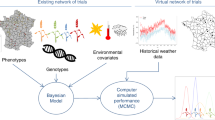
Abstract
Global climate change necessitates crop varieties with good environmental adaptability. As a proxy for climate adaptation, crop breeders could select for adaptability to different latitudes, but the lengthy procedures for that slow development. Here, we combined molecular technologies with a streamlined in-house screening method to facilitate rapid selection for latitude adaptation. We established the daylength-sensing-based environment adaptation simulator (DEAS) to assess rice latitude adaptation status via the transcriptional dynamics of florigen genes at different latitudes. The DEAS predicted the florigen expression profiles in rice varieties with high accuracy. Furthermore, the DEAS showed potential for application in different crops. Incorporating the DEAS into conventional breeding programmes would help to develop cultivars for climate adaptation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Information about indica hybrid rice varieties grown in East Asia, including days to heading and geographic distribution, was obtained from the China Rice Data Center (http://www.ricedata.cn/). Daylength data for different latitudes were collected using the Rise and Set Times app developed by S. Vdovenko (http://www.lifewaresolutions.com/). The haplotype information of flowering-time genes for sterile and restorer lines can be obtained from MBKBASE (http://www.mbkbase.org)33. Hd3a and RFT1 expression data for a rice paddy field at Tsukuba, Japan, grown under normal agricultural conditions can be obtained from https://ricexpro.dna.affrc.go.jp/category-select.php47.
Code availability
In this study, we did not generate any custom code. All mathematical algorithms can be implemented through MATLAB (MathWorks; licence of Xiamen University) and R. All maps were drawn using libraries mapdata, maptools and ggplot2 in R software.
References
Sloat, L. L. et al. Climate adaptation by crop migration. Nat. Commun. 11, 1243 (2020).
Rosenzweig, C. et al. Assessing agricultural risks of climate change in the 21st century in a global gridded crop model intercomparison. Proc. Natl Acad. Sci. USA 111, 3268–3273 (2014).
Teixeira, J. E. et al. Hallauer’s Tuson: a decade of selection for tropical-to-temperate phenological adaptation in maize. Heredity 114, 229–240 (2015).
Eshed, Y. & Lippman, Z. B. Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science 366, eaax0025 (2019).
Kloosterman, B. et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495, 246–250 (2013).
Lu, S. et al. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779 (2017).
Lu, S. et al. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436 (2020).
Guo, L. et al. Stepwise cis-regulatory changes in ZCN8 contribute to maize flowering-time adaptation. Curr. Biol. 28, 3005–3015 (2018).
Hung, H. Y. et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl Acad. Sci USA 109, E1913–E1921 (2012).
Yang, Q. et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl Acad. Sci. USA 110, 16969–16974 (2013).
Zhang, J. et al. Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 208, 1056–1066 (2015).
Gao, H. et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl Acad. Sci. USA 111, 16337–16342 (2014).
Yan, W. et al. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 23, 969–971 (2013).
Koo, B. H. et al. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6, 1877–1888 (2013).
Dai, X. et al. LHD1, an allele of DTH8/Ghd8, controls late heading date in common wild rice (Oryza rufipogon). J. Integr. Plant Biol. 54, 790–799 (2012).
Yan, W. H. et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330 (2011).
Xue, W. et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767 (2008).
Zhang, Z. et al. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci. Rep. 7, 5388 (2017).
Du, A. et al. The DTH8-Hd1 module mediates day-length-dependent regulation of rice flowering. Mol. Plant 10, 948–961 (2017).
Nemoto, Y., Nonoue, Y., Yano, M. & Izawa, T. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 86, 221–233 (2016).
Zhu, S. et al. The OsHAPL1-DTH8-Hd1 complex functions as the transcription regulator to repress heading date in rice. J. Exp. Bot. 68, 553–568 (2017).
Goretti, D. et al. Transcriptional and post-transcriptional mechanisms limit Heading Date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 13, e1006530 (2017).
Yano, M. et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484 (2000).
Wei, X. et al. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153, 1747–1758 (2010).
Itoh, H. & Izawa, T. The coincidence of critical day length recognition for florigen gene expression and floral transition under long-day conditions in rice. Mol. Plant 6, 635–649 (2013).
Simpson, G. G. & Dean, C. Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289 (2002).
Salazar, J. D. et al. Prediction of photoperiodic regulators from quantitative gene circuit models. Cell 139, 1170–1179 (2009).
Sawa, M., Nusinow, D. A., Kay, S. A. & Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265 (2007).
Osugi, A., Itoh, H., Ikeda-Kawakatsu, K., Takano, M. & Izawa, T. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol. 157, 1128–1137 (2011).
Itoh, H., Nonoue, Y., Yano, M. & Izawa, T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42, 635–638 (2010).
Vince-Prue, D. Photoperiodism in Plants (McGraw-Hill, 1975).
Cheng, S. H., Zhuang, J. Y., Fan, Y. Y., Du, J. H. & Cao, L. Y. Progress in research and development on hybrid rice: a super-domesticate in China. Ann. Bot. 100, 959–966 (2007).
Li, X. et al. Analysis of genetic architecture and favorable allele usage of agronomic traits in a large collection of Chinese rice accessions. Sci. China Life Sci. 63, 1688–1702 (2020).
Wu, W. et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl Acad. Sci. USA 110, 2775–2780 (2013).
Kojima, S. et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105 (2002).
Izawa, T. et al. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16, 2006–2020 (2002).
Takahashi, Y., Shomura, A., Sasaki, T. & Yano, M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc. Natl Acad. Sci. USA 98, 7922–7927 (2001).
Doi, K. et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936 (2004).
Hori, K. et al. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 76, 36–46 (2013).
Matsubara, K. et al. Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 53, 709–716 (2012).
Saito, H. et al. Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol. 53, 717–728 (2012).
Yang, Y., Peng, Q., Chen, G. X., Li, X. H. & Wu, C. Y. OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol. Plant 6, 202–215 (2013).
Zhao, J. et al. OsELF3-1, an ortholog of ARABIDOPSIS EARLY FLOWERING 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS ONE 7, e43705 (2012).
Shibaya, T. et al. Hd18, encoding histone acetylase related to Arabidopsis FLOWERING LOCUS D, is involved in the control of flowering time in rice. Plant Cell Physiol. 57, 1828–1838 (2016).
Zhang, B. et al. Genetic interactions among Ghd7, Ghd8, OsPRR37 and Hd1 contribute to large variation in heading date in rice. Rice 12, 48 (2019).
Wang, P., Gong, R., Yang, Y. & Yu, S. Ghd8 controls rice photoperiod sensitivity by forming a complex that interacts with Ghd7. BMC Plant Biol. 19, 462 (2019).
Nagano, A. J. et al. Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 151, 1358–1369 (2012).
Jaeger, P. A., Doherty, C. & Ideker, T. Modeling transcriptome dynamics in a complex world. Cell 151, 1161–1162 (2012).
Araki, M. G., Gyokusen, K. & Kajimoto, T. Vertical and seasonal variations in temperature responses of leaf respiration in a Chamaecyparis obtusa canopy. Tree Physiol. 37, 1269–1284 (2017).
Hu, Y. N. et al. Rice production and climate change in northeast China: evidence of adaptation through land use shifts. Environ. Res. Lett. 14, 024014 (2019).
Lv, Z. F. et al. Climate change impacts on regional rice production in China. Clim. Change 147, 523–537 (2018).
Liu, Z. H. et al. Shifts in the extent and location of rice cropping areas match the climate change pattern in China during 1980-2010. Reg. Environ. Change 15, 919–929 (2015).
Li, Z. G. et al. Chinese rice production area adaptations to climate changes, 1949-2010. Environ. Sci. Technol. 49, 2032–2037 (2015).
Zhao, C. et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl Acad. Sci. USA 114, 9326–9331 (2017).
Lobell, D. B., Schlenker, W. & Costa-Roberts, J. Climate trends and global crop production since 1980. Science 333, 616–620 (2011).
Ma, X. et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8, 1274–1284 (2015).
Naito, Y., Hino, K., Bono, H. & Ui-Tei, K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 (2015).
Acknowledgements
This work was supported by the National Key R&D Program of China (2017YFA0506100), the National Natural Science Foundation (31671378) and the Fundamental Research Funds for the Central Universities (20720170068 and 20720190085). We thank Y. Liu (South China Agricultural University) for providing the pYLCRISPR/Cas9-MTmono vectors. We thank the breeder L. Wang (Sichuan Agricultural University) for providing the indica hybrid rice seeds. We thank X. Wang Deng (Peking University), H. Wang (South China Agricultural University) and F. Kong (Guangzhou University) for reading and commenting on the manuscript. We thank Y. Cui (Xiamen University), Z. Zeng (Sichuan Agricultural University), Y. Wang (Sichuan Agricultural University), W. Hu (Xiamen University) and X. Liu (Xiamen University) for their technical assistance.
Author information
Authors and Affiliations
Contributions
X.O. designed the research; L.Q., Q.W., X.W., G.Z., Z.S., J.H., H.W., W.T., Q.L., J.R., J.X., C.L., Y.L., S.L., R.H., X.C., C.Z., M.L., X.H., S.L. and X.O. performed the research; L.Q., P.Q., Z.C., Z.L., H.H., J.H., X.W., C.L. and X.O. analysed the data; X.O. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Food thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–25, and Tables 1 and 2.
Rights and permissions
About this article
Cite this article
Qiu, L., Wu, Q., Wang, X. et al. Forecasting rice latitude adaptation through a daylength-sensing-based environment adaptation simulator. Nat Food 2, 348–362 (2021). https://doi.org/10.1038/s43016-021-00280-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43016-021-00280-2