Abstract
To conserve biodiversity, it is imperative to maintain and restore sufficient amounts of functional habitat networks. Therefore, the location of the remaining forests with natural structures and processes over landscapes and large regions is a key objective. Here we integrated machine learning (Random Forest) and open landscape data to scan all forest landscapes in Sweden with a 1 ha spatial resolution with respect to the relative likelihood of hosting High Conservation Value Forests. Using independent spatial stand- and plot-level validation data, we confirmed that our predictions correctly represent different levels of forest naturalness, from degraded to those with high and associated biodiversity conservation values. Given ambitious national and international conservation objectives and increasingly intensive forestry, our model and the resulting wall-to-wall mapping fill an urgent gap for assessing the achievement of evidence-based conservation targets, spatial planning, and designing forest landscape restoration.
Similar content being viewed by others
Introduction
Remnants of naturally dynamic forest landscapes are key biodiversity hotspots providing habitats to a large number of species, forming intact and resilient ecosystems, and providing multiple ecosystem-level services1,2. The scarcity and continuing loss of such forest areas3 has raised awareness about the need for spatially effective conservation and landscape restoration4. In particular, primary and old-growth forests with low human impact form important local biodiversity hotspots5,6,7,8. Hence, there is a growing need for effective mapping of such areas to safeguard their existence through conservation and nature restoration to ensure habitat network functionality9,10,11,12,13.
Forest species have adapted to a diversity of naturally occurring disturbance regimes14, which create structural patterns at multiple scales15 and form diverse habitats. The level of forest naturalness16,17 reflects a transition from naturally dynamic forests as complex ecosystems to simplified cropping systems managed for wood biomass production18. Globally, such transitions are creating expanding frontiers of naturalness loss19,20.
Green infrastructure (GI) is an established concept addressing the urgency of conserving and restoring sufficient areas of functional and representative habitat networks21,22,23 through a multi-scale approach supporting biodiversity conservation, ecosystem resilience, biodiversity and ecosystem services24. Planning and maintaining functional GI networks requires knowledge about the existence and location of necessary amounts of high conservation value areas able to maintain structural and functional connectivity across landscapes and regions23,24,25. In regions dominated by intensive cropping systems for the production of wood biomass, the provisioning of other ecosystem services and biodiversity conservation become limited. The need to identify remnant natural forest patches of importance for biodiversity conservation triggered the use of the term forest naturalness16, and the establishment and mapping of High Conservation Value Forests (HCVF)26, generically capturing forests with high levels of naturalness27. Typically, HCVF harbour native tree species, have a long history of forest continuity, vertical and horizontal structural complexity, and low levels of anthropogenic influence7,28,29. Such HCVF mapping data can be used for to assess the practical achievement of evidence-based conservation targets23,30, spatial planning25,28, and designing forest landscape and nature restoration31.
With a long history of intensive wood biomass production, Sweden is a globally important producer of wood products with a rotational forest management system that is applied systematically across the forest landscape. However, this has caused an overall transformation of naturally dynamic forests into effective wood production cropping systems32,33. Outside the foothill forests of the Scandinavian mountain ranges, only fragments of such naturally dynamic forests remain12. In Sweden, the first national-scale HCVF dataset was compiled in 2016, based on known and registered forest biodiversity hotspots within and outside formally protected areas at the time. The HCVF were mapped by field surveys during several decades and without a predefined sampling scheme34. Thus, the background information, based on costly and time-consuming field work, is neither up to date nor comprehensive and is limited in terms of its spatial and habitat representativeness.
The increasing availability of wall-to-wall spatial datasets describing multiple dimensions of landscapes with unprecedented thematic resolution and spatial precision, creates an opportunity to overcome these limitations35,36. Examples of such datasets range from raw remote sensing data collected globally by modern, civil satellite missions such as Landsat 837 and Sentinel-238; sophisticated data products such as the high-resolution maps of global forest cover change39 and forest canopy height40; to LIDAR-based forest structure measurements41 and high-spatial and thematic resolution land cover/land-use maps42. Furthermore, rapid developments in applying machine learning within ecology and conservation science43,44,45 provide an opportunity to map the naturalness and thus conservation values of forests46. It is challenging to provide direct mechanistic meanings to multi-scale spatial proxy variables describing forest naturalness, as they interact in complex and highly non-linear ways. However, machine learning, with its ability to flexibly and automatically detect the best predictive patterns that explain the data, is promising for providing a robust solution to this problem. Big spatial data can be processed with machine learning to develop a contiguous and complex measure of the naturalness of forest ecosystems, accounting not only for simple tree cover dynamics, but also for forest structural properties and composition of surrounding landscapes at different spatial scales46.
Given the high pressure on forest ecosystems and the urgent need for spatially explicit information on remaining HCVF, the aim of this study was to provide validated mapping of forest areas crucial for biodiversity conservation at landscape, regional and national levels. Implemented and tested on Sweden’s forests, the methods we developed are potentially applicable to other forest regions. We used Sweden as a case study for the following reasons: (1) Sweden’s forest area is the largest in the European Union (c. 280,000 km2), ranges from temperate deciduous, through boreal to subalpine ecoregions, and covers wide gradients in forest type, landscape configuration, and ownership pattern; (2) Its natural forests and forest landscapes have been transformed into wood biomass cropping systems to a high degree, which makes identifying HCVF remnants critical12,47,48; (3) There is strong political pressure to further intensify wood biomass oriented forestry49, and (4); The systematic public HCVF field surveys have been terminated50.
We present a comprehensive framework for predicting the occurrence of forests with different levels of naturalness using a data mining and predictive modelling approach. More specifically, we used the Random Forest (RF) machine learning algorithm and publicly available high-resolution spatial datasets describing landscape configuration, topography, forest structural properties, and various socio-economic factors affecting landscape patterns and processes at multiple scales. We trained our models and tested their performance using the only existing, yet incomplete, national-scale HCVF database. Finally, using a comprehensive set of independent spatial datasets, we validated the extent to which the predicted relative likelihoods of HCVF occurrence capture the gradient of forest naturalness and conservation values. Our focus was on predicting the continuous values of the relative likelihood of HCVF occurrence. This is more informative than binary output, and is crucial for identifying areas that represent different levels of forest naturalness, ranging from degraded forests to those with high biodiversity conservation values. The derived prediction map can support landscape conservation planning by providing crucial information on where to locate conservation measures, such as protected areas and/or forest restoration, or alternatively, where to prioritise forestry-oriented management practices.
Results
Predicting naturalness as conservation value
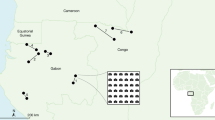
We used data mining and predictive modelling51 to scan forest landscapes in Sweden (Fig. 1, Table 1) for the occurrence of HCVF. This modelling approach was motivated by the inherent properties of HCVF, which are complex and high-dimensional with respect to, among other factors, the physical landscape, biodiversity, socio-economics, intensity of management, and history of use. This complexity means that a complete set of wall-to-wall spatial data describing all relevant dimensions is unavailable, and one has to rely on available spatial proxy variables. Focusing on the level of naturalness as a proxy for HCVF, our approach was to integrate many different data sources describing multiple dimensions of forest landscapes at multiple spatial scales, including the physical landscape, tree stands’ bio-physical structure, and socio-economic factors related to current and historical anthropogenic pressure on forests in Sweden (Table 2). We used the machine learning classifier RF52 to train our models and generate predictions. We predicted the relative likelihood of HCVF occurrence in c. 21.85 million 1 ha pixels that are dominated by forest (i.e., forest cover >0.5) in the four study regions (Fig. 1), representing c. 78% of the total forest area in Sweden (Table 1). The results based on a 10-fold spatial cross-validation (SCV)53,54,55 show that all four RF models, trained and validated independently for each of the four study regions, fit the data well and have high predictive capabilities. The model performance metrics fall into the following ranges (metric name [range]): ROC AUC [0.89–0.90], PR AUC [0.84–0.89], Pearson R [0.66–0.68], Brier’s score [0.14–0.15] and MCC [0.61–0.62] (see Fig. 2 and Table 3 for detailed statistics for each model).
The study area (Sweden) divided into 4 regions (1: North boreal, 2: South boreal, 3: Hemiboreal, 4: Nemoral) and the illustration of the 1 ha forest pixel sampling density used for Random Forest models training and validation (red: HCVF presence, black: HCVF pseudo-absence). The barplots show the sampling distribution of presences (True) and pseudo-absences (False) for each region. The minimum distance between all sampling locations from the same category was set to 5 km, while the minimum distance between categories was set to 1 km. The forest cover is based on the Swedish national land cover data (NMD), but excluding temporarily non-forest areas (i.e., recently logged forest or young plantations).
The performance of four Random Forest models trained independently for each study region as resulted from the 10-fold spatial cross-validation (SCV)53,54,55, and visualised as ROC Curve96 (right) and Precision-Recall Curve96 (left). For SCV we overlaid on the study area a grid of 20 × 20 km2 and randomly assigned all grid cells to different spatial subsets (i.e., folds) for the cross-validation procedure. The bold lines are based on the mean values of model performance metrics (calculated for each k-fold), while shaded areas indicate ±1 SD. AUC is the area under the curve. The dashed-lines indicate a reference to no-skilled classifiers (for the PR Curve they correspond to the proportion of HCVF presence samples in each region).
The variable selection procedure to decrease the level of co-linearity between the initial 128 explanatory variables, applied before the 10-fold SCV and training the final models, reduced the number of variables to 49, 50, 53 and 48 for the North boreal, South boreal, Hemiboreal and Nemoral regions, respectively (Supplementary Figs. 2–5). Interestingly, although a unique combination of variables and spatial scales was selected in each model, we observed the same variables (and similar spatial scales) among the most influential for each model (Fig. 3; Supplementary Figs. 2–5; see Table 2 for all variable acronyms). Moreover, the Partial Dependencies Plots43,56 clearly indicated consistent patterns of the variables’, often non-linear, relationships with the relative likelihood of HCVF occurrence across all independently trained models. This included (1) the variables describing forest structural properties, such as the uncorrected and regionally-corrected forest height calculated at a target 1 ha scale (HEIGHT, HEIGHTc), as well as at 0.3 (HEIGHTc003), 0.5 (HEIGHTc005) and 0.11 (HEIGHTc011) spatial scales (expressed in km as the length of a squared window placed at the centre of a 1 ha target pixel), which were some of the most important variables for all regions, and the variation in forest height within each 1 ha pixel (HEIGHTv), a highly important variable for the North boreal and South boreal regions; (2) the variables describing the multi-scale landscape patterns of forest management intensity based on Hansen’s data39 (GFC; combined layers of forest loss and gain, HANSEN003, HANSEN005 and HANSEN011) and the Swedish national land cover data (NMD; logged forest, FOPEN003 and FOPEN011), which were the most influential variables for all regions; and (3) the variables describing the physical structure of a landscape (also a proxy for its accessibility) as DEM011 and SLOPE011 (dominant drivers for North boreal, South boreal and Nemoral regions).
The partial dependence plots (PDPs)43,56 for each study region (a−d) of the six most influential variables in the Random Forest models (top row in each panel) and the corresponding maps (bottom row in each panel). PDPs show the mean response with 95% confidence intervals obtained from 500 bootstrap replicates. All variables were scaled to 0 mean before the model training. The vertical dashed-line, together with the corresponding number in grey, indicates the mean value in the original variable units. The vertical black ticks on the x-axis represent the distribution of samples used in model fitting for each variable. The maps are visualised in the original variable units. See Table 2 for the full list and detailed descriptions of all explanatory variables used in this study. The multi-scale variables are coded using the following template: {variable_acronym}{spatial_scale_code}; the codes for spatial scales are as follows: 003: 0.3 km, 005: 0.5 km, 011: 1.1 km, 051: 5.1 km and 101: 10.1 km.
In all regions, HCVF were more likely to be found in areas with more complex topography (SLOPE, SLOPEv), higher elevation (DEM), in tree stands that are taller than the regional average tree height (HEIGHTc), and structurally more diverse (HEIGHTv). All the above variables are consistent with higher levels of forest naturalness, and showed monotonically increasing non-linear relationships with the relative likelihood of HCVF occurrence, in most cases with a form of sigmoid-shaped function (Fig. 3). In the Hemiboreal and Nemoral regions, the proportion of broadleaf tree stands also followed the same pattern. The opposite, i.e., monotonically decreasing relationships, were observed for all variables representing different aspects of a priori negative human impact on landscapes surrounding the target 1 ha forest pixels. These were multi-scale and distance-based variables related to both direct forest management practices and indirect measures of landscape accessibility for forestry aimed at wood production (e.g., HANSEN, FOPEN, ROADS). These patterns were consistent among regions.
The multi-scale variables, acting as proxies for forest management intensity (e.g., HANSEN or FOPEN), were important predictors not only in forest dominated landscapes, but also captured isolated HCVF patches surrounded by a non-forest matrix (e.g., forest patches on islands, along the sea coast, or surrounded by agricultural fields). Here, a low value for forest management intensity in the immediately surrounding pixels increased the relative likelihood of HCVF presence.
Validation of predictions
Two independent sources of relevant spatially explicit information were used to validate the model. The stand-level validation of the predicted relative likelihood of HCVF occurrence, based on data from the Sveaskog forest company (n = 57548 tree stand polygons), showed consistent patterns across all four study regions (Fig. 4, Supplementary Tables 1–4). The mean predictions per category clearly followed the ordinal scale of forest management types. The highest values were associated with tree stands set-aside for conservation, i.e., without any management NF (mean values for regions 1−4: 0.64, 0.67, 0.59, 0.72) and NF_NM (0.72, 0.70, 0.67, NA), followed by tree stands with conservation-oriented management NM (0.57, 0.53, 0.58, 0.63) and production forests with enhanced conservation concern PE (0.42, 0.40, 0.43, 0.59). The lowest values were associated with production forests of general concern PG (0.29, 0.28, 0.32, 0.37). The pairwise numerical differences between conservation-oriented (NF, NF_NM and NM) and production-oriented (PE, PG) management objectives were highly statistically significant (Tukey’s posthoc test, p < 0.001) in all regions except the Nemoral region, where there were no significant differences between PE, NM and NF. Similarly, for the forest naturalness variable in all regions, tree stands labelled as natural had significantly higher mean predicted values (0.65, 0.63, 0.58, 0.65) than tree stands without this label (0.30, 0.30, 0.33, 0.39).
The results of external validation with the Sveaskog forest management compartment dataset (stand-level; n = 57548 polygons; a, b), and the NFI (plot-level; n = 13775 plots; c, d) datasets. The boxplots show the median and the quartiles of the dataset, while the whiskers extend to show the rest of the distribution, except for points that are determined to be “outliers” using a method that is a function of the inter-quartile range. Additionally, the boxplots are overlaid with the mean values (blue dots) with 95% confidence intervals. Four categorical variables related to forest naturalness and conservation values verified in the field were used for validation: (a) Forest management objectives for individual compartments (Sveaskog): “NF” (Nature conservation, non-intervention), “NF_NM” (Nature conservation, not yet specified), “NM” (Nature conservation-oriented active management), “PF” (Production with enhanced conservation concern), and “PG” (Production with general conservation concern); (b) Forest naturalness (Sveaskog, binary); (c) Forest naturalness (NFI): “plantation”, “normal”, “natural”; (d) Natura 2000 habitat qualities (NFI, binary). The horizontal red dashed-line indicates the HCVF relative likelihood threshold at 0.5.
The results of the second validation, based on plot-level data from the Swedish National Forest Inventory (NFI; n = 13,775 plots) were in accordance with the Sveaskog stand-level validation and were also consistent between regions (Fig. 4, Supplementary Tables 5–8). In the North boreal, South boreal and Hemiboreal regions, NFI plots that had already been recognised as having a high level of forest naturalness (category “natural”; mean values for regions 1−3: 0.84, 0.76, 0.63) had significantly higher values of predicted relative likelihood of HCVF occurrence than average (category “normal”; 0.36, 0.37, 0.38) and forest plots labelled as “plantation” (0.20, 0.24, 0.27). In the Nemoral region there was no difference between the naturalness categories; however, there were only two data points available for the “natural” category. Comparison of predicted values between NFI plots classified as having Natura 2000 habitat qualities showed clear and highly statistically significant differences in all regions. These plots had much higher values of predicted relative likelihood of HCVF occurrence (mean values for regions 1−4: 0.83, 0.83, 0.72, 0.69) compared to areas without such qualities (0.35, 0.36, 0.37, 0.41).
Discussion
This study explored integrating machine learning and landscape data mining to scan forest landscapes across all forestland in Sweden with respect to the relative likelihood of hosting biodiversity hotspots in the form of HCVF. The application of the RF model generated high-accuracy predictions, resulting in a thematic map that ranks forests and landscapes based on their levels of naturalness, representing the HCVF relative likelihood surface (Fig. 5). We validated the models against different independent datasets representing forest naturalness at both the forest stand and the plot scales. This confirmed that the predicted relative likelihoods of HCVF occurrence actually represent forests with different levels of naturalness and conservation values. Therefore, we demonstrated that publicly available spatial datasets and current machine learning-based predictive modelling can generate urgently needed mapping of forests with high conservation values, as well as identify forests with low risks of conflicts between intensive forestry and biodiversity conservation57. Our predictions can be used as the first step in the process of making informed strategic conservation decisions and forest management planning. Obviously, before making any final tactical and operational decisions about the area, field validation should always be performed.
The map can be used for ranking landscapes with respect to forest naturalness. An independent Random Forest model was trained for each study region, and predictions for all regions were compiled into the final map. The online interactive version of this map is available at: https://bubnicki.users.earthengine.app/view/swedentest. Background map attribution: Imagery©2023 TerraMetrics, Map data ©2023 Google.
Comparison with other studies
Attempts to map forests with high levels of naturalness and conservation value encompass many different approaches ranging from systematic field inventories (e.g., woodland key habitats schemes in northern Europe)58, analyses of historical and contemporary databases29,30,59, and more recently, remote sensing based analyses using multiple sensors and increasingly advanced mapping methods, including the application of machine learning3,11,28,60,61. However, in contrast to our work, most other recent studies using high-resolution and wall-to-wall spatial datasets to map HCVF either apply a global or continental perspective, as is the case with the maps of global Intact Forest Landscapes3 and European primary forests11, or encompass only local areas of general conservation interest28,60. In the case of global or continental models, the spatial and thematic mapping resolution is usually too coarse to be useful for strategic landscape-scale spatial planning. For example, models generating large-scale predictions are trained using limited and spatially clustered data, often extrapolating beyond the feature (predictors) space covered by reference data62. The latter usually results in weak predictive performance, especially when assessed using spatial k-fold cross-validation strategies55,62 (but See ref. 63). In addition, mapping efforts focused exclusively on target areas of general conservation interest are obviously limited by their spatial coverage.
Consequently, global mapping approaches and those targeting local conservation areas are not well suited for strategic landscape-scale spatial planning and prioritising areas for conservation and restoration. This lack of “actionable” maps has been identified as a serious obstacle in the development of national climate and biodiversity strategies and in the implementation of effective actions in the conservation and restoration of biodiversity64. Our study, presenting detailed information on the relative likelihood of HCVF occurrence across several eco-regions for an entire country, supports informed conservation planning by identifying patches of high conservation value and the spatial opportunities to expand or link such patches through area protection or active forest landscape restoration13. For example, it can be inferred that areas with intermediate likelihood values that are spatially connected to existing protected areas can support their functionality through restoration that over time advances their conservation values and contributes to functional connectivity. Moreover, our modelling approach also identifies landscapes with a low level of forest naturalness and conservation value. Two alternative future management trajectories could be considered for such areas; forestry oriented to wood biomass production can be continued, or depending on local environmental priorities, forest conservation and restoration plans can be developed and implemented13,57,65.
Global models, usually trained using limited and spatially clustered data62, tend to oversimplify local-scale relationships between explanatory and response variables in attempts to generalise patterns over large spatial domains. This often leads to poor predictive performance at local (landscape) scales, even when robust machine learning algorithms such as RF or Boosted Regression Trees are used11. A potential solution would be to apply a spatially-explicit machine learning algorithm able to cope with spatial heterogeneity in explanatory variables and their interactions over large spatial domains, but these methods are currently either at a early stage of development (e.g., Spatial RF)66,67 or rarely applied in the ecological and conservation context68. The practical workaround used in this study was the “regionalisation” of the study area, i.e., dividing it into four regions characterised by different climatic, anthropogenic, and historical forest conditions and fitting of an independent RF model to each.
The recent studies by Munteanu et al.28 in the Romanian Carpathians and by Ørka et al.61 in Norway are conceptually and technically the most similar to our work. Munteanu et al. used Maxent software, satellite images, and information on current potential anthropogenic pressure to map HCVF in the Carpathians in Romania. However, it is difficult to assess the real predictive power of this model, as the training data were highly spatially clustered and the model performance assessment did not take into account spatial correlation in the training data that had not been generated by a random probabilistic sampling process55,62. Moreover, they excluded historically disturbed forests, contrary to our work, and targeted only forests with the highest level of naturalness, thus making the output map less suitable for landscape-scale spatial planning applications including both wood production and biodiversity conservation. Furthermore, while Munteanu et al. proposed an interesting conceptual framework for mapping HCVF, they neither explicitly included the interactions between structural and human-related HCVF dimensions, nor explored the effects of landscape variables at multiple scales, as we did in our study.
Ørka et al.61 recently presented a framework for a remote sensing-based forest ecological base map of Norway. One of the mapped variables was forest naturalness classified as a binomial variable using generalised boosted regression modelling. Similarly to our work, Ørka et al. stratified the study area into five regions and fitted an independent model to each region and provided the maps with predictions for the whole of Norway, although at a much coarser spatial resolution (10 × 10 km2) than in our study. This makes the results less suitable for landscape-scale spatial planning and conservation prioritisation. However, the most evident difference between our study and the forest naturalness model of Ørka et al. is that they focused only on LiDAR-derived structural metrics as one dimension of HCVF, but not on direct drivers affecting the level of naturalness.
Limitations of our approach
As training data, we applied a comprehensive compilation of identified HCVF as included in the national Swedish database34 with updates in 2019 and 2020. This database includes all types of forests in Sweden and is representative in terms of its spatial coverage of different biomes and landscapes throughout the country. However, the database does not formally represent a random probabilistic sample of existing HCVF. Being identified over decades, for different purposes, using different (but comparable) protocols over many years and including formally protected and voluntarily set aside forests as well as non-protected forests, their actual level of naturalness and conservation value may differ. This variability could not be controlled for in the modelling and represents a priori expected noise in the predicted relative likelihood of HCVF occurrence. Moreover, we were unable to track the cases where a field inventory failed to confirm that a candidate forest is actual HCVF, because this information was not saved in the database. We accounted for this while arranging the training data (presence-only) and interpreting the final results (relative likelihood instead of an actual probability), as described in detail in the Methods section. Still, by building our models on more than 6000 1 ha pixels selected out of a total area of c. 3.44 million ha of confirmed HCVF by applying our sampling strategy (including a minimum distance of 5 km between sampled pixels and filtering out areas <10 ha), we compiled a representative training dataset of HCVF occurrences covering different landscapes and distributed uniformly across the entire country and the four study regions.
All our models use Global Forest Change (GFC)39 data as one of the variables predicting the relative likelihood of HCVF occurrence. Recognising issues related to the use of these data in calculating the absolute level of forest loss69, we argue that our models use these data in a correct way. In particular, we used the GFC as one of many potential proxies of human-related pressure on forest areas that, in most cases, correctly reflects the spatial patterns of forest management intensity at the local landscape level. By combining the “loss” and “gain” layers of the GFC, we provided the model with information about the continuity of forest cover (since 2001) at multiple scales. However, this information does not take into account the cause of the detected change (i.e., natural disturbance, final felling or thinning).
The applicability of our approach in other countries and regions (in terms of its transferability and scalability) may be limited by the availability of similar spatially-extensive datasets as used in this study. However, as we observe a rapid technological breakthrough in “sensing” the environment generating an enormous amount of new accumulated data, we expect this to change in the near future. Even today, most of the source datasets (or their counterparts) used in this work as spatial predictors are globally or nationally available (GFC, night-time lights, high-resolution land cover maps, road networks, etc.). On the other hand, the availability of LiDAR-derived proxies describing forest biophysical structural properties is still limited in many countries and regions. An interesting alternative to airborne LiDAR for obtaining similar information can, at least to some extent, be the use of satellite data and statistical extrapolation of point-based measurements of forest canopy height as proposed by Potapov et al.40.
However, we argue that the most limiting factor to apply our approach in other areas is still the availability of HCVF training data. Potential useful sources of national level reference data on HCVF occurrence can include, for example, databases of forest monitoring systems assessing forest naturalness in the field (as, e.g., National Forest Inventories in Sweden, Norway and Finland), retrospective remote sensing analysis of forest cover dynamics12,28,47, or citizen-science projects addressing high conservation value areas64 and involving different groups of forest landowners. At the international level, initiatives such as the “European primary forest database v2.0” seem to be especially promising59. Moreover, rapidly growing information on the occurrence of species of conservation interest such as that in the Global Biodiversity Information Facility (GBIF) will increasingly allow for additional validation of identified (potential) HCVF areas.
Making use of the prediction map
In spite of national and international policies on biodiversity conservation, the favourable conservation status of species populations, habitat network functionality, and resilience of forest ecosystems is deteriorating70. Consistent with the quantitative conservation targets of the Convention of Biological Diversity (CBD)71, the EU Biodiversity Strategy for 203072 proposed to protect at least 30% of the EU land area, of which a third should be under strict legal protection. In contrast to this, the actual area of legally protected forests in Sweden is 8.9% if including mountain areas and only 3.9% if not (based on Official Statistics of Sweden). This calls for filling of the gaps between the targets and currently set-aside area shares, both through the conservation of existing HCVF73, management and restoration of near-natural forest remnants13. Our approach to mapping the relative likelihood of hosting HCVF can contribute to addressing both these challenges, provided that field validation will precede actual conservation decisions.
In addition to providing quantitative conservation targets, establishing functional GI networks also requires that qualitative targets are satisfied23,71. An important principle for assessing and planning functional habitat networks is the acronym BBMJ74, which stands for Better, Bigger, More and Joined. This approach is a key principle for ranking local landscapes with respect to where to focus on establishing protected areas, or initiating landscape and nature restoration23.
The model we provide constitutes a systematic and consistent first-step conservation and restoration priority mapping, based on which a second-step field validation can be designed for final selection of additional areas that strengthen GI functionality. Accordingly, we provide both a national-scale filter for spatial GI-planning, which currently does not exist but which is urgently needed, and opportunities for cost-efficient and precise HCVF field surveys. Furthermore, the model provides spatially explicit identification of areas in a continuous gradient from the lowest to the highest relative likelihood of HCVF occurrence. Therefore, the areas where our models predicted the intermediate values (that is, the green, orange and yellow colour codes in Figs. 5, 6) represent “crossroad” entities that, depending on the governance and management choices, can be placed on a path to support GI or not. For supporting GI, nature conservation values can develop naturally over time if the focal forest patch is set aside for natural (free) development, or be enhanced through active restoration management75. Based on habitat functionality assessments, restoration management can be oriented towards improving currently unfavourable ecological status by, for example, increasing quantities of dead wood, increasing structural complexity or favouring broadleaf trees34. These forest attributes are insufficient in large areas of northern European boreal forests13,76. Additionally, the continuous gradient of predicted values of HCVF relative likelihood, provides opportunities at regional scales to enlarge previously known conservation hotspot areas, such as expanding the Scandinavian mountain intact forest landscape77 eastwards, or identifying previously unknown clusters of forests that in the future may develop intact forest landscape qualities and improve landscape connectivity over larger scales.
a histograms with fitted kernel density estimates (upper row) and (b) barplots of the fraction of total forestland estimated per region and including temporary non-forest areas (bottom row). The colours correspond to the six classes of HCVF relative likelihood visualised in the prediction map (Fig. 5).
On the contrary, forest landscapes dominated by low values of HCVF relative likelihood may identify areas where conservation ambitions may be lower and that can support continued forestry for wood biomass production, climate smart forestry approaches or closer to nature management alternatives78 depending on the local premises. In addition to configuring forests into efficient spatial management units, however, it is also important to consider factors such as proximity to road networks and industry, as well as the forestland ownership status, particularly when concerning non-industrial private and private forest company ownership. Thus, we see a potential for our model to provide the input needed to design a landscape-scale zoning of management as proposed, for example, in the TRIAD approach78.
Conclusions
To conclude, in the era of big data and technological revolution, access to free and open evidence-based knowledge is key to democratic and efficient governance and operational decision-making towards sustainable forest landscapes. Our approach uses open access spatial data including remote sensing and LiDAR data, open source software tools, and machine learning to assist ranking forest landscapes at multiple scales with respect to their level of naturalness and conservation values. We provide an “actionable” map with sufficient details for regional to local-landscape spatial planning, which fills a critical gap for implementing national, EU and international biodiversity conservation policy.
Methods
Study area
The study area covers the whole country of Sweden, of which 70% is forested (Table 1). We divided Sweden into four regions (Table 1, Fig. 1) based on biogeography (from nemoral to north boreal) delimited by county borders, which represent units for statistical reporting and regional spatial planning. Although forest is the main land cover in all regions, the fraction is lowest in the Nemoral (49.2%) and highest in the South boreal region (78.0%). Scots pine (Pinus sylvestris) forests dominate (39.8%), followed by Norway spruce (Picea abies) forests 27.7%, mixed coniferous forests 12.8% and mixed coniferous and deciduous forests 7.0%48. Broadleaf forests cover 7.4% of the entire forestland, including 1.1 million ha of subalpine mountain birch (Betula pubescens ssp. czerepanovii) tree line forests79.
The Swedish forest landscape has been the subject of clearing forests for agriculture on fertile soils for millennia. Commercial harvesting of wood has occurred since medieval times80, and has re-shaped the forest landscape from the 1800s onwards, with forestry expanding from the south, north- and westwards throughout the country81. Initially, logging targeted large diameter trees for saw timber, and regionally even-aged managed forests were exploited to provide wood and bioenergy for the mining and iron industries, which reduced growing stocks significantly up to the early 20th century82. In the 1920s, silvicultural measures to increase wood production were introduced, and since then the growing stock has increased significantly. Clearcutting became the dominant harvesting method from the 1950s and onwards. Currently, approximately 70% of Swedish forestland has been clear-cut at least once83. In the 1990s, some measures were introduced to ameliorate the impact of intensive silvicultural practices on biodiversity. Current management practices include soil scarification, planting, pre-commercial and commercial thinning, and in many cases also draining and fertilisation84. This has created forests with high growth rates, dominated by cohorts of single tree species (mainly Picea abies or Pinus sylvestris) void of old-growth characteristics48. Generally, historical land clearing for agriculture has had a larger impact in the Hemiboreal and Nemoral regions than in the North and South boreal regions, where human expansion and settlement are less pronounced, being limited to fertile soils and favourable local climates, particularly along river valleys. Nationally, the largest share of protected forests is in the Scandinavian Mountains Green Belt12 of the North and South boreal regions, in which also the presence of indigenous Sami culture and reindeer husbandry is an important characteristic of forest landscapes.
Data sources
We used the publicly available national Swedish HCVF database (see Data availability section for more information about all data sources used in this study) with 641,095 polygons delineating c. 3.44 Mha of forests with known high levels of naturalness and conservation values34. This data is a comprehensive compilation of ten different data sources describing forests known to have high conservation value identified through field surveys and delineated based on the forest cover of the national topographic terrain (1:50,000) and road maps (1:100,000). The database documents the conservation value of HCVFs as the level of naturalness assessed in the field, indicated by dead wood in different stages of decay, multi-layered old-growth vegetation structure, and presence of indicator species. This database originally provided the status up to 2016 but was updated in 2019 and 2020 with the new areas in the mountain region added. The database includes different categories of long- and short-term formally protected, voluntarily set aside, and unprotected areas. We used the Swedish National Land Cover data (NMD) as a source of information on forests type, height, and productivity, as well as non-forest land cover (e.g., open land, water, agricultural land). NMD is provided in raster format with a spatial resolution of 10 × 10 m2 and was produced based on a combination of data sources including existing digital maps (from 2018), satellite images (from 2015 to 2018) and airborne LiDAR (from 2009 to 2018). The height of trees and their coverage were derived from the NMD auxiliary layers based on airborne LiDAR data.
The other publicly available datasets used in this study were (1) a digital elevation model of Sweden (DEM) with a spatial resolution of 50 × 50 m2, (2) GFC maps with data on global forest loss (2000−2020) and gain (2000−2012) with a spatial resolution of 30 × 30 m2 (GFC)39, (3) a harmonised global night time light dataset with a spatial resolution of 1 × 1 km2 (1992−2018; LIGHTS)85 and (4) a database of total human population in Sweden with spatial resolution of 1 × 1 km2 (POP).
Data pre-processing
All raster and vector datasets (see Tables 2 and 3) were organised into a country-wide GIS database using the GRASS GIS software v8.2.186. We defined the minimum mapping unit as 1 ha (100 × 100 m2) for our predictive modelling, and re-sampled all imported raster and rasterised vector layers to this target resolution. We used dataset-specific re-sampling procedures that, in most cases, preserved the spatial information from the original dataset’s resolution (see below).
The original 10 × 10 m2 NMD raster was decomposed into multiple target land cover classes (or a combination of classes; see Table 2) using the parallelised version of the GRASS GIS “r.resamp.stats” resampling algorithm, which aggregates raster values at coarser resolutions using statistical functions. For each target class, its area proportion within 1 ha raster cells was computed. The same resampling algorithm was used to process the other categorical raster layers (e.g., the forest productivity layer). The categorical maps of forest cover loss (2000–2020) and gain (2000–2012) (GFC) were converted into binary maps (loss/gain vs no change) and merged prior to re-sampling.
The distance-based explanatory proxy variables for high forest management intensity (e.g., distance to roads and built-up areas) were calculated using the original resolution and the GRASS GIS algorithm “r.grow.distance” generating raster maps containing distances to the nearest target features and then re-sampled using the average as an aggregation function. A similar re-sampling approach was used to compute the variables originating from continuous rasters (i.e., maps of landscape or forest structure attributes expressed as arrays of float numbers, e.g., elevation, tree height, or night-time lights). In the latter cases, in addition to an average, we also calculated variance among pixels aggregated at a coarser resolution (e.g., variation in forest height).
As tree height is determined by numerous factors, including climate (varying along a latitudinal gradient, from temperate to boreal forests), site productivity, and elevation, we developed a simple procedure to compute a corrected (or regionalised) version of the original LiDAR-based tree height variable. For each forest type (derived from NMD) we computed the height deviation from an average value calculated within a 10 × 10 km2 moving window (the variable H5c in Table 3).
The other explanatory variables derived from the “decomposed” NMD layer were two Shannon’s indices expressing the diversity of (1) different forest types (SHAFOR) and (2) all land cover classes we considered natural elements of a landscape (i.e., not containing human-made features; SHANAT). Both indices were calculated at multiple scales.
In the next step, similar to Cushman et al.87, we computed multi-scale versions of selected variables (see Table 2, Supplementary Fig. 1) to represent information about the neighbourhood of each target 1 ha forest pixel. Hence, we strengthened our model with information about patterns of landscape configuration and composition across multiple spatial scales88,89. We used the moving window algorithm (MW) available in the ndimage module from the scipy v1.6.0 Python package for scientific computing90. We ran MW with a variable-specific aggregation function (see Table 2) for five different spatial scales (expressed here as the length of a squared window placed at the centre of a target 1 ha pixel): 0.3, 0.5, 1.1, 5.1, and 10.1 km.
Finally, to focus our predictive modelling of HCVF on 1 ha pixels dominated by forest, we developed a forest mask by applying a threshold of 0.5 to the proportion of total forest cover. All pixels below this threshold were not taken into account when preparing the training and validation datasets and making the final prediction maps.
Modelling approach
The RF classifier is an ensemble model (or meta-classifier) that fits many decision tree classifiers (individual models) on various sub-samples of a dataset and then combines predictions from all decision trees to improve the predictive accuracy and to control for over-fitting43. It uses bagging (bootstrap aggregation) as an ensemble method. The RF has several advantages over other statistical classifiers, including the ability to model complex interactions among predictor variables and its known robustness in generating useful predictions from noisy, non-normal data43,52,87. Another useful built-in feature of RF is that, by design, it provides a probability-like unbiased individual estimate that a given sample belongs to a certain class. However, this estimate is just a fraction of decision trees that vote for a certain class, and thus it is not a true probability (i.e., there is no strong theoretical foundation for such an interpretation).
The national HCVF database contains information about forest areas verified as HCVF (i.e., true presences) but not about forests classified as non-HCVF (i.e., true absences). Hence, this is a presence-only (or presence-background) data type91,92 for which training and validation subsets need to be selected even more carefully than in the other classification tasks so that they are representative and reflect the composition of an actual landscape. Moreover, this type of data influences how the output of a model can be interpreted. The HCVF detection process could not be explicitly modelled and there was likely spatial sampling bias when identifying new HCVF areas introduced by various regional socio-economic factors and/or land-use history. Thus, we interpreted and ranked the predictions of the model as a relative likelihood of occurrence, rather than an actual probability of occurrence92. We minimised spatial sampling bias and ensured that landscape sampling was representative with our procedure of generating training and validation datasets as described below. When designing this procedure, our aim was to mimic (to the largest possible extent) a random probabilistic sampling process sensu Meyer & Pebesma62.
The HCVF training samples (true presences) were generated using both formally protected and unprotected HCVF areas distributed throughout Sweden. First, the HCVF vector layer was rasterised and overlaid with the forest mask based on the original NMD dataset (10 × 10 m2). Next, we re-sampled the HCVF layer to 1 ha resolution (the result was a proportion of HCVF pixels) and applied a threshold of 0.5 to select only those 1 ha forest pixels that are dominated by HCVF. To reduce noise in the training samples, we excluded HCVF areas smaller than 10 ha (i.e., areas consisting of less than 10 connected 1 ha pixels). Finally, we used the GRASS GIS “r.random.cells” algorithm to randomly distribute sampling locations over these areas, assuring a minimum distance of 5 km between each two selected pixels to minimise the effect of any potential spatial bias that may have been present in the original HCVF database.
To generate pseudo-absences (background data) we applied a 1 km buffer around HCVF areas to lower the chance of unrecognised HCVFs nearby being used as pseudo-absence samples used for model training, thus minimising the number of false negative predictions (or increasing model sensitivity/recall). We then subtracted the buffered areas from the forest mask raster and excluded areas smaller than 10 ha. In the last step, we used the same algorithm as for the HCVF presence samples to distribute sampling locations over these subtracted areas, keeping a minimum distance of 5 km between selected pixels. After this procedure the number of HCVF pseudo-absences was larger than the number of presences of HCVF (Fig. 1). This caused the training dataset to be moderately imbalanced.
We trained all RF models and computed performance metrics using the Python library scikit-learn v1.2.156 and imbalanced-learn v0.10.1, and following the recommendation of Valavi et al.93, used a balanced RF94 implementation, which is more robust in dealing with imbalanced datasets. In this implementation, each tree of RF is provided with a balanced bootstrap sample (i.e., sampling with replacement) using a random down-sampling procedure at the level of an individual tree. Initially, we trained and evaluated all models with default values for all hyper-parameters (see the official scikit-learn documentation), except that the number of decision tress (estimators) was set to 500 (default value was 100). RF are known to work reasonably well with default parameters93, and an expected potential gain in performance metrics is usually only around 1–2%95. Moreover, our final model validation procedure (see the next section) was based on external independent spatial datasets (of different structure and properties than the data used for model training), and it is not clear how this could be integrated into the tuning procedure. However, to ensure that default hyper-parameters are robust, we ran the tuning procedure for the best-performing models using Bayesian optimisation with Gaussian processes implemented in the Python package scikit-optimise v0.9.0. For all models and regions, the difference in performance of tuned models compared to models ran with default hyper-parameters was less than 0.5% as measured by ROC AUC (see Supplementary Table 10 and Supplementary Data). This confirmed that the default values were robust for our application.
We evaluated the performance of each model (internal validation) using a 10-fold SCV53,54,55 and a set of complementary model performance metrics. For the SCV, we overlaid a 20 × 20 km2 grid on the study area and randomly assigned grid cells to different spatial subsets (i.e., folds) during each model fitting, including the finally selected models for the four regions and the alternative models presented below.
Since we were more interested in predicting the continuous relative likelihood of HCVF occurrence rather than a binary categorisation of target forest pixels, which reduces the content of information compared with using the full range of values92, we put more emphasis on threshold-independent metrics evaluating continuous patterns of predicted values, such as the area under ROC curve (ROC AUC)96, area under Precision-Recall curve (PR AUC)96 and Brier’s score97. The ROC AUC can be interpreted as the overall probability that a classifier will predict a higher probability for true positive cases than true negative cases. The PR AUC was computed to better understand the behaviour and performance of our models in predicting the positive class. As both precision and recall focus on the positive class (that is, the presence of HCVF), the PR AUC score gives a general evaluation of the model performance related to this class. Brier’s score is the mean squared difference between the predicted probability and the actual outcome. Additionally, we computed two correlation-like metrics: the Pearson correlation coefficient and Matthews Correlation Coefficient (MCC)98, which is regarded as a robust metric of the quality of binary classification, especially for imbalanced datasets. For completeness, we provided two threshold-based metrics (calculated for the 0.5 threshold): accuracy and True Skill Statistic (TSS)99. The latter is commonly used in the ecological literature.
To decrease the level of co-linearity between explanatory variables used for model training and to enhance the interpretability of the final model, we followed the procedure proposed by McGarigal et al.88 and Cushman et al.87 with some modifications. We fit univariate RF models for all 125 variables (including multi-scale variables), assessed their performance with the 10-fold SCV and computed the average values of two metrics: ROC AUC and Pearson correlation between predicted and test HCVF datasets (as computed for each k-fold). In the next step we ranked variables based on the ROC AUC and iteratively checked all pairs of variables for severe co-linearity (Pearson correlation between variables > 0.7)100. Where co-linearity was found, we selected the variable with a higher ROC AUC if a difference was >= 0.01 or with a higher Pearson correlation (the threshold also set at 0.01) if the ROC AUC difference was <0.01. If there was no difference in both metrics we kept the variable with the higher absolute value of ROC AUC.
To test the robustness of the specification of our model (M1, which we refer to as “selected”), we compared its performance against seven alternative models trained and validated using the same data: (M2) the “global” model using data from the entire Sweden (i.e., without regional stratification; (M3) the “full” model trained with all spatial predictors, including all derived multi-scale features, and ignoring a strong (>0.7) pairwise correlation between some variables; (M4) the “onescale” model trained with all spatial predictors but excluding all multi-scale features; (M5) the “baseline” model trained using only 4 key spatial predictors, all hypothetically having a strong effect on the probability of HCVF occurrence (elevation, tree height, distance to roads and % of logged areas within 1 ha); Additionally, the “full”, “onescale” and “baseline” models were trained with and without longitude and latitude as the auxiliary variables. The results of 10-fold SCV confirmed the robustness of our model’s specification as it performed best or equally well as the “full” model for all regions. (Supplementary Table 9). Moreover, we have not noticed any significant differences when comparing the results of the external validation (as described in the next section; Supplementary Data). The “global” model performed equally well but, based on pixel-to-pixel pairwise Pearson correlation coefficients, produced different predictions for the Hemiboreal and Nemoral regions when compared to the best “regionalised” models (Supplementary Fig. 7), suggesting that global predictions were mainly driven by the patterns that the model learnt for the northern Sweden. As expected, the weakest performance was observed for the “baseline” model followed by the “onescale” model, but interestingly the latter performed almost as well as the best models in all regions.
Finally, we compared the predictive performance of our RF models against the Logistic Regression (LR) as implemented in the Python library scikit-learn v1.2.1. We fitted six additional LR models for each study region (i.e., 24 models in total; Supplementary Table 9). The overall difference between RF and LR for the North & South Boreal and Nemoral study regions was approximately 2–3% for the ROC AUC and 3–4% for the PR AUC metrics in favour of RF for all models. The exception was the Hemiboreal region, where LR outperformed RF by less than 1%, meaning that both models performed equally well. These results are in line with the results of the two largest (to our knowledge) comparative studies of predictive performance of RF and LR. The study by Couronné, Probst & Boulesteix101 was a large-scale benchmark experiment run on 243 real datasets from various domains provided by the OpenML online database. The authors reported that the mean difference between RF and LR was ~3% for accuracy and ~4% for ROC AUC in favour of RF. Another large-scale benchmark study, more similar to our application, was done by Valavi et al.102, where the authors compared the predictive performance of multiple ML and statistical models, including RF and LR, in predicting species distributions using presence-only data. Again, on average, the difference between RF and LR was around 2–3% in both ROC and PR AUC in favour of RF (see Fig. 3 and Fig. 10 in ref. 102). The authors also reported that the difference between models was not consistent across study regions, which is in line with our results. This can indicate that for some regions, non-linear responses to predictors can play a bigger role than in others. This is how we interpret equal performance of RF and LR in the Hemiboreal region, where the response of HCVF to the most important predictors seems to be more linear than in the other regions (Fig. 3). In the next step, we checked the pixel-to-pixel Pearson correlations between maps predicted by RF and LR for different regions (Supplementary Fig. 7). Despite the similar predictive performance scores, the maps produced by LR differ spatially from those produced by RF, as indicated by correlation coefficients ranging from 0.9 for the North Boreal region to 0.82 for the Nemoral region. This is not very surprising, as the two models are based on different assumptions and use different algorithms to fit the data. However, this indicates some spatial differences, confirmed by the visual inspection of prediction maps. The most interesting case was for the Hemiboreal region, where both models performed equally well, but the Pearson correlation was “only” 0.84. The validation of LR predictions against the external independent datasets (described in the next section) reproduced similar patterns as for RF predictions, but the variation of predicted values for most of the categorical variables related to forest naturalness and conservation values was higher than for RF predictions. This is indicated by larger interquartile ranges (IQR) of boxplots (Fig. 4, Supplementary Data). Lastly, we noticed a similarity in the qualitative interpretation of LR and RF results for our selected model (M1). That is, approximately the same set of factors had the strongest effect on the relative likelihood of HCVF occurrence in both cases (Supplementary Data). This indicates that non-linear responses captured by RF can be approximated by linear responses captured by LR, and the cost of this simplification is around 2–3%. In summary, although LR is a viable alternative to RF in our application, RF outperforms LR for three out of four study regions and seems to be more precise, as indicated by the validation against external datasets. Taking all these results together, we conclude that our approach is robust and the choice of RF is reasonable, although alternative methods, such as LR, may provide similar results.
Model validation with external data
Two independent sources of relevant spatially explicit information were used for model validation. The first is the forest management data from Sweden’s largest forest owner, the state company Sveaskog. Their forest management data provide information at the tree stand (compartment) level. It is based on field sampling that assesses variables such as total wood volume (living trees and deadwood), proportion of wood volume of living trees divided into different species, stand age, site type, and many other characteristics. We used two categorical variables from the Sveaskog dataset expressing different levels of forest conservation and naturalness values on an ordinal scale: (1) forest management type (5 levels) and (2) forest naturalness (binary). The forest management type is set for each tree stand using the following categories103: (1) NF = Nature conservation, non-intervention; (2) NF_NM = Nature conservation, not yet specified; (3) NM = Nature conservation-oriented active management, often implying restoration measures; (4) PE = Production with enhanced conservation concern; (5) PG = Production with general conservation concern (green-tree and deadwood retention for biodiversity at harvest). For each stand polygon that had a minimum of 10 pixels, we calculated the mean value of predicted HCVF relative likelihoods. In total, we used data from 57,548 Sveaskog tree stand polygons.
The second source of information for model validation was the Swedish National Forest Inventory (NFI), which uses a randomly planned regular sampling grid104, including around 4500 permanent tracts with each tract being surveyed once every 5 years. The tracts have a rectangular shape of different sizes in different parts of the country and consist of 4–8 circular sample plots (each plot 314 m2). We used two NFI categorical variables relevant to conservation and forest naturalness values: (1) the level of forest naturalness (three levels: natural, normal, plantation) and (2) a binary indicator if plots meet the minimum requirements to be considered Natura 2000 habitat according to the EU Habitats Directive based on its interpretation in Sweden. The NFI data used in this study originated from the 2015 to 2019 inventories. For each NFI plot selected for validation, we extracted the average predicted HCVF relative likelihood from the pixel spatially overlapping with the plot and its four nearest neighbours. In total, we used data from 13,775 NFI plots spread across all study regions.
We summarised both datasets for each study region using boxplots produced with the Python package Seaborn v0.11.1. Finally, we used Tukey’s posthoc test to check the statistical significance of the differences between the different levels of all validation variables. The motivation for these comparisons was to test if our predictions are consistent with the expected patterns of HCVF occurrence in forests characterised by different management practices and representing varying levels of naturalness, as measured on the ground.
Final predictions
After the 10-fold SCV validation procedure, to achieve the highest prediction performance (see ref. 53), we produced the final predictions using all available training data to fit new models for all four regions. Here, we assume that the model performance metrics estimated from the 10-fold SCV are conservative, i.e., the final models can still perform better53. These final predictions were used for model validation with external independent spatial datasets as described above.
To understand which variables were the main drivers of the predicted values for each model, we estimated the impurity-based variable importance43,56. Furthermore, to visualise and explore the relationships between the top six most important variables and predict the relative likelihood of HCVF occurrence, we generated plots of partial dependence, which is the dependence of the relative likelihood of HCVF presence on one predictor variable after averaging the effects of the other predictor variables in the model43,56. The 95% confidence intervals of the mean response were obtained from 500 bootstrap replicates.
All resulting maps were created using QGIS v3.22 “Białowieża” (QGIS Development Team, 2022) and the Python package matplotlib v3.3.3. The interactive visualisation of the prediction map was implemented using the Google Earth Engine platform and is available online at https://bubnicki.users.earthengine.app/view/swedentest.
Data availability
The spatial datasets used in this study are publicly available: DEM. Terrain Model Download, grid 50 + . Lantmateriet, Swedish Ministry of Finance. Available online at https://www.lantmateriet.se/en/maps-and-geographic-information/geodataprodukter/produktlista/terrain-model-download-grid-50/ (accessed April 28, 2022). GFC. Global Forest Change. Global Land Analysis and Discovery, Department of Geographical Sciences, University of Maryland. Available online at https://glad.earthengine.app (accessed April 28, 2022). HCVF. A database of High Conservation Value Forests in Sweden. Swedish Environmental Protection Agency. Available online at https://geodata.naturvardsverket.se/nedladdning/land/skogliga_vardekarnor_2016.zip (accessed April 28, 2022). [Please note that this database originally provides the status up to 2016 but has been updated in 2019 and 2020 with the new areas in the mountain region which were not publicly available at the moment of writing this manuscript but are available on request from the Swedish Environmental Protection Agency]. LIGHTS. A harmonised global nighttime light dataset 1992–2018. Available online at https://doi.org/10.6084/m9.figshare.9828827.v2 (accessed April 28, 2022). NMD. National Land Cover Data. Swedish Environmental Protection Agency. Available online at https://www.naturvardsverket.se/en/services-and-permits/maps-and-map-services/national-land-cover-database/ (accessed April 28, 2022). POP. Total Population in Sweden. Statistics Sweden. Available online at https://www.scb.se/en/services/open-data-api/open-geodata/grid-statistics/ (accessed April 28, 2022). The original independent spatial datasets used for validation (Sveaskog and NFI) are not publicly available. However, we have included the minimal dataset required to interpret and replicate our results in this repository: https://gitlab.com/oscf/hcvf-model-sweden/-/tree/main/src/config/validation.
Code availability
The source code to reproduce the spatial data processing pipeline, training and validation of the Random Forest models, and all other analyses presented in this paper is available at https://gitlab.com/oscf/hcvf-model-sweden.
References
Brockerhoff, E. G. et al. Forest biodiversity, ecosystem functioning and the provision of ecosystem services. Biodivers. Conserv. 26, 3005–3035 (2017).
Mori, A. S., Lertzman, K. P. & Gustafsson, L. Biodiversity and ecosystem services in forest ecosystems: a research agenda for applied forest. Ecol. J. Appl. Ecol. 54, 12–27 (2016).
Potapov, P. et al. The last frontiers of wilderness: tracking loss of intact forest landscapes from 2000 to 2013. Sci. Adv. 3, e1600821 (2017).
UN. United Nations Decade on Ecosystem Restoration (2021-2030): resolution /. 6 p. http://digitallibrary.un.org/record/3794317 (2019).
Morales-Hidalgo, D., Oswalt, S. N. & Somanathan, E. Status and trends in global primary forest, protected areas, and areas designated for conservation of biodiversity from the Global Forest Resources Assessment 2015. For. Ecol. Manag. 352, 68–77 (2015).
Betts, M. G. et al. Global forest loss disproportionately erodes biodiversity in intact landscapes. Nature 547, 441–444 (2017).
Watson, J. E. M. et al. The exceptional value of intact forest ecosystems. Nat. Ecol. Amp Evol. 2, 599–610 (2018).
European Commission. Guidelines for Defining, Mapping, Monitoring and Strictly Protecting EU Primary and Old-Growth Forests. https://environment.ec.europa.eu/publications/guidelines-defining-mapping-monitoring-and-strictly-protecting-eu-primary-and-old-growth-forests_en (2023).
Potapov, P. et al. Mapping the World’s intact forest landscapes by remote sensing. Ecol. Soc. 13, art51 (2008).
Mikoláš, M. et al. Primary forest distribution and representation in a central european landscape: results of a large-scale field-based census. For. Ecol. Manag. 449, 117466 (2019).
Sabatini, F. M. et al. Where Are Europe’s last primary forests? Divers. Distrib. 24, 1426–1439 (2018).
Svensson, J., Bubnicki, J. W., Jonsson, B. G., Andersson, J. & Mikusiński, G. Conservation significance of intact forest landscapes in the scandinavian mountains green belt. Landsc. Ecol. 35, 2113–2131 (2020).
Svensson, J., Mikusiński, G., Bubnicki, J. W., Andersson, J. & Jonsson, B. G. Boreal forest landscape restoration – in the face of extensive forest fragmentation and loss. In: (eds. Girona, M. M., Morin, H., Gauthier, S. & Bergeron, Y.) Boreal Forests in the Face of Climate Change - Sustainable Management (Springer, 2022).
Kuuluvainen, T. et al. Natural Disturbance-Based Forest Management: Moving Beyond Retention and Continuous-Cover Forestry. Front. For. Glob. Change 4, 629020 (2021).
Turner, M. G. Landscape ecology: the effect of pattern on process. Annu. Rev. Ecol. Syst. 20, 171–197 (1989).
Peterken, G. F. Natural Woodland: Ecology and Conservation in Northern Temperate Regions. (Cambridge University Press, 1996).
Winter, S. Forest naturalness assessment as a component of biodiversity monitoring and conservation management. Forestry 85, 293–304 (2012).
Williams, M. Deforesting the Earth: From Prehistory to Global Crisis. (University of Chicago Press, 2003).
Zanon, M., Davis, B. A. S., Marquer, L., Brewer, S. & Kaplan, J. O. European Forest Cover During the Past 12,000 Years: a Palynological Reconstruction Based on Modern Analogs and Remote Sensing. Front. Plant Sci. 9, 331707 (2018).
Angelstam, P. et al. Frontiers of protected areas versus forest exploitation: assessing habitat network functionality in 16 case study regions globally. Ambio 50, 2286–2310 (2021).
Wickham, J. D., Riitters, K. H., Wade, T. G. & Vogt, P. A National assessment of green infrastructure and change for the conterminous united states using morphological image processing. Landsc. Urban Plan. 94, 186–195 (2010).
Snäll, T., Lehtomäki, J., Arponen, A., Elith, J. & Moilanen, A. Green infrastructure design based on spatial conservation prioritization and modeling of biodiversity features and ecosystem services. Environ. Manage. 57, 251–256 (2015).
Angelstam, P. et al. Sweden does not meet agreed national and international forest biodiversity targets: a call for adaptive landscape planning. Landsc. Urban Plan. 202, 103838 (2020).
Commission, E. & Environment, D.-G. for. Building a Green Infrastructure for Europe. (Publications Office of the European Union, 2014). https://doi.org/10.2779/54125.
Virkkala, R. et al. Developing fine‐grained nationwide predictions of valuable forests using biodiversity indicator bird species. Ecol. Appl. 32, e2505 (2021).
Jennings, S. et al. The high conservation value forest toolkit. Ed. ProForest Oxf. OX 12, 1–62 (2003).
Buchwald, E. A hierarchical terminology for more or less natural forests in relation to sustainable management and biodiversity conservation. In Proceedings: Third expert meeting on harmonizing forest-related definitions for use by various stakeholders.Food and Agriculture Organization of the United Nations. Rome. 17–19 (2005).
Munteanu, C. et al. Leveraging historical spy satellite photographs and recent remote sensing data to identify high conservation value forests. Conserv. Biol. 36, e13820 (2021).
Pătru-Stupariu, I., Angelstam, P., Elbakidze, M., Huzui, A. & Andersson, K. Using forest history and spatial patterns to identify potential high conservation value forests in Romania. Biodivers. Conserv. 22, 2023–2039 (2013).
Kurlavicius, P. et al. Identifying high conservation value forests in the baltic states from forest databases. Ecol. Bull. 51, 351–366 (2004).
Mansuy, N. et al. Scaling up forest landscape restoration in Canada in an era of cumulative effects and climate change. For. Policy Econ. 116, 102177 (2020).
Angelstam, P. et al. Tradition as asset or burden for transitions from forests as cropping systems to multifunctional forest landscapes: Sweden as a case study. For. Ecol. Manag. 505, 119895 (2022).
Hertog, I. M., Brogaard, S. & Krause, T. Barriers to expanding continuous cover forestry in Sweden for delivering multiple ecosystem services. Ecosyst. Serv. 53, 101392 (2022).
Mikusiński, G., Orlikowska, E. H., Bubnicki, J. W., Jonsson, B. G. & Svensson, J. Strengthening the network of high conservation value forests in boreal landscapes. Front. Ecol. Evol. 8, 595730 (2021).
Kennedy, R. E. et al. Bringing an ecological view of change to landsat-based remote sensing. Front. Ecol. Environ. 12, 339–346 (2014).
Runting, R. K., Phinn, S., Xie, Z., Venter, O. & Watson, J. E. M. Opportunities for big data in conservation and sustainability. Nat. Commun. 11, 2003 (2020).
Roy, D. P. et al. Landsat-8: science and product vision for terrestrial global change research. Remote Sens. Environ. 145, 154–172 (2014).
Drusch, M. et al. Sentinel-2: Esa’s optical high-resolution mission for Gmes operational services. Remote Sens. Environ. 120, 25–36 (2012).
Hansen, M. C. et al. High-resolution Global Maps of 21st-century forest cover change. Science 342, 850–853 (2013).
Potapov, P. et al. Mapping global forest canopy height through integration of gedi and landsat data. Remote Sens. Environ. 253, 112165 (2021).
Davies, A. B. & Asner, G. P. Advances in animal ecology from 3D-LiDAR ecosystem mapping. Trends Ecol. Evol. 29, 681–691 (2014).
Malinowski, R. et al. Automated production of a land cover/use Map of Europe Based on Sentinel-2 Imagery. Remote Sens. 12, 3523 (2020).
Cutler, D. R. et al. Random forests for classification in ecology. Ecology 88, 2783–2792 (2007).
Tabak, M. A. et al. Machine learning to classify animal species in camera trap images: applications in ecology. Methods Ecol. Evol. 10, 585–590 (2018).
Tuia, D. et al. Perspectives in machine learning for wildlife conservation. Nat. Commun. 13, 792 (2022).
Chiarucci, A. & Piovesan, G. Need for a Global Map of forest naturalness for a sustainable future. Conserv. Biol. 34, 368–372 (2020).
Svensson, J., Andersson, J., Sandström, P., Mikusiński, G. & Jonsson, B. G. Landscape trajectory of natural boreal forest loss as an impediment to green infrastructure. Conserv. Biol. 33, 152–163 (2018).
Angelstam, P. & Manton, M. Effects of forestry intensification and conservation on green infrastructures: a spatio-temporal evaluation in Sweden. Land 10, 531 (2021).
Chapron, G. Sweden threatens European biodiversity. Science 378, 364–364 (2022).
Swedish Forest Agency. Skogsstyrelsens arbete med nyckelbiotoper och objekt med naturvärden. https://www.skogsstyrelsen.se/miljo-och-klimat/biologisk-mangfald/nyckelbiotoper/arbete-med-nyckelbiotoper/ (2021).
Kuhn, M. & Johnson, K. Applied Predictive Modeling. (Springer, 2013).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Roberts, D. R. et al. Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 40, 913–929 (2017).
Meyer, H., Reudenbach, C., Wöllauer, S. & Nauss, T. Importance of spatial predictor variable selection in machine learning applications - moving from data reproduction to spatial prediction. Ecol. Model. 411, 108815 (2019).
Ploton, P. et al. Spatial validation reveals poor predictive performance of large-scale ecological mapping models. Nat. Commun. 11, 4540 (2020).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Andersson, K., Angelstam, P., Elbakidze, M., Axelsson, R. & Degerman, E. Green infrastructures and intensive forestry: need and opportunity for spatial planning in a Swedish rural–urban gradient. Scand. J. For. Res. 28, 143–165 (2013).
Timonen, J. et al. Woodland key habitats in Northern Europe: concepts, inventory and protection. Scand. J. For. Res. 25, 309–324 (2010).
Sabatini, F. M. et al. European primary forest database V2.0. Sci. Data 8, 220 (2021).
Koskikala, J., Kukkonen, M. & Käyhkö, N. Mapping natural forest remnants with multi-source and multi-temporal remote sensing data for more informed management of global biodiversity hotspots. Remote Sens. 12, 1429 (2020).
Ørka, H. O., Jutras-Perreault, M.-C., Næsset, E. & Gobakken, T. A framework for a forest ecological base map - an example from Norway. Ecol. Indic. 136, 108636 (2022).
Meyer, H. & Pebesma, E. Machine learning-based global maps of ecological variables and the challenge of assessing them. Nat. Commun. 13, 2208 (2022).
Wadoux, A. M. J.-C., Heuvelink, G. B. M., de Bruin, S. & Brus, D. J. Spatial cross-validation is not the right way to evaluate map accuracy. Ecol. Model. 457, 109692 (2021).
Schmidt-Traub, G. National climate and biodiversity strategies are hamstrung By a Lack of Maps. Nat. Ecol. Amp Evol. 5, 1325–1327 (2021).
Sarr, D. A. & Puettmann, K. J. Forest management, restoration, and designer ecosystems: Integrating strategies for a crowded planet. Ecoscience 15, 17–26 (2008).
Georganos, S. et al. Geographical random forests: a spatial extension of the random forest algorithm to address spatial heterogeneity in remote sensing and population modelling. Geocarto Int. 36, 121–136 (2019).
Talebi, H., Peeters, L. J. M., Otto, A. & Tolosana-Delgado, R. A truly spatial random forests algorithm for geoscience data analysis and modelling. Math. Geosci. 54, 1–22 (2021).
Domisch, S. et al. Spatially explicit species distribution models: a missed opportunity in conservation planning? Divers. Distrib. 25, 758–769 (2019).
Palahí, M. et al. Concerns about reported harvests in European forests. Nature 592, E15–E17 (2021).
Scholes, R. J. et al. IPBES (2018): Summary for Policymakers of the Assessment Report on Land Degradation and Restoration of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services. 44 (2018).
CBD. Convention on Biological Diversity. Kunming-Montreal Global Biodiversity Framework. Conference of the Parties to the Convention on Biological Diversity, Montreal, Canada (2022). https://www.cbd.int/doc/decisions/cop-15/cop-15-dec-04-en.pdf.
European Commission & Directorate-General Environment. EU Biodiversity Strategy for 2030: Bringing Nature Back into Our Lives. Publications Office of the European Union (2021). https://doi.org/10.2779/677548.
Barredo, J. I. et al. Mapping and assessment of primary and old-growth forests in Europe. Publ. Off. Eur. Union Luxemb. JRC124671 https://doi.org/10.2760/13239 (2021).
Lawton, J. Making Space for Nature: A Review of England’s Wildlife Sites and Ecological Networks. (Defra, 2010).
Crouzeilles, R. et al. A Global meta-analysis on the ecological drivers of forest restoration success. Nat. Commun. 7, 11666 (2016).
Kyaschenko, J. et al. Increase in dead wood, large living trees and tree diversity, yet decrease in understory vegetation cover: the effect of three decades of biodiversity-oriented forest policy in Swedish Forests. J. Environ. Manage. 313, 114993 (2022).
Svensson, J., Bubnicki, J. W., Angelstam, P., Mikusiński, G. & Jonsson, B. G. Spared, shared and lost-routes for maintaining the scandinavian mountain foothill intact forest landscapes. Reg. Environ. Change 22, 31 (2022).
Larsen, J. B. et al. Closer-to-Nature Forest Management. https://efi.int/publications-bank/closer-nature-forest-management (2022) https://doi.org/10.36333/fs12.
Hedenås, H., Christensen, P. & Svensson, J. Changes in vegetation cover and composition in the Swedish mountain region. Environ. Monit. Assess. 188, 452 (2016).
Berglund, B. Landscape reconstructions in South Sweden for the past 6000 years. In Proceedings of the British Academy. 17, 25–31 (1992).
Östlund, L., Zackrisson, O. & Axelsson, A.-L. The history and transformation of a Scandinavian boreal forest landscape since the 19th century. Can. J. For. Res. 27, 1198–1206 (1997).
Angelstam, P. et al. Learning about the history of landscape use for the future: consequences for ecological and social systems in Swedish Bergslagen. Ambio 42, 146–159 (2013).
Nilsson, P., Roberge, C. & Fridman, J. Skogsdata 2021: aktuella uppgifter om de svenska skogarna från SLU Riksskogstaxeringen. https://publications.slu.se/?file=publ/show&id=113145 (2021).
Elbakidze, M. et al. Sustained yield forestry in Sweden and Russia: how does it correspond to sustainable forest management policy? Ambio 42, 160–173 (2013).
Li, X., Zhou, Y., Zhao, M. & Zhao, X. A Harmonized Global Nighttime Light Dataset 1992-2018. Sci. Data 7, 168 (2020).
Neteler, M., Bowman, M. H., Landa, M. & Metz, M. GRASS GIS: a multi-purpose open source GIS. Environ. Model. Softw. 31, 124–130 (2012).
Cushman, S. A., Macdonald, E. A., Landguth, E. L., Malhi, Y. & Macdonald, D. W. Multiple-Scale prediction of forest loss risk across Borneo. Landsc. Ecol. 32, 1581–1598 (2017).
McGarigal, K., Wan, H. Y., Zeller, K. A., Timm, B. C. & Cushman, S. A. Multi-scale habitat selection modeling: a review and outlook. Landsc. Ecol. 31, 1161–1175 (2016).
Remmel, T. K. & Perera, A. H. Mapping Forest Landscape Patterns. (Springer, 2017).
Virtanen, P. et al. Scipy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Barbet-Massin, M., Jiguet, F., Albert, C. H. & Thuiller, W. Selecting pseudo-absences for species distribution models: how, where and how many? Methods Ecol. Evol. 3, 327–338 (2012).
Guillera-Arroita, G. et al. Is My species distribution model fit for purpose? Matching data and models to applications. Glob. Ecol. Biogeogr. 24, 276–292 (2015).
Valavi, R., Elith, J., Lahoz‐Monfort, J. J. & Guillera‐Arroita, G. Modelling species presence‐only data with random forests. Ecography 44, 1731–1742 (2021).
Chen, C., Liaw, A. & Breiman, L. Using random forest to learn imbalanced data. Univ. Calif. Berkeley 110, 24 (2004).
Probst, P., Boulesteix, A.-L. & Bischl, B. Tunability: importance of hyperparameters of machine learning algorithms. J. Mach. Learn. Res. 20, 1934–1965 (2019).
Fawcett, T. An Introduction To Roc Analysis. Pattern Recognit. Lett. 27, 861–874 (2006).
Brier, G. W. Verification of forecasts expressed in terms of probability. Mon. Weather Rev. 78, 1–3 (1950).
Chicco, D. & Jurman, G. The Matthews correlation coefficient (MCC) should replace the ROC AUC as the standard metric for assessing binary classification. BioData Min 16, 4 (2023).
Allouche, O., Tsoar, A. & Kadmon, R. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 43, 1223–1232 (2006).
Dormann, C. F. et al. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36, 27–46 (2013).
Couronné, R., Probst, P. & Boulesteix, A.-L. Random forest versus logistic regression: a large-scale benchmark experiment. BMC Bioinform. 19, 270 (2018).
Valavi, R., Guillera-Arroita, G., Lahoz-Monfort, J. J. & Elith, J. Predictive performance of presence-only species distribution models: a benchmark study with reproducible code. Ecol. Monogr. 92, e01486 (2022).
Angelstam, P. & Bergman, P. Assessing actual landscapes for the maintenance of forest biodiversity: a pilot study using forest management data. Ecol. Bull. 51, 413–425 (2004).
Fridman, J. et al. Adapting National Forest Inventories to changing requirements–the case of the Swedish National Forest Inventory at the turn of the 20th century. Silva Fenn. 48, 1095 (2014).
Acknowledgements
We thank Prof. Carsten Dormann for his valuable feedback and critical comments on an earlier version of the manuscript. We thank seven anonymous reviewers for their important feedback during the review process. We thank Tom Diserens for his great help with improving the English language in this manuscript. This study was funded by the Swedish Environmental Protection Agency, grant NV-03728–17, to Bengt Gunnar Jonsson.
Author information
Authors and Affiliations
Contributions
B.G.J., P.A., J.S., G.M. and J.W.B. originally conceived the idea. J.W.B. developed the methodological approach with help from G.M., B.G.J., P.A., and J.S. J.W.B. collected spatial data with help from J.S., G.M. and B.G.J. J.W.B. processed the spatial data, developed the computer code, trained and validated the machine learning models and produced all figures and maps. J.W.B. led the writing with substantial help from P.A., B.G.J., J.S. and G.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Earth & Environment thanks Werner Rammer, Mihai D. Nita and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Erika Buscardo and Aliénor Lavergne. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bubnicki, J.W., Angelstam, P., Mikusiński, G. et al. The conservation value of forests can be predicted at the scale of 1 hectare. Commun Earth Environ 5, 196 (2024). https://doi.org/10.1038/s43247-024-01325-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-024-01325-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.