Abstract
Other than the Methanobacteriales and Methanomassiliicoccales, the characteristics of archaea that inhabit the animal microbiome are largely unknown. Methanimicrococcus blatticola, a member of the Methanosarcinales, currently reunites two unique features within this order: it is a colonizer of the animal digestive tract and can only reduce methyl compounds with H2 for methanogenesis, a increasingly recognized metabolism in the archaea and whose origin remains debated. To understand the origin of these characteristics, we have carried out a large-scale comparative genomic analysis. We infer the loss of more than a thousand genes in M. blatticola, by far the largest genome reduction across all Methanosarcinales. These include numerous elements for sensing the environment and adapting to more stable gut conditions, as well as a significant remodeling of the cell surface components likely involved in host and gut microbiota interactions. Several of these modifications parallel those previously observed in phylogenetically distant archaea and bacteria from the animal microbiome, suggesting large-scale convergent mechanisms of adaptation to the gut. Strikingly, M. blatticola has lost almost all genes coding for the H4MPT methyl branch of the Wood–Ljungdahl pathway (to the exception of mer), a phenomenon never reported before in any member of Class I or Class II methanogens. The loss of this pathway illustrates one of the evolutionary processes that may have led to the emergence of methyl-reducing hydrogenotrophic methanogens, possibly linked to the colonization of organic-rich environments (including the animal gut) where both methyl compounds and hydrogen are abundant.
Similar content being viewed by others
Introduction
Methanogenic archaea are common components of the intestinal microbiota of animals ranging from insects to humans [1,2,3,4]. However, archaea are generally overlooked in intestinal microbiome studies, leaving their ecology and diversity largely undescribed in this context. Additionally, only a small number of gut-associated archaeal isolates exists outside of the Methanobrevibacter and Methanosphaera genera (both belonging to the order Methanobacteriales), resulting in little knowledge concerning the adaptations that allowed archaea to colonize the animal gut. The physical/chemical conditions of a microbe’s niche exert selective pressures that can influence its genome content [5]. The gut of animals is characterized by a high and almost constant intake of fresh organic matter that distinguishes it from most aquatic and terrestrial environments. At the same time, gut microbes have to adapt to stressors such as potential removal from the host by the peristaltic movement of the digestive tract [5]. Host-associated archaea lineages have not only distinct genetic background due to their distant evolutionary relationships but also different histories of adaptation to the gut, and they can cover a wide variety of hosts. This poses the question of whether these distantly related archaea may display convergent adaptations or not. Genomic adaptations to the intestinal microbiome have been proposed in some methanogens: these include the gain of genes coding for cell surface proteins facilitating interactions or adhesion and possibly genomic streamlining [3, 6,7,8,9,10].
Within the order Methanosarcinales, two host-associated methanogens have been isolated, Methanimicrococcus blatticola PA from a cockroach (Periplaneta americana) [11] and Methanosarcina barkeri CM1 from the rumen of a cow [12]. However, the genome of M. barkeri CM1 is very similar to that of its freshwater close relative M. barkeri fusaro [13] and Methanosarcina members do not regularly occur in the rumen microbiota, suggesting a generalist lifestyle or a transient presence in the gut. In contrast, Methanimicrococcus has been reported multiple times in the gut of animals, and in some cases, it was found to represent a large majority of the methanogenic community in termites and some ruminants [14, 15]. Based on this apparent niche specificity, M. blatticola should display specific genomic adaptations to a host-associated lifestyle.
In addition to being an interesting model to study adaptations of archaea to the gut environment, M. blatticola is the only known member of Methanosarcinales that obligately uses H2 to reduce methyl compounds [11, 16, 17] (i.e., methyl-reducing hydrogenotrophic methanogenesis), a metabolism that has been recently reported in a growingly large number of newly discovered methanogen lineages [18,19,20,21,22,23]. Here we have analyzed the genome of M. blatticola PA, currently the only available isolated Methanimicrococcus strain, and carried out a large-scale comparative genomic analysis to understand the emergence of its unique characteristics. Our results provide new insights into the processes leading to the adaptation of archaea to the animal digestive tract and highlight one of the possible paths that led to the emergence of methyl-reducing hydrogenotrophic methanogenesis.
Materials and methods
Genome sequencing and annotation
The M. blatticola PA genome was sequenced from DNA ordered at the DSMZ German culture collection (DSM 13328), using Illumina MiSeq Nano V2 (2 ×250 PE). Reads were assembled using Spades 3.11 [24]. A total of 12 contigs >1 kb were obtained, with an average coverage of 157×. Almost all reads were assembled, as 99.97% of them aligned on these contigs. Genes were predicted using prodigal [25]. The genome and protein sequences are available in GenBank, PRJNA731512, and in Supplementary Dataset 1 and 2. Gene functions were annotated with Kyoto Encyclopedia of Genes and Genomes (KEGG) [26] and Eggnog [27]. Genes associated with the methanogenesis pathways were specifically targeted with HMM searches using PFAM [28], TIGRFAMs [29], and custom HMM profiles. Presence of transmembrane domains was determined using TMHMM 2.0 [30]. In the course of our analysis, this strain was also sequenced by JGI (2756170388) and released in GenBank (GCA_004363215.1). Unsurprisingly, the sequences and statistics of the two genomes are practically identical (size difference of <1 kb, average nucleotide identity (ANI) of 99.99%, 13 contigs). The analyses presented here were conducted on the genome we sequenced.
Dataset selection
Phylogenetic and comparative genomic analyses were carried out using 21 Methanosarcinales genomes, including 15 Methanosarcinaceae (Table S1). We prioritized the selection of high-quality genomes, a majority of which came from cultured and well-characterized species. Genomes from metagenomes were only included when no genome from an isolated strain was available and only if the estimated completeness level was >90% and the contamination <5%, as estimated with CheckM [31]. Four additional genomes were used as outgroups, three Methanocellales and one “Ca. Methanoflorentaceae” genomes, corresponding to close relatives to the Methanosarcinales.
Glycosyltransferase (GT) and PGF-CTERM annotation
GTs were annotated using dbCAN2 [32]. dbCAN2 was run on 507 archaeal genomes from a local database (Table S2), covering all major lineages of archaea and having >75% of completeness and <5% contamination. GTs in Methanosarcinales of the selected genome dataset described above was determined using the annotation found in the CAZy database [33].
Proteins containing a PGF-CTERM domain, which are associated with the PGF-CTERM/archaeosortase A system and possibly N-glycosylated, were first identified in the 21 Methanosarcinales plus the M. blatticola PA genomes using TIGR04126 HMM profile. Because this domain is short, new HMM profiles were generated from the PGF-CTERM domains of Methanosarcinales and Methanimicrococcus proteins (Supplementary Dataset 3). These new HMM profiles allowed the identification of extra proteins with a PGF-CTERM domain.
Phylogenetic analyses
The phylogenetic position of M. blatticola PA was established using three different datasets: (i) the concatenation of 40 universal markers as described in [18] that correspond to 36 proteins of the Phylosift dataset [34], plus the alpha and beta subunits of the RNA polymerase and two universal ribosomal proteins (L30, S4), (ii) the concatenation of the three MCR subunits (McrABG), and (iii) the 16S rRNA gene. The protein sequences for the phylogenetic analysis were retrieved from M. blatticola PA and the other 25 selected genomes using HMMer [35]. Single-protein datasets were aligned using MAFFT [36] (mafft-linsi), trimmed with BMGE [37] (BLOSUM30 substitution matrix) and concatenated. The 16S rRNA genes of M. blatticola PA and the 25 selected genomes were also aligned with MAFFT. A Bayesian phylogeny was built for the concatenation of the 40 universal markers using PhyloBayes [38] under the CAT + GTR + Γ4 model with four independent MMC chains, until convergence (maxdiff < 0.05). For the concatenation of the McrABG and the 16S rRNA gene sequences, Maximum Likelihood phylogenies were calculated with IQ-Tree [39] with the NEWTEST option for best model selection (McrABG: LG + F + Γ4; 16S: GTR + R3) and 1000 ultrafast bootstrap iterations. To investigate the environmental distribution of Methanimicrococcus, 16S rRNA gene sequences were searched in GenBank (Nucleotide collection nr/nt) and in all assembled metagenomes of the IMG databases using BLAST. Sequences >750 bp and with >91% identity with M. blatticola PA were downloaded. This identity cutoff was based on the minimal distance between M. blatticola PA and Methanosarcina spp. 16S rRNA gene sequences. A phylogeny of these sequences was built as described above for the 16S rRNA genes. Fasta files of the sequences used for phylogeny are available in Supplementary Dataset 4.
Gene flow reconstruction
This analysis was based on the phylogeny of the 40 concatenated markers described above and on the distribution of protein families among the 26 taxa used in this phylogeny. The protein families were determined using Silix with a 40% identity and 80% coverage cut-off [40], generating 21,377 protein families whose distribution was mapped on the reference phylogeny. The events of gene gains and losses that occurred during the evolution of the Methanosarcinales, and in particular on the branch leading to M. blatticola PA, were assessed using a death and birth model implemented in Count [41]. The rate of variation across families were optimized iteratively from uniform rates of gain, loss, and duplication to three discrete categories for the gamma distribution, using default parameters. Family history was calculated using posterior probabilities. M. blatticola PA protein sequences and representative of Methanosarcinaceae protein families that have been lost in M. blatticola PA were compared with three recently released Methanimicrococcus metagenome-assembled genomes (MAGs; i.e., GCA_009784005.1, GCA_009783635.1, and GCA_009776675.1, having 92.1/0%, 82.5/2.3%, 75.1/4.6% of completeness/contamination, respectively, according to CheckM [31], Table S3). A 40% identity and 50% coverage cut-off was used to determine whether proteins present or predicted to be lost in M. blatticola PA were coded in these MAGs. A lower coverage cut-off than in the main analysis was chosen because two of these MAGs are sheared (138 and 239 contigs), and as a consequence, a number of their proteins can be cut and split between different contigs.
Results and discussion
Widespread association of the Methanimicrococcus genus with gut microbiomes and extensive genome reduction in M. blatticola
We sequenced the genome of M. blatticola PA from the DSMZ German culture collection (DSM 13328). The draft genome (12 contigs) is 1.78 Mb long with a GC content of 42% and is predicted to encode 1569 proteins (Table S1). It is practically identical to the one sequenced by JGI (2756170388) in the course of our analysis and released in GenBank (GCA_004363215.1) (size difference of <1 kb, ANI of 99.99%, 13 contigs). The analyses presented hereafter were conducted on the genome we sequenced.
We first established the phylogenetic position of M. blatticola PA using three different datasets: the concatenation of 40 universal markers as described in [18] (Fig. 1a), the 16S rRNA gene (Fig. S1), and a concatenation of the three MCR subunits (McrABG; Fig. S2). In all three resulting phylogenies, M. blatticola PA consistently and robustly branches within Methanosarcinaceae, at the base of Methanosarcina. This placement is different from the one previously proposed, at the base of the Methanosarcinaceae, but the phylogeny was poorly resolved [11].
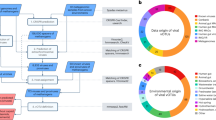
a Bayesian phylogeny (PhyloBayes, CAT + GTR + Γ4) of the Methanosarcinales based on a concatenation of 40 markers universal markers (10,126 positions). Numbers on the nodes indicate the genes present (black), gained (green, preceded by “G”), and lost (red preceded by “L”) along the evolution of the Methanosarcinales, as predicted by Count program [41]. The color of the circles at the nodes indicates the posterior probability values (black, >0.95; gray, between 0.70 and 0.95). b Maximum likelihood phylogeny (GTR + I + Γ4) based on the 16S rRNA gene sequences (1497 positions), displaying the environments where Methanimicrococcus occurs, mainly insect and mammalian digestive tract. The soil clade is followed by a question mark as all sequences of this poorly supported clade come from a single unpublished study, and their origin is therefore not totally reliable. The color of the circles at the nodes indicates ultrafast bootstrap values (black, >95; gray, between 70 and 95). The supports of the branches are only displayed for the main clades. c For each COG category, proportion of protein families in Methanimicrococcus blatticola PA relative to those present in the last common ancestor (LCA) of Methanimicrococcus + Methanosarcina (Msar).
Investigation of the environmental distribution of Methanimicrococcus members based on 16S rRNA gene sequences retrieved from GenBank and IMG databases revealed that they commonly occur in the animal digestive tract (Fig. 1b), in contrast to other members of the Methanosarcinales that are generally found in wetland soils and sediments [42]. Three distinct clades can be observed within the Methanimicrococcus genus: one is formed by sequences mainly from the digestive tract of insects (including M. blatticola PA), the second corresponds to sequences from mammalian digestive tracts, and the third consists of a few sequences from a single unpublished study on soil (Fig. 1b). These three clusters may indicate either one ancestral or two independent specializations to the animal digestive tract (mammals and insects) in Methanimicrococcus.
To investigate how genome content was affected during the transition from an open environment to the animal gut, we inferred the gene gains and losses that occurred between the last common ancestor (LCA) of Methanosarcina/Methanimicrococcus and M. blatticola PA. This analysis revealed an important shift in the gene content (Fig. 1a and Table S4). Indeed, one-third of M. blatticola PA genes (604 genes) were not present in Methanosarcina/Methanimicrococcus LCA (Fig. 1a) whereas two-thirds of the genes present in Methanosarcina/Methanimicrococcus LCA were lost (1522 genes), corresponding by far to the largest genome reduction across all Methanosarcinales (Fig. 1a). The small number of proteins encoded in the M. blatticola genome (1569) compared to Methanosarcina (3400 in average) or most other Methanosarcinaceae (2300 in average) further supports this prediction (Table S1). All Clusters of Orthologous Genes (COG) categories display a net loss in M. blatticola PA compared to Methanosarcina/Methanimicrococcus LCA (Fig. 1c) but some are more impacted than others. Several core cellular processes, such as “Translation, ribosomal structure and biogenesis (J),” “Lipid transport and metabolism (I),” or “Nucleotide transport and metabolism (F)” are only weakly affected, while others, such as cell envelope biogenesis, display a massive reduction in the number of protein families involved (Fig. 1c). A majority of the acquired genes have only a general or no predicted function (60% have no COG category and 80% no KEGG annotation), in contrast with those that were lost (4% have no COG category and 57% no KEGG annotation; Table S4).
Extensive modification of the cell–host interaction surface
An expected trait of adaptation to a host-associated lifestyle is modification of the repertoire of membrane-bound proteins, situated at the interface between the cell and its environment. Indeed, we observed that two-thirds of the proteins inferred to be membrane bound in M. blatticola PA were not present in Methanosarcina/Methanimicrococcus LCA and were therefore specifically acquired in the lineage leading to this archaeon. Similar to the cytoplasmic proteins acquired by M. blatticola PA, most of the membrane-associated proteins have an unknown function (Table S4). However, among them, several correspond to putative adhesin-like proteins (ALPs) (Tables S4 and 1). They are annotated as cell wall-binding repeat-containing protein or collagen-binding protein, and two contain collagen or cellulose (CBM44) binding domains. Sixteen acquired proteins contain one or several Listeria-Bacteroidetes repeats/Flg_New (PF09479) domains that display structural similarities with β-grasp folds, having various binding functions [43]. In Archaea, proteins with these domains were previously only known from Methanomassiliicoccales of the human gut (up to 38 in “Ca. Methanomassiliicoccus intestinalis”) where they have been suggested to be involved in the attachment to specific sites in the digestive tract [3]. Bacteria with a high number of genes coding for Listeria-Bacteroidetes repeats/Flg_New domains are also associated with an animal host [3]. The presence of these putative ALPs in two phylogenetically distant gut-associated archaeal lineages (belonging to Methanomassiliicoccales and Methanosarcinales) and in host-associated bacteria further supports the hypothesis that they are specific adaptations to the digestive tract [3] and indicate convergent adaptation. Among the acquired genes coding for membrane-bound proteins, 15 contain a GLUG motif of unknown function (2 of them also have Flg_new domains) and may also have a role in adhesion. In the absence of flagella, these proteins may prevent the washout of M. blatticola PA cells from the insect gut and help them to remain at specific sites where they are most competitive for substrates [14]. It was shown in fact that many methanogens associated with cockroaches are loosely associated with chitin bristles present in the hindgut [1].
While M. blatticola PA has kept the potential to express major S-layer proteins present in other Methanosarcinales, its cell surface was likely highly modified through the loss of many enzymes involved in the synthesis of nucleotide-activated sugars that are precursors of cell-surface glycoconjugates/extracellular polysaccharides (Figs. 1c and 2). Moreover, M. blatticola PA has lost up to 35 glycosyl transferases (GT), including 12 with transmembrane domains. Within Archaea, this is the genome that currently encodes the lowest number of GTs (2) (Table S2) and it also contrasts with other Methanosarcinales that have 33 GTs genes on average (Table S5). Even if not to the same extent, other host-associated methanogens (Methanobacteriales and Methanomassiliicoccales) also tend to have a lower number of GTs than their closest free-living relatives (Fig. 3), suggesting that the loss of GTs is another convergent characteristic related to the adaptation to gut conditions in archaea. The two GTs conserved in M. blatticola PA are involved in N-glycosylation of cell surface proteins by (i) transferring the first nucleotide-activated sugar on the lipid carrier (AglJ) and (ii) transferring the (oligo)saccharide from the lipid carrier to the nascent protein (AglB). Considering the almost complete loss of GTs and nucleotide-activated sugars synthesis genes, the nature of the glycans on the N-glycosylated proteins remains unclear (Fig. 2). Four membrane-bound proteins of unknown function and one with Flg_new domains have a PGF-CTERM domain, indicating that they are recognized by the archeosortase A present in M. blatticola PA and possibly N-glycosylated [44]. The number of such proteins is much lower in M. blatticola PA compared to other Methanosarcinales, which on average encode 20 proteins with this signal, suggesting a lower number of N-glycosylated proteins in M. blatticola PA (Table S5). Moreover, M. blatticola PA has lost two archaeosortases systems (PEF-CTERM and VPXXXP-CTERM) that may be involved in export and posttranslational modification of additional membrane-bound proteins [44].
The enzyme names in orange indicate that the corresponding genes were lost in M. blatticola PA, while those in blue correspond to genes that have been kept in M. blatticola PA. The genes lost in M. blatticola PA are also absent in the three Methanimicrococcus MAGs and those present in M. blatticola PA are also present in the three Methanimicrococcus MAGs. In other organisms, GlmM could perform three different reactions, albeit with different level of activity. Methanosarcinales genomes have two copies of glmM, but M. blatticola PA has only one. This enzyme is likely involved in the conversion of glucose-6-P into glucose-1-P. The accession numbers below the protein names provide an example of sequences in the Methanosarcinaceae.
For Methanosarcinales and Methanobacteriales, number of glycosyltransferases genes was compared between host-associated genera and the closest related genus (i.e., Methanosarcina for Methanimicrococcus; Methanobacterium for Methanobrevibacter and Methanosphaera). The three Methanimicrococcus MAGs were added to this analysis. For Methanomassiliicoccales, “Ca. M. intestinalis” and Methanomethylophilaceae MAGs were pooled in “Mmassilii gut” and all other MAGs in “Mmassilii env”. Stars represent the significant differences between groups, determined using a Wilcoxon test (continuity correction: ***p.adj < 1e−4, **p.adj < 0.001, *p.adj < 0.05). Methanimicro Methanimicrococcus, Msarcinales Methanosarcinales, Mmassilii Methanomassiliicoccales, Mbacteriales Methanobacteriales.
Glycans originating from commensal microorganisms cover many roles in the digestive tract, including persistence in this environment, microbe–microbe interactions, protection from phages, and host–microbe interactions, such as host-immune system stimulation [45]. However, there are no data on the role of archaeal glycans in the host microbiome and very few on the cell wall of the Methanosarcinaceae members outside the Methanosarcina genus. When they are aggregated, Methanosarcina members are surrounded by methanochondroitin [46], a fibrillar polymer that is analogous to the chondroitin present in tissues of animals, including cockroaches [47]. The ability to synthesize two of the sugars that constitute methanochondroitin (N-acetylgalactosamine and glucuronic acid) was lost in M. blatticola PA (Fig. 2). To our knowledge, such a large loss of genes associated with cell-surface glycoconjugates or exopolysaccharide synthesis has never been reported in the archaea, and how this event is linked to adaptation of M. blatticola to the digestive tract remains to be elucidated.
Adaptations to the gut as a nutrient/substrate rich environment
Several other gene losses reflect an adaptation to the gut as a nutrient/substrate-rich environment (Table 1), relative to non-gut environments that are essentially oligotrophic [48]. M. blatticola PA has lost a high-affinity phosphate transporter (pstABC) and an alkaline phosphatase (phoA) for organo-phosphate utilization, both of which are known to be upregulated under low phosphate concentrations in Methanosarcina mazei [49]. In contrast, the PitA low-affinity phosphate transporter [50] was kept in M. blatticola PA. Moreover, M. blatticola PA has lost the capacity to fix atmospheric nitrogen (nitrogenase nifKDU and regulatory genes P-II) and carbon dioxide (cdhAB) for autotrophic growth. The loss of these genes has also been previously reported in other gut methanogens belonging to Methanosphaera and Methanobrevibacter [51], and a loss of the nitrogenase was also suggested in host-associated Methanomassiliicoccales [52]. In contrast, termite-associated Methanobrevibacter spp. have not lost these genes [51], which may reflect adaptation to a different gut compartment or a lesser degree of specialization to the gut environment. The dependence of M. blatticola PA to a nutrient-rich environment is also reflected by the loss of several genes involved in amino acid synthesis (e.g., asnB for asparagine, trpABDFGE for tryptophan). The gene cstA was acquired in M. blatticola PA, possibly allowing the uptake of organic carbon and nitrogen in the form of pyruvate and peptides, as reported in gut-associated bacteria [53, 54]. This gene was independently acquired in a gut-associated Methanomassiliicoccales (Ca. Methanomassiliicoccus intestinalis Mx-01) suggesting convergent adaptation (Table 1 and Fig. S3). All these characteristics fit well with the high growth requirements of M. blatticola PA (acetate, yeast extract, tryptic soy, and vitamins [11]), as compared to most other Methanosarcinaceae members [42]. With the loss of comDE genes coding for coenzyme-M, the dependency of M. blatticola PA on other members of the gut microbiota includes this key cofactor of methanogenesis, consistently with the previously reported need of M. blatticola PA for an external source of this coenzyme [11]. The absence of comDE genes has been previously reported in several other gut methanogens such as Methanobrevibacter ruminantium [8] and several Methanomassiliicoccales (ISO4-H4 [55], ISO4-G1 [56]), including the termite-associated Methanoplasma termitum [9].
Seven markers recently identified as co-occurring with MCR and that are mostly conserved in Class I/Class II methanogens [18] were lost in M. blatticola PA. Three of them (m19, m21, and m24) are also missing in methanogens from nutrient-rich environments, including the host-associated Methanomassiliicoccales (m19 and m21) and host-associated Methanobacteriales (m21 and m24) (Tables 1 and S6; [18]). The function of these genes is currently unknown, but they have been suggested to be involved in regulatory processes of methanogenesis related to changes in substrate/nutrient availability [18]. Various methanogens, including Methanosarcinaceae [57, 58], are able to store carbon/energy as glycogen when substrates are available and to use it in periods of starvation. Genes involved in glycogen synthesis and degradation and those involved in glycolysis were lost in M. blatticola PA (Tables 1 and S4), indicating an adaptation to more stable conditions in terms of substrate availability.
The lower variability in the conditions faced by M. blatticola PA are also reflected in a drastically reduced capacity to sense its environment and move to more favorable conditions (Fig. 1C), with the loss of the cheABCFDRWY chemotaxis genes, 19 histidine kinase/PAS sensor genes, and the whole motility machinery (flaBCEFGH) (Tables 1 and S4). In addition, 43 genes coding for transcriptional regulators were lost. This matches previous observations reported for gut-associated Methanobrevibacter/Methanosphaera and Methanomassiliicoccales, which have a significantly smaller number of genes involved in chemotaxis, signal transduction, and transcriptional regulation, as compared to non-gut methanogens [6, 52] (Table 1). These characteristics are also consistent with what has been reported from gut bacteria, which have generally a smaller repertoire of genes involved in motility, chemotaxis [59], and transcriptional regulation [60], relative to non-gut bacteria (Table 1). These results reveal the existence of largely shared traits of adaptation to a host-associated lifestyle in both archaea and bacteria.
A unique case of loss of the H4MPT methyl-branch of the Wood–Ljungdahl pathway among Class I/II methanogens and evolution toward methyl-reducing hydrogenotrophy
Among the enzymes needed for energy conservation, the most notable losses correspond to several genes involved in methanogenesis, particularly those coding for the H4MPT methyl branch of the Wood–Ljungdahl (H4MPT mWL) pathway (fwdABCDEFG, fmdBD, ftr, mch, and mtd; Fig. 4 and Table 1). Moreover, these losses also include the genes coding for the biosynthesis of two cofactors of Fwd/Fmd, (methanofuran (mfnBDEF) and molybdopterin (moaABC/mobAB/moeA)) as well as those used to import the ions associated with these complexes (molybdate (modABCDE, wtpAB) and tungstate (tupAB, wtpAB)). Altogether, this explains the inability of M. blatticola PA to grow by reducing CO2 with H2 or by the disproportionation of methyl compounds (methylotrophic methanogenesis) as reported by Sprenger et al. [11]. This pathway was previously suggested to be absent in M. blatticola PA [16], but it was only inferred indirectly by the low enzymatic activity of the F420-reducing hydrogenase, which did not provide information on the presence/absence of the genes of the H4MPT mWL pathway. The almost complete absence of the genes of the H4MPT mWL pathway is remarkable, as it has never been previously observed in a member of Class I (Methanobacteriales, Methanopyrales, Methanococcales) and Class II (Methanosarcinales, Methanomicrobiales, Methanocellales) methanogens, even in those that do not use it for methanogenesis, such as Methanosphaera stadtmanae [61].
Enzymes in gray were lost in M. blatticola PA. The Ech complex (in white) was not present in the last common ancestor of Methanimicrococcus and Methanosarcina but likely acquired in Methanosarcina. Enzymes/enzymatic complexes within the beige frame (MCR, VhtAGC, HdrDE, MtsAB, MtaABC, MtbA, MttBC, MtbBC, MtmBC, and ATP synthase) are involved in energy conservation. Other enzymes/enzymatic complexes that can be involved in methane metabolisms in other archaea but predicted to be involved in anabolism in M. blatticola PA: MTR complex, Frh, Mer, AckA/Pta, Acs, CdhCDE, and the putative HdrABC/MvhD/FdhAB-like complex. CdhCDE may also have a role in CO detoxification. The full names of the proteins are displayed in Table S4.
Interestingly, one of the genes of the H4MPT mWL pathway (mer) and those coding for the MTR complex were kept (Fig. 4). In the absence of the remaining components of the H4MPT mWL pathway, their implication in methanogenesis is unlikely. However, these enzymes are presumably functional, as M. blatticola PA has also retained the capacity to synthesize their associated cofactors, H4MPT (Table S4; [62]) and F420 (Fig. 4). The presence of the F420 cofactor is supported by the previously reported observation of fluorescence at 420-nm excitation in M. blatticola PA [11]. Mer and MTR may have been kept in M. blatticola PA for their anabolic role, as suggested by the presence of MetE and ThyA homologs, possibly involved in the synthesis of methionine using H4MPT-CH3 [63] and thymidylate (dTMP) using H4MPT-CH2 [64] (Fig. 4). Consistently with this hypothesis, it was shown that M. barkeri growing on methanol and H2 is dependent on Na+ for growth, but not for ATP and methane formation [65]. This further supports the potential anabolic role of MTR (that is Na+ dependent) during the growth of Methanosarcinaceae spp. on methyl compounds with H2. In M. blatticola PA, the methyl and methylene groups needed for these anabolic reactions would thus be mainly provided by the methyl compounds used for methanogenesis (Fig. 4). The methylene groups may additionally be derived from formaldehyde produced by the pentose phosphate pathway [66], because a bifunctional Fae-Hps enzyme (a fused formaldehyde-activating/3-hexulose-6-phosphate synthase) is encoded by M. blatticola PA.
The specialization of M. blatticola PA on methyl-reducing hydrogenotrophic methanogenesis was also likely associated with loss of the membrane-bound Fpo and Rnf complexes (Fig. 4), which are involved in energy conservation in other Methanosarcinales [67]. The absence of the Fpo complex and of an Ech complex in M. blatticola PA was previously reported [68]. The Ech complex, which was likely acquired in Methanosarcina, is notably used to reduce ferredoxin for anabolic purpose in species growing by reduction of methanol with H2 [69]. One can therefore wonder how M. blatticola PA reduces ferredoxin for anabolic purposes (e.g., pyruvate synthesis; Fig. 4). An HdrABC/MvhD/Fd(r)hB complex may play this role by reducing ferredoxin and CoM-S-S-CoB using F420H2 (generated during the oxidation of H4MPT-CH3 by Mer, or by FrhABG using H2 as electron donor) through electron bifurcation, in a similar way as the HdrABC/MvhADG complex [70] but using F420H2 instead of H2 (Fig. 4). This potential complex was first reported in a methanotrophic member of the Methanosarcinales (ANME-2d) where it was referred as HdrABC/MvhD/FrhB and proposed to perform electron confurcation (the reverse reaction of what we propose to occur in M. blatticola) for ferredoxin and CoM-S-S-CoB recycling [71]. This putative complex is encoded by a gene cluster that was reported in many methane and short-chain alkane-oxidizing archaea [18].
Except for M. blatticola PA, all other Methanosarcinaceae representatives have the capacity to disproportionate methyl compounds. Thus, the specialization of M. blatticola PA on methyl-reducing hydrogenotrophic methanogenesis likely emerged from this type of metabolism. For methyl-compound disproportionation, Methanosarcinaceae spp. use the H4MPT mWL pathway to oxidize one methyl group into CO2, producing three reducing equivalents for the reduction of three additional methyl groups into CH4 [72]. However, when M. barkeri grows on methanol in the presence of H2, methanol is totally reduced by H2 and no longer oxidized into CO2 [65], suggesting that the H4MPT mWL pathway is no longer used for methanogenesis under such conditions. It is thus possible that an ancestor of M. blatticola PA lost the genes encoding the H4MPT mWL pathway after stable colonization of the animal gut environment where both H2 and methyl compounds are available. Indeed, the concentrations of H2 in the gut (4 μM in cockroaches [17]; 100 nM–50 μM in ruminants [73]; 168 μM in mice [74]; 5–156 μM in humans [75, 76]) are generally 10–1000-folds higher than those in anoxic soils or sediments (10–30 nM [77, 78]). This is likely due to the constant load of fresh organic matter making H2 production rates much higher in the animal gut (5 μM h−1 and 2 mM h−1 [79]) than in the environment (1 μM h−1 in eutrophic lake sediments [77]). For similar reasons, methyl-compound concentrations in the gut (e.g., for methanol, 10 μM in cockroaches [17]; 23–72 μM in the rumen [80]; 70 μM in humans [3]) are also higher than those in sediments, where they are generally around or below the micromolar level [81, 82]. The availability of methanol and hydrogen also potentially drove the specialization on methyl-reducing hydrogenotrophic methanogenesis in the members of the genus Methanosphaera (Class I methanogen, Methanobacteriales). Several Methanobacteriales have the mtaABC genes for methanol utilization, but except Methanosphaera spp. only a few have been shown to grow on methanol + H2 [83, 84]. It is therefore possible that the ancestors of Methanosphaera spp. evolved stepwise from (i) CO2-reducing methanogenesis (which is shared by all Methanobacteriales) to (ii) facultative methyl-reducing methanogenesis and then to (iii) obligate methyl-reducing methanogenesis. The transition from facultative to obligate methyl-reducing methanogenesis likely occurred when molybdopterin biosynthesis genes (absent in Methanosphaera spp. [51, 61] were lost. All the main enzymes of the H4MPT mWL pathway are present in Methanosphaera and involved in anabolic reactions [61].
With M. blatticola PA, the only other known methanogens missing part or all of the H4MPT mWL pathway belong to recently discovered lineages branching all over the tree of archaea: Methanomassiliicoccales [22] Methanofastidiosa [19], Verstraetearchaeota [21], Methanonatronarchaeia [20], and NM3 Acherontia [18]. All these methanogens have been experimentally characterized or predicted to be methyl-reducing hydrogenotrophic methanogens. The origin of this metabolism in these evolutionarily distant lineages has been discussed and investigated previously [18, 85]. Briefly, methyl-reducing hydrogenotrophic methanogenesis could have originated early in the evolution of archaea and been vertically inherited in these lineages or rather have been acquired by horizontal gene transfers and/or emerged through the loss of the H4MPT mWL pathway and the MTR complex. These hypotheses are not mutually exclusive, and according to phylogenetic analyses of the MCR complex, the acquisition of this metabolism via horizontal gene transfer is very likely at least for some of these lineages [18]. The hypothesis of the emergence of methyl-reducing hydrogenotrophic methanogenesis from methanogenesis involving the H4MPT mWL pathway is difficult to test in these deep-branching lineages. The case of M. blatticola therefore represents the first concrete proof of a partial loss of the H4MPT mWL pathway in a methanogen. When methanogens stop using the H4MPT mWL pathway for methanogenesis, they tend to keep these enzymes for anabolic purposes, but whether those enzymes are replaced by other enzymes over long evolutionary periods remains unknown.
Three novel MAGs support the patterns of gene gains/losses in Methanimicrococcus
During the course of this analysis, three Methanimicrococcus MAGs from a termite gut sample were released in GenBank [86] (Table S3 and Fig. S4). By comparing the protein sequences of these MAGs with those of M. blatticola PA, we observe that most of the genes (93.3%) that we predicted to have been lost in M. blatticola PA are also absent in these three MAGs (Table S4). This shows that most of these losses occurred before the LCA of these four Methanimicrococcus species. The proteins absent in the three Methanimicrococcus MAGs cover those discussed above and are involved in methanogenesis (mWL pathway and MCR-associated markers), glycogen synthesis/utilization, chemotaxis, nucleotide-activated sugar synthesis, GTs (1–3 only are present, Fig. 3), and 86% (37) of the transcriptional regulators lost in M. blatticola PA (Table 1). Only 0.01% of genes predicted to be lost in M. blatticola PA are present in all 3 MAGs and could therefore represent losses specific to this species. A smaller fraction of the genes that were gained in M. blatticola PA are also present in the 3 Methanimicrococcus MAGs: 50% are present in at least 1 of them and only 19.4% are present in all 3 MAGs, suggesting that there is a weaker pattern of gene acquisition than gene loss. However, some of the gained proteins—in particular ALPs—may have evolved faster than average following acquisition and could have been missed by our identity threshold. For example, no genes coding for homologs of the M. blatticola PA proteins with a Flg_new domain were found in the 3 MAGs using a 40% identity threshold, but we detected 3 (in the least complete MAG) to 13 proteins with a Flg_new domain in the proteome of these MAGs using a specific HMM search.
Concluding remarks
Our results uncover how the transition from the open environment to the gut can deeply modify the physiology of an archaeon, including its central energy metabolism. Moreover, it reveals one of the possible paths toward the emergence of methyl-reducing hydrogenotrophic methanogenesis. In contrast to Methanomassiliicoccales, M. blatticola PA and Methanosphaera spp. have specialized relatively recently (at the genus level) on methyl-reducing hydrogenotrophic methanogenesis. This probably occurred after gut colonization, being triggered by the conditions encountered in this environment. After they became obligate methyl-reducing hydrogenotrophic methanogens, M. blatticola PA and Methanosphaera spp. kept one or all of the H4MPT mWL enzymes, respectively, for the synthesis of amino acids and purines. This may represent an intermediate state toward the complete loss of H4MPT mWL pathway and the MTR complex, as observed in the deep-branching lineages of obligate methyl-reducing hydrogenotrophic methanogens that were recently discovered [18,19,20,21,22]. There are multiple other (non-exclusive) explanations for the presence of this metabolism in free-living methanogens [18]: (i) it could have arisen from horizontal gene transfer to previously non-methanogenic archaea, (ii) the mWL pathway could have been lost in CO2-reducing methanogens thriving in some specific non-gut environment where the conditions are favorable to this type of methanogenesis, and (iii) the methyl-reducing hydrogenotrophic methanogenic pathway may be the ancestral type of methanogenesis. In this latter case, the specialization of Methanimicrococcus and Methanosphaera on this type of metabolism because of gut colonization would correspond to a step back to the original type of methanogenesis.
Our analysis also highlights multiple adaptations to the digestive tract of animals, several of which have also been previously proposed in phylogenetically distant host-associated archaea belonging to the Methanobacteriales and Methanomassiliicoccales. These convergent adaptations include the loss of the ability to grow autotrophically, to fix nitrogen, and of specific methanogenesis/MCR-associated markers. They also indicate a reduced capacity to sense and respond to variations in the environment, and an important shift in cell surface elements, notably by reduction in the number of GTs and gain of ALPs such as the Listeria-Bacteroidetes repeats/Flg_New domain proteins. The gain and loss of some of these genes also fit patterns of gene depletion or enrichment observed in gut bacteria, revealing common adaptation mechanisms across the two prokaryotic domains. Several gene losses observed in M. blatticola PA, and suggested in other gut methanogens, are also similar to those reported for two methanogens endosymbionts of ciliates, Methanobrevibacter sp. NOE and Methanocorpusculum sp. MCE, affiliated to Methanobacteriales and Methanomicrobiales, respectively [87]. Indeed, these two endosymbionts have also lost transcriptional regulators, aromatic amino acid biosynthesis, and cell surface modification enzymes [87]. Beyond the gut environment, some of these losses could thus be more generally indicative of host adaptation.
Currently, the availability of archaeal genomes associated with a host is essentially restricted to humans, ruminants, and, to a lesser extent, termites and sponges. Sequencing of new archaeal genomes from a wide variety of animal hosts will therefore provide key information to draw a comprehensive picture of host-adaptation mechanisms in the archaea.
References
Hackstein JH, Stumm CK. Methane production in terrestrial arthropods. Proc Natl Acad Sci USA. 1994;91:5441–5.
Hackstein JHP, van Alen TA. Fecal methanogens and vertebrate evolution. Evolution. 1996;50:559–72.
Borrel G, McCann A, Deane J, Neto MC, Lynch DB, Brugère JF, et al. Genomics and metagenomics of trimethylamine-utilizing archaea in the human gut microbiome. ISME J. 2017;11:2059–74.
Raymann K, Moeller AH, Goodman AL, Ochman H. Unexplored archaeal diversity in the great ape gut microbiome. mSphere. 2017;2:e00026-17.
Douglas AE. Multiorganismal insects: diversity and function of resident microorganisms. Annu Rev Entomol. 2015;60:17–34.
Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA. 2007;104:10643–8.
Gaci N, Borrel G, Tottey W, O'Toole PW, Brugère JF. Archaea and the human gut: new beginning of an old story. World J Gastroenterol. 2014;20:16062–78.
Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM, et al. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS ONE. 2010;5:e8926.
Lang K, Schuldes J, Klingl A, Poehlein A, Daniel R, Brunea A. New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of ‘Candidatus Methanoplasma termitum’. Appl Environ Microbiol. 2015;81:1338–52.
Borrel G, Brugère JF, Gribaldo S, Schmitz RA, Moissl-Eichinger C. The host-associated archaeome. Nat Rev Microbiol. 2020;18:622–36.
Sprenger WW, van Belzen MC, Rosenberg J, Hackstein JH, Keltjens JT. Methanomicrococcus blatticola gen. nov., sp. nov., a methanol- and methylamine-reducing methanogen from the hindgut of the cockroach Periplaneta americana. Int J Syst Evol Microbiol. 2000;50:1989–99.
Jarvis GN, Strömpl C, Burgess DM, Skillman LC, Moore ER, Joblin KN. Isolation and identification of ruminal methanogens from grazing cattle. Curr Microbiol. 2000;40:327–32.
Lambie SC, Kelly WJ, Leahy SC, Li D, Reilly K, McAllister TA, et al. The complete genome sequence of the rumen methanogen Methanosarcina barkeri CM1. Stand Genomic Sci. 2015;10:57.
Brune, A. Methanogens in the digestive tract of termites. In: Hackstein JHP, editor. (Endo)symbiotic methanogenic archaea. Berlin: Springer; 2018. p. 81–101.
Li Z, Wang X, Alberdi A, Deng J, Zhong Z, Si H, et al. Comparative microbiome analysis reveals the ecological relationships between rumen methanogens, acetogens, and their hosts. Front Microbiol. 2020;11:1311.
Sprenger WW, Hackstein JHP, Keltjens JT. The energy metabolism of Methanomicrococcus blatticola: physiological and biochemical aspects. Antonie van Leeuwenhoek. 2005;87:289–99.
Sprenger WW, Hackstein JHP, Keltjens JT. The competitive success of Methanomicrococcus blatticola, a dominant methylotrophic methanogen in the cockroach hindgut, is supported by high substrate affinities and favorable thermodynamics. FEMS Microbiol Ecol. 2007;60:266–75.
Borrel G, Adam PS, McKay LJ, Chen LX, Sierra-García IN, Sieber C, et al. Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea. Nat Microbiol. 2019;4:603–13.
Nobu MK, Narihiro T, Kuroda K, Mei R, Liu WT. Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J. 2016;10:2478–87.
Sorokin DY, Makarova KS, Abbas B, Ferrer M, Golyshin PN, Galinski EA, et al. Discovery of extremely halophilic, methyl-reducing euryarchaea provides insights into the evolutionary origin of methanogenesis. Nat Microbiol. 2017;2:17081.
Vanwonterghem I, Evans PN, Parks DH, Jensen PD, Woodcroft BJ, Hugenholtz P, et al. Methylotrophic methanogenesis discovered in the novel archaeal phylum Verstraetearchaeota. Nat Microbiol. 2016;1:16170.
Borrel G, O'Toole PW, Harris HM, Peyret P, Brugère JF, Gribaldo S. Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol Evol. 2013;5:1769–80.
Söllinger A, Urich T. Methylotrophic methanogens everywhere—physiology and ecology of novel players in global methane cycling. Biochem Soc Trans. 2019;47:1895–907.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119.
Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–31.
Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, et al. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–14.
El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–32.
Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–3.
Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. DbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–101.
Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–17.
Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, Eisen JA. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014;2:e243.
Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:431.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210.
Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–8.
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74.
Miele V, Penel S, Duret L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics. 2011;12:116.
Csurös, M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26:1910–2.
Oren, A. The family methanosarcinaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes: other major lineages of bacteria and the archaea. Berlin: Springer; 2014. p. 259–81.
Ebbes M, Bleymüller WM, Cernescu M, Nölker R, Brutschy B, Niemann HH. Fold and function of the InlB B-repeat. J Biol Chem. 2011;286:15496–506.
Haft DH, Payne SH, Selengut JD. Archaeosortases and exosortases are widely distributed systems linking membrane transit with posttranslational modification. J Bacteriol. 2012;194:36–48.
Porter NT, Martens EC. The critical roles of polysaccharides in gut microbial ecology and physiology. Annu Rev Microbiol. 2017;71:349–69.
Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–26.
Ashhurst DE, Costin NM. Insect mucosubstances. III. Some mucosubstances of the nervous systems of the wax-moth (Galleria mellonella) and the stick insect (Carausius morosus). Histochem J. 1971;3:379–87.
Morita, RY. Bacteria in oligotrophic environments. New York, NY: Chapman & Hall; 1997.
Paula FS, Chin JP, Schnürer A, Müller B, Manesiotis P, Waters N, et al. The potential for polyphosphate metabolism in archaea and anaerobic polyphosphate formation in Methanosarcina mazei. Sci Rep. 2019;9:17101.
Harris RM, Webb DC, Howitt SM, Cox GB. Characterization of PitA and PitB from Escherichia coli. J Bacteriol. 2001;183:5008–14.
Poehlein A, Schneider D, Soh M, Daniel R, Seedorf H. Comparative genomic analysis of members of the genera methanosphaera and methanobrevibacter reveals distinct clades with specific potential metabolic functions. Archaea. 2018;2018:609847.
Borrel G, Parisot N, Harris HM, Peyretaillade E, Gaci N, Tottey W, et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics. 2014;15:679.
Hwang S, Choe D, Yoo M, Cho S, Kim SC, Cho S, et al. Peptide transporter CstA imports pyruvate in Escherichia coli K-12. J Bacteriol. 2018;200:e00771-17.
Rasmussen JJ, Vegge CS, Frøkiær H, Howlett RM, Krogfelt KA, Kelly DJ, et al. Campylobacter jejuni carbon starvation protein A (CstA) is involved in peptide utilization, motility and agglutination, and has a role in stimulation of dendritic cells. J Med Microbiol. 2013;62:1135–43.
Li Y, Leahy SC, Jeyanathan J, Henderson G, Cox F, Altermann E, et al. The complete genome sequence of the methanogenic archaeon ISO4-H5 provides insights into the methylotrophic lifestyle of a ruminal representative of the Methanomassiliicoccales. Stand Genomic Sci. 2016;11:59.
Kelly WJ, Li D, Lambie SC, Jeyanathan J, Cox F, Li Y, et al. Complete genome sequence of methanogenic Archaeon ISO4-G1, a member of the Methanomassiliicoccales, isolated from a sheep rumen. Genome Announc. 2016;4:e00221-16.
Maitra PK, Bhosale SB, Kshirsagar DC, Yeole TY, Shanbhag AN. Metabolite and enzyme profiles of glycogen metabolism in Methanococcoides methylutens. FEMS Microbiol Lett. 2001;198:23–9.
Santiago-Martínez MG, Encalada R, Lira-Silva E, Pineda E, Gallardo-Pérez JC, Reyes-García MA, et al. The nutritional status of Methanosarcina acetivorans regulates glycogen metabolism and gluconeogenesis and glycolysis fluxes. FEBS J. 2016;283:1979–99.
Dobrijevic D, Abraham AL, Jamet A, Maguin E, van de Guchte M. Functional comparison of bacteria from the human gut and closely related non-gut bacteria reveals the importance of conjugation and a paucity of motility and chemotaxis functions in the gut environment. PLoS ONE. 2016;11:e0159030.
Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct. 2009;4:13.
Fricke WF, Seedorf H, Henne A, Krüer M, Liesegang H, Hedderich R, et al. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol. 2006;188:642–58.
Adam PS, Borrel G, Gribaldo S. An archaeal origin of the Wood–Ljungdahl H4MPT branch and the emergence of bacterial methylotrophy. Nat Microbiol. 2019;4:2155–63.
Schröder I, Thauer RK. Methylcobalamin:homocysteine methyltransferase from Methanobacterium thermoautotrophicum. Identification as the metE gene product. Eur J Biochem. 1999;263:789–96.
Krone UE, McFarlan SC, Hogenkamp HPC. Purification and partial characterization of a putative thymidylate synthase from Methanobacterium thermoautotrophicum. Eur J Biochem. 1994;220:789–94.
Muller V, Blaut M, Gottschalk G. Utilization of methanol plus hydrogen by Methanosarcina barkeri for methanogenesis and growth. Appl Environ Microbiol. 1986;52:269–74.
Kato N, Yurimoto H, Thauer RK. The physiological role of the ribulose monophosphate pathway in bacteria and archaea. Biosci Biotechnol Biochem. 2006;70:10–21.
Welte C, Deppenmeier U. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta Bioenerg. 2014;1837:1130–47.
Kurth JM, den Camp HJMO, Welte CU. Several ways one goal—methanogenesis from unconventional substrates. Appl Microbiol Biotechnol. 2020;104:6839–54.
Meuer J, Kuettner HC, Zhang JK, Hedderich R, Metcalf WW. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc Natl Acad Sci USA. 2002;99:5632–7.
Wagner T, Koch J, Ermler U, Shima S. Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science. 2017;357:689–703.
Arshad A, Speth DR, de Graaf RM, Op den Camp HJ, Jetten MS, Welte CU. A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front Microbiol. 2015;6:6.
Hedderich R, Whitman WB. Physiology and biochemistry of the methane-producing archaea. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes - prokaryotic biology and symbiotic associations. Berlin: Springer; 2013. p. 1050–79.
Morgavi DP, Martin C, Jouany JP, Ranilla MJ. Rumen protozoa and methanogenesis: not a simple cause-effect relationship. Br J Nutr. 2012;107:388–97.
Maier RJ, Olczak A, Maier S, Soni S, Gunn J. Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect Immun. 2004;72:6294–9.
Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat Rev Gastroenterol Hepatol. 2012;9:504–18.
Kalantar-Zadeh K, Berean KJ, Ha N, Chrimes AF, Xu K, Grando D, et al. A human pilot trial of ingestible electronic capsules capable of sensing different gases in the gut. Nat Electron. 2018;1:79–87.
Conrad R, Phelps TJ, Zeikus JG. Gas metabolism evidence in support of the juxtaposition of hydrogen-producing and methanogenic bacteria in sewage sludge and lake sediments. Appl Environ Microbiol. 1985;50:595–601.
Lovley DR, Goodwin S. Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochim Cosmochim Acta. 1988;52:2993–3003.
Hackstein JHP. Genetic and evolutionary constraints for the symbiosis between animals and methanogenic bacteria. Environ Monit Assess. 1996;42:39–56.
Ametaj BN, Zebeli Q, Saleem F, Psychogios N, Lewis MJ, Dunn SM, et al. Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics. 2010;6:583–94.
Mausz MA, Chen Y. Microbiology and ecology of methylated amine metabolism in marine ecosystems. Curr Issues Mol Biol. 2019;33:133–48.
King GM, Klug MJ, Lovley DR. Metabolism of acetate, methanol, and methylated amines in intertidal sediments of Lowes Cove, Maine. Appl Environ Microbiol. 1983;45:1848–53.
Borrel G, Joblin K, Guedon A, Colombet J, Tardy V, Lehours AC, et al. Methanobacterium lacus sp. nov., isolated from the profundal sediment of a freshwater meromictic lake. Int J Syst Evol Microbiol. 2012;62:1625–9.
Krivushin KV, Shcherbakova VA, Petrovskaya LE, Rivkina EM. Methanobacterium veterum sp. nov., from ancient Siberian permafrost. Int J Syst Evol Microbiol. 2010;60:455–9.
Wang Y, Wegener G, Williams TA, Xie R, Hou J, Tian C, et al. A methylotrophic origin of methanogenesis and early divergence of anaerobic multicarbon alkane metabolism. Sci Adv. 2021;7:eabj1453.
Hervé V, Liu P, Dietrich C, Sillam-Dussès D, Stiblik P, Šobotník J, et al. Phylogenomic analysis of 589 metagenome-assembled genomes encompassing all major prokaryotic lineages from the gut of higher termites. PeerJ. 2020;8:e8614.
Lind AE, Lewis WH, Spang A, Guy L, Embley TM, Ettema T. Genomes of two archaeal endosymbionts show convergent adaptations to an intracellular lifestyle. ISME J. 2018;12:2655–67.
Acknowledgements
We thank the computational and storage services (TARS cluster) provided by the IT department at Institut Pasteur, Paris.
Funding
CMT is supported by a PhD fellowship from Paris Sorbonne Université Science and by funds from the doctoral school Bio Sorbonne Paris Cité “BioSPC”. GB acknowledges funding from the French National Agency for Research Grant Methevol (ANR-19-CE02-0005-01) and SG acknowledges Archaevol (ANR-16-CE02-0005-01). This study has received funding from the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant no. ANR-10-LABX-62-IBEID).
Author information
Authors and Affiliations
Contributions
G.B. conceived the study. G.B., C.M.T. and N.T. performed the analyses. G.B., C.M.T. and S.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Thomas, C.M., Taib, N., Gribaldo, S. et al. Comparative genomic analysis of Methanimicrococcus blatticola provides insights into host adaptation in archaea and the evolution of methanogenesis. ISME COMMUN. 1, 47 (2021). https://doi.org/10.1038/s43705-021-00050-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43705-021-00050-y
This article is cited by
-
Presence and role of viruses in anaerobic digestion of food waste under environmental variability
Microbiome (2023)
-
Factors shaping the abundance and diversity of the gut archaeome across the animal kingdom
Nature Communications (2022)