percentage, total protein, mean corpuscular volume, mean corpuscular hemoglobin, reticulocyte count, glycated hemoglobin (HbA1c), lymphocyte count, cholesterol, red blood cell (erythrocyte) count, and creatinine. The number of genome-wide significant independent loci for each trait is indicated by SNPs (n), with SNPs within 10,000 kilobase pairs and R2 ≥ 0.001 removed.
Figure 2 presents the significant bidirectional MR results for TRS and candidate biomarkers. The IVW MR analyses indicated that TRS was positively associated with mean corpuscular volume (estimate (SE) = 0.025 (0.010), P = 0.009), while TRS was negatively associated with total protein (estimate (SE) = −0.027 (0.009), P = 0.002) (Fig. 2). In contrast, MR-Egger analyses showed an opposite causal direction between TRS and total protein (estimate (SE) = 0.137 (0.058), P = 0.027). Notably, MR-Egger analyses demonstrated a causal relationship between TRS and cholesterol (estimate (SE) = −0.363 (0.122), P = 0.008). The weighted median MR analyses showed the same causal direction as IVW analyses for the associations between mean corpuscular volume, total protein and TRS (Fig. 2). Additionally, the weighted median MR analysis showed a slightly weaker causal relationship between TRS and lymphocyte count (estimate (SE) = 0.015 (0.007), P = 0.019), and mean corpuscular hemoglobin (estimate (SE) = 0.019 (0.007), P = 0.007). Supplementary Data 6 provides detailed MR estimates of genetic instruments for TRS and candidate biomarkers based on Pearson correlation results.
The error bars indicate 95% confidence intervals (CIs). An asterisk (*) indicates the presence of pleiotropy in this MR analysis, which results in more precise estimates from MR-Egger. A hashtag (#) indicates the presence of heterogeneity in this MR analysis, which results in more precise estimates from the weighted median. TRS treatment-resistant schizophrenia, IVW inverse variance weighted, WM weighted median, MR Mendelian randomization.
To evaluate the potential for horizontal pleiotropy, we assessed heterogeneity and conducted sensitivity analyses that are more robust to pleiotropy, including weighted median MR and MR-Egger regression. Heterogeneity tests did not indicate heterogeneity in the IVW and MR-Egger estimates for the association of TRS with cholesterol, lymphocyte count, and total protein (P range, 0.089–0.960) (Supplementary Data 7). Notably, heterogeneity was observed in mean corpuscular volume, mean corpuscular hemoglobin, and TRS, so the weighted median method was more accurate for these two biomarkers36. The MR-Egger intercept analysis indicates that horizontal pleiotropy exists between TRS and cholesterol and total protein, indicating that the MR-Egger is more accurate in the MR analysis of TRS and these two biomarkers36 (Supplementary Data 8).
Discussion
In this study, we conducted a large observational and genetic PRS analysis to comprehensively evaluate the associations between blood/urine biomarkers and TRS using the UKB cohort. We then performed bidirectional MR analysis to validate the candidate causal relationship. Our findings revealed several potential correlations in blood and urine biomarkers. Further analysis provided tentative evidence for sex differences in the characteristics between TRS with blood and urine biomarkers.
Currently, there is no objective biological measurement available for the diagnosis of TRS. Phosphate is an essential mineral for humans and plays numerous functions in the body. The blood level of phosphate is tightly regulated within a narrow range. Both hyperphosphatemia and hypophosphatemia can lead to the development of diseases39. The disturbed integrity of myelin and white matter, along with dysregulation of lipid metabolism, may be involved in schizophrenia pathophysiology40. Previous studies have reported increased high-energy phosphate metabolism in the basal ganglia of neuroleptic-naive schizophrenia patients using magnetic resonance spectroscopy (MRS)41,42. However, there are very few studies investigating the relationship between phosphate concentration and TRS in schizophrenia patients. Our study provides evidence for a correlation between phosphate and TRS, suggesting that blood phosphate may be a potential biomarker in TRS.
Previous studies have identified some biomarkers associated with TRS or schizophrenia, but not with TRS PRS in our study, such as bilirubin and creatinine. Bilirubin, the final product of heme metabolism in the body, is an endogenous antioxidant with an anti-inflammatory effect43. Unbound bilirubin has been studied as a promising molecule that could be used as a possible biological marker for schizophrenia44. In a prospective study, blood levels of unbound bilirubin were higher in patients with schizophrenia than in patients with bipolar disorder45. Serum creatinine is one of the most commonly measured products in clinical chemistry laboratories worldwide46 and has been linked with many neurodegenerative diseases47. The evidence linking schizophrenia and creatinine is primarily from observational studies. Researchers have identified multiple potential metabolite biomarkers of schizophrenia, such as reduced levels of essential polyunsaturated fatty acids and creatinine48. Additionally, an association with schizophrenia was also found in urine. Previous studies have shown that urinary creatinine concentrations were reduced in patients with schizophrenia compared with healthy controls49,50. These studies, together with our results, suggest that these biomarkers may not be influenced by individual genetic inclination.
Epidemiological data has revealed sex differences in the prevalence of schizophrenia51. Similarly, sex-specific blood biomarkers were observed in this study. Glycated hemoglobin (HbA1c), lymphocyte count, and red blood cell (erythrocyte) count showed potential correlation signals in male samples, while creatinine and reticulocyte percentage showed potential correlation signals in female samples. Sex-specific differences associated with age of onset, duration, and antipsychotic response in schizophrenia may be reflected in sex-related differences in the underlying molecular pathways52. Researchers have identified the structural and neurophysiological sex characteristics of schizophrenia by focusing on specific biomarkers53. A study found that males with schizophrenia had higher glycated hemoglobin (HbA1c), lower high-density lipoprotein, and an earlier age of onset compared to females54. Neurosteroids, including DHEA, DHEA-S, and pregnenolone, are involved in the pathophysiology of schizophrenia in male patients, but not in female patients55. Therefore, our results emphasize the importance of considering sex-specific differences in TRS research and highlight the clinical potential of these blood biomarkers for predicting TRS. Although the TRS PRS demonstrates potential as a tool for identifying individuals with an elevated predisposition to developing TRS, it is not a conclusive diagnostic measure. There may be instances where individuals with a high TRS-PRS do not ultimately develop TRS, while those with a low TRS-PRS may indeed manifest symptoms of TRS. Thus, it is crucial to recognize that TRS-PRS requires complementary clinical assessments for precise diagnosis and effective treatment.
Our bidirectional MR analyses demonstrated a negative association between TRS and cholesterol. In terms of metabolic alterations, Francesca et al. observed lower levels of high-density lipoprotein (HDL)-cholesterol among male treatment-resistant patients treated with clozapine56. The study selected a total of 33 patients previously diagnosed with TRS, who were prescribed clozapine followed by PP1M and PP3M when available, and the data showed a decrease in cholesterol57. These findings are consistent with the results of our MR analysis. Furthermore, we found that TRS was positively associated with red blood cell parameters, including mean corpuscular volume and mean corpuscular hemoglobin. Studies compared differences in mean corpuscular volume and mean corpuscular hemoglobin (MCH) in patients with schizophrenia, and found differences for all red cell parameters between study groups58. Age and sex may affect various erythrocyte parameters58. Another study investigated changes in various biochemical parameters in schizophrenia patients using clozapine and found that MCH and red blood cell count (RBC) levels were lower in the clozapine group compared with healthy volunteers59. Future studies should verify our results and further explore the biological confounding factors that could explain associations between red blood cell parameters and TRS.
This study comprehensively investigates the association between blood/urine biomarkers and TRS. Our sample size maximizes power for genetic analyses. This study also has some limitations. Firstly, the TRS PRSs were computed utilizing a subgroup of the UKB cohort consisting of individuals with white British ancestry, primarily of middle-aged demographics. Therefore, caution should be exercised when applying our results to young people and other ethnic populations. Secondly, the use of residuals may have impacted the size of the effects reported, and future studies should consider alternative methods to further validate our findings. The use of independent clinical specimens or cohorts to validate our findings and investigate the underlying biological mechanisms of the observed association between candidate blood and urine biomarkers with TRS will be necessary in future studies. Thirdly, the method and accuracy of the UKB biomarker measurement have potential implications for our results. Finally, due to the highly polygenic and pleiotropic nature of the genetic architecture of psychiatric phenotypes, the core MR assumptions are easily violated60. Pleiotropy will bias estimates by reintroducing confounding, and heterogeneity in the outcome will reduce the precision of our causal estimates, making it harder to identify a true causal effect should one exist61. Therefore, all analyses should be carefully considered and cautiously interpreted.
In conclusion, our study systematically analyzed the associations between TRS and blood/urine biomarkers using UKB individual-level traits and genotype data and TRS GWAS summary data. Our study highlights the sex-related differences in the underlying blood biomarkers, identifies the associations between TRS with phosphate and cholesterol, and may provide insights to reveal the roles of blood/urine biomarkers in the development of TRS. Our findings of potential blood biomarkers for TRS may be useful for diagnostic purposes, as well as for drug development and monitoring disease progression.
Data availability
Access to the UK Biobank data can be obtained by applying to the UK Biobank Access Management System, details are at https://www.ukbiobank.ac.uk/. We will return the derived data fields following UK Biobank policy; in due course, they will be available through the UK Biobank Access Management System. The source data underlying Figs. 1 and 2 are in Supplementary Data 3–5 and 6, respectively.
Code availability
The R-based custom code for statistical analysis for this work is publicly available at https://github.com/DrBolunCheng/PRSMR/62.
References
McGrath, J., Saha, S., Chant, D. & Welham, J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol. Rev. 30, 67–76 (2008).
Owen, M. J., Sawa, A. & Mortensen, P. B. Schizophrenia. Lancet (London, England) 388, 86–97 (2016).
Howes, O. D. et al. Treatment-resistant schizophrenia: Treatment Response and Resistance in Psychosis (TRRIP) working group consensus guidelines on diagnosis and terminology. Am. J. Psychiatry 174, 216–229 (2017).
Pardiñas, A. F. et al. Interaction testing and polygenic risk scoring to estimate the association of common genetic variants with treatment resistance in schizophrenia. JAMA Psychiatry 79, 260–269 (2022).
Nucifora, F. C. Jr., Woznica, E., Lee, B. J., Cascella, N. & Sawa, A. Treatment resistant schizophrenia: clinical, biological, and therapeutic perspectives. Neurobiol. Disease 131, 104257 (2019).
Iasevoli, F. et al. Treatment resistant schizophrenia is associated with the worst community functioning among severely-ill highly-disabling psychiatric conditions and is the most relevant predictor of poorer achievements in functional milestones. Prog. Neuro-psychopharmacol. Biol. Psychiatry 65, 34–48 (2016).
Sinnott-Armstrong, N. & Tanigawa, Y. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 53, 185–194 (2021).
Goetz, R. L. & Miller, B. J. Total and differential white blood cell counts, cocaine, and marijuana use in patients with schizophrenia. J. Nerv. Mental Disease 207, 633–636 (2019).
van Buel, E. M. et al. Major depressive disorder is associated with changes in a cluster of serum and urine biomarkers. J. Psychosom. Res. 125, 109796 (2019).
Huan, Y. et al. Label-free liquid chromatography-mass spectrometry proteomic analysis of the urinary proteome for measuring the escitalopram treatment response from major depressive disorder. Front. Psychiatry 12, 700149 (2021).
Likhitweerawong, N. et al. Profiles of urine and blood metabolomics in autism spectrum disorders. Metab. Brain Disease 36, 1641–1671 (2021).
Vismara, M., Girone, N. & Cirnigliaro, G. Peripheral biomarkers in DSM-5 anxiety disorders: an updated overview. Brain Sci. 10, https://doi.org/10.3390/brainsci10080564 (2020).
Fiandaca, M. S. et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimer’s Dement. 11, 600–607.e601 (2015).
An, M. & Gao, Y. Urinary biomarkers of brain diseases. Genom. Proteom. Bioinform. 13, 345–354 (2015).
Lai, C. Y. et al. Biomarkers in schizophrenia: A focus on blood based diagnostics and theranostics. World J. Psychiatry 6, 102–117 (2016).
Chan, M. K. et al. Converging evidence of blood-based biomarkers for schizophrenia: an update. Int. Rev. Neurobiol. 101, 95–144 (2011).
Miller, B. J., Culpepper, N. & Rapaport, M. H. C-reactive protein levels in schizophrenia: a review and meta-analysis. Clin. Schizophr. Relat. Psychoses 7, 223–230 (2014).
Pillai, A. et al. Decreased BDNF levels in CSF of drug-naive first-episode psychotic subjects: correlation with plasma BDNF and psychopathology. Int. J. Neuropsychopharmacol. 13, 535–539 (2010).
Emanuele, E. et al. Elevated urine levels of bufotenine in patients with autistic spectrum disorders and schizophrenia. Neuro Endocrinol. Lett. 31, 117–121 (2010).
Fusar-Poli, L. & Rutten, B. P. F. Polygenic risk scores for predicting outcomes and treatment response in psychiatry: hope or hype? Int. Rev. Psychiatry 34, 663–675 (2022).
Fang, Y., Fritsche, L. G., Mukherjee, B., Sen, S. & Richmond-Rakerd, L. S. Polygenic liability to depression is associated with multiple medical conditions in the electronic health record: phenome-wide association study of 46,782 individuals. Biol. Psychiatry 92, 923–931 (2022).
Liang, X. et al. Assessing the genetic correlations between blood plasma proteins and osteoporosis: a polygenic risk score analysis. Calcif. Tissue Int. 104, 171–181 (2019).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Mortimer, A. M., Singh, P., Shepherd, C. J. & Puthiryackal, J. Clozapine for treatment-resistant schizophrenia: National Institute of Clinical Excellence (NICE) guidance in the real world. Clin. Schizophr. Relat. Psychoses 4, 49–55 (2010).
Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. GigaScience 8, https://doi.org/10.1093/gigascience/giz082 (2019).
Tiihonen, J. et al. Real-world effectiveness of antipsychotic treatments in a nationwide cohort of 29/823 patients with schizophrenia. JAMA Psychiatry 74, 686–693 (2017).
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G. R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012).
Delaneau, O., Zagury, J. F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Altman, D. G. & Bland, J. M. Interaction revisited: the difference between two estimates. BMJ 326, 219 (2003).
Euesden, J., Lewis, C. M. & O’Reilly, P. F. PRSice: polygenic risk score software. Bioinformatics 31, 1466–1468 (2015).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
Hemani, G. & Zheng, J. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, https://doi.org/10.7554/eLife.34408 (2018).
Hemani, G., Bowden, J. & Davey Smith, G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum. Mol. Genet. 27, R195–r208 (2018).
Burgess, S., Small, D. S. & Thompson, S. G. A review of instrumental variable estimators for Mendelian randomization. Stat. Methods Med. Res. 26, 2333–2355 (2017).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted Median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Greco, M. F., Minelli, C., Sheehan, N. A. & Thompson, J. R. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat. Med. 34, 2926–2940 (2015).
Metz, C. E. Some practical issues of experimental design and data analysis in radiological ROC studies. Investig. Radiol. 24, 234–245 (1989).
Takashi, Y. & Fukumoto, S. Phosphate-sensing. Adv. Exp. Med. Biol. 1362, 27–35 (2022).
Esaki, K. et al. Evidence for altered metabolism of sphingosine-1-phosphate in the corpus callosum of patients with schizophrenia. Schizophr. Bull. 46, 1172–1181 (2020).
Jayakumar, P. N. et al. High energy phosphate abnormalities normalize after antipsychotic treatment in schizophrenia: a longitudinal 31P MRS study of basal ganglia. Psychiatry Res. 181, 237–240 (2010).
Gangadhar, B. N., Jayakumar, P. N., Subbakrishna, D. K., Janakiramaiah, N. & Keshavan, M. S. Basal ganglia high-energy phosphate metabolism in neuroleptic-naive patients with schizophrenia: a 31-phosphorus magnetic resonance spectroscopic study. Am. J. Psychiatry 161, 1304–1306 (2004).
Thakkar, M., Edelenbos, J. & Doré, S. Bilirubin and ischemic stroke: rendering the current paradigm to better understand the protective effects of bilirubin. Mol. Neurobiol. 56, 5483–5496 (2019).
Pommerening Dornelles, E. & Gama Marques, J. Unconjugated bilirubin and schizophrenia: a systematic review. CNS Spectr. 24, 577–588 (2019).
Pradeep, J. R., Acharya, M. S., Radhakrishnan, R. & Srinivasan, K. Elevated unconjugated bilirubin in schizophrenia compared to bipolar affective disorder. Prim. Care Companion CNS Disord. 21, https://doi.org/10.4088/PCC.19m02448 (2019).
Delanaye, P., Cavalier, E. & Pottel, H. Serum creatinine: not so simple! Nephron 136, 302–308 (2017).
Wang, N. et al. High serum creatinine is associated with reduction of psychiatric symptoms in female patients with anti-NMDAR encephalitis. Neurosci. Lett. 746, 135650 (2021).
Davison, J., O’Gorman, A., Brennan, L. & Cotter, D. R. A systematic review of metabolite biomarkers of schizophrenia. Schizophr. Res. 195, 32–50 (2018).
Cai, H. L. et al. Metabolomic analysis of biochemical changes in the plasma and urine of first-episode neuroleptic-naïve schizophrenia patients after treatment with risperidone. J. Proteome Res. 11, 4338–4350 (2012).
Karoum, F., Karson, C. N., Bigelow, L. B., Lawson, W. B. & Wyatt, R. J. Preliminary evidence of reduced combined output of dopamine and its metabolites in chronic schizophrenia. Arch. Gen. Psychiatry 44, 604–607 (1987).
Dion-Albert, L. et al. Sex differences in the blood-brain barrier: implications for mental health. Front. Neuroendocrinol. 65, 100989 (2022).
Ramsey, J. M. et al. Distinct molecular phenotypes in male and female schizophrenia patients. PLoS ONE 8, e78729 (2013).
Leung, A. & Chue, P. Sex differences in schizophrenia, a review of the literature. Acta Psychiatr. Scand. Suppl. 401, 3–38 (2000).
Zhang, B. H. et al. Gender differences in cognitive deficits in schizophrenia with and without diabetes. Compr. Psychiatry 63, 1–9 (2015).
Huang, Y. C. et al. Gender differences in susceptibility to schizophrenia: potential implication of neurosteroids. Psychoneuroendocrinology 84, 87–93 (2017).
Martini, F. et al. Clozapine tolerability in treatment resistant schizophrenia: exploring the role of sex. Psychiatry Res. 297, 113698 (2021).
Martínez-Andrés, J. A. & García-Carmona, J. A. Switching from clozapine to paliperidone palmitate-3-monthly improved obesity, hyperglycemia and dyslipidemia lowering antipsychotic dose equivalents in a treatment-resistant schizophrenia cohort. Int. Clin. Psychopharmacol. 35, 163–169 (2020).
Wysokiński, A. & Szczepocka, E. Red blood cells parameters in patients with acute schizophrenia, unipolar depression and bipolar disorder. Psychiatr. Danub. 30, 323–330 (2018).
Kamil Gharab, K. M. et al. The relationship between serum clozapine concentrations and hematological parameters by a validated mass spectrometric method. J. Pharm. Biomed. Anal. 180, 113056 (2020).
Davies, N. M., Holmes, M. V. & Davey Smith, G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362, k601 (2018).
Wootton, R. E. & Jones, H. J. Mendelian randomisation for psychiatry: how does it work, and what can it tell us? Mol Psychiatry 27, 53–57 (2022).
Bolun Cheng, Y. B. et al. Code for “Mendelian randomization study of the relationship between blood and urine biomarkers and schizophrenia in the UK Biobank cohort”. Zenodo https://doi.org/10.5281/zenodo.10453112 (2024).
Acknowledgements
This work was funded by the Natural Science Basic Research Plan in Shaanxi Province of China (2021JCW-08), and the Fundamental Research Funds for the Central Universities (xzy012023092). The funders had no role in study design, data collection and analysis, preparation of the paper, or the decision to publish. This research has been conducted using the UK Biobank Resource under Application Number 46478.
Author information
Authors and Affiliations
Contributions
All authors agree to the submission of this manuscript and are responsible for the integrity and ethics of the methods or data they contributed. B.C. and Y.B. contributed to the study design, the writing of original and revised manuscripts. L.L. and P.M. assisted with the study design, calculated the polygenic risk score, and reviewed the literature. S.C., X.Y., and C.P. interpreted the data. W.W. and H.L. performed bidirectional two-sample Mendelian randomization analyses. Y.J. and Y.W. made the figures and tables. F.Z. was the Principal Investigator for the study. All authors provided critical comments, edited the manuscript, and approved its final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Mulong Du and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cheng, B., Bai, Y., Liu, L. et al. Mendelian randomization study of the relationship between blood and urine biomarkers and schizophrenia in the UK Biobank cohort. Commun Med 4, 40 (2024). https://doi.org/10.1038/s43856-024-00467-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-024-00467-1